Background: The mammalian formin FMNL3 influences actin dynamics and promotes filopodia formation.
Results: The C terminus of FMNL3 synergizes with the FH2 domain to accelerate actin polymerization, binds actin monomers, and enhances filament severing.
Conclusion: The C terminus of FMNL3 contains a WH2-like sequence that binds filament barbed ends and monomers.
Significance: The contribution of the C terminus to actin dynamics differs significantly between formins.
Keywords: Actin, Cell Motility, Cytoskeleton, Formin, Protein Chemistry, Bundling, Elongation, FH2, FRL2, Nucleation
Abstract
Formin proteins are actin assembly factors that accelerate filament nucleation then remain on the elongating barbed end and modulate filament elongation. The formin homology 2 (FH2) domain is central to these activities, but recent work has suggested that additional sequences enhance FH2 domain function. Here we show that the C-terminal 76 amino acids of the formin FMNL3 have a dramatic effect on the ability of the FH2 domain to accelerate actin assembly. This C-terminal region contains a WASp homology 2 (WH2)-like sequence that binds actin monomers in a manner that is competitive with other WH2 domains and with profilin. In addition, the C terminus binds filament barbed ends. As a monomer, the FMNL3 C terminus inhibits actin polymerization and slows barbed end elongation with moderate affinity. As a dimer, the C terminus accelerates actin polymerization from monomers and displays high affinity inhibition of barbed end elongation. These properties are not common to all formin C termini, as those of mDia1 and INF2 do not behave similarly. Interestingly, mutation of two aliphatic residues, which blocks high affinity actin binding by the WH2-like sequence, has no effect on the ability of the C terminus to enhance FH2-mediated polymerization. However, mutation of three successive basic residues at the C terminus of the WH2-like sequence compromises polymerization enhancement. These results illustrate that the C termini of formins are highly diverse in their interactions with actin.
Introduction
Actin filaments are integral to a large number of cellular structures (1). Due to the widespread use of actin, cells must rigorously control when and where new filaments assemble. Spontaneous actin nucleation is unfavorable, but filament assembly is promoted by three classes of assembly factors: Arp2/3 complex, formin proteins, and compound WH2 domain proteins (COWs) such as Spire, Cordon Bleu (Cobl), and VopL (2–4).
The large number of formins (15 mammalian formin genes) provides the potential for diverse regulatory mechanisms (5). Formins are also diverse in their effects on actin. All formins possess a formin homology 2 (FH2)2 domain that mediates actin nucleation and then remains at the elongating filament barbed end, thus modulating elongation (6). The nucleation abilities of FH2 domains vary considerably from highly potent nucleators like mDia1 and mDia2 to extremely weak nucleators like DAAM1 (7–10). Formin effects on filament elongation also vary significantly, with mDia1 having little effect and mDia2 slowing elongation by >85% (11). For all formins tested, elongation rate is increased by profilin binding to the FH1 domain, which is N-terminal to the FH2 (see Fig. 1A) (11, 12). In addition to their effects on actin dynamics, the FH2 domains of some formins, including mDia2, FMNL1, FMNL2, and FMNL3, can bind filament sides and bundle actin filaments (7, 13).
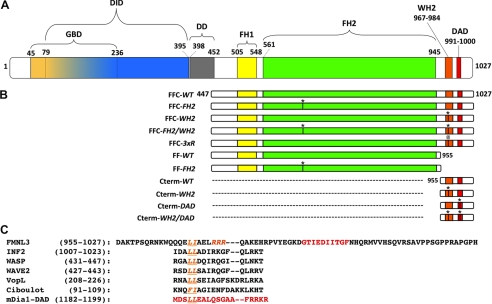
FIGURE 1.
FMNL3 protein. A, FMNL3 domain organization. Mouse FMNL3 (1027 amino acids) contains the following domains: GBD (amino acids 45–236), DID (79–395), DD (dimerization domain, 398–452), FH1 (505–548), FH2 (561–945), and DAD (991–1000). B, FMNL3 constructs used in this study. * denotes the point mutations FH2-I649A, WH2-L970/I971A, and DAD I996/I997A, and @ denotes the R975–977A mutation. C, the amino acid sequence of the FMNL3 C terminus and comparison with WH2 sequences or C termini from other proteins. Indicated in orange are conserved WH2 residues required for actin binding. Residues in red denote the DAD in FMNL3 and mDia1.
In addition to the FH1 and FH2 domains, the C-terminal regions of formins can also affect actin polymerization. For several formins, including mDia1 and DAAM1, the C terminus enhances the nucleation activity of the FH2 domain (12). For mDia1, basic residues at the C terminus of the diaphanous autoinhibitory domain (DAD) are important for this enhancement. Somewhat differently, the INF2 C terminus contains an actin monomer binding WASp homology 2 (WH2) domain that overlaps its DAD sequence (14). This WH2 domain is necessary for the unique depolymerization activity of INF2 when tethered to the FH1 and FH2 domains. Thus, the C termini of formins can differentially influence effects of the FH2 domain on actin dynamics.
WH2 domains, present in many actin assembly proteins, bind actin monomers in a groove between subdomains 1 and 3 on the barbed end side and make additional lateral contacts with monomers (15). Alone, a WH2 domain can either sequester an actin monomer or block its addition to pointed ends (16, 17). When combined with other sequences, WH2 domains can have more intricate effects. Activators of Arp2/3 complex either contain one (WASp, WAVE proteins, ActA) or two (N-WASP) WH2 domains (18). COWs contain multiple (3 or more) WH2 domains that are used in combination with other sequences to nucleate, sever, or modulate the elongation of filaments (19–24). For formins, INF2 has a bona fide WH2 domain, and the FMNL family is predicted to have a WH2 domain (13).
Although WH2 domains are mostly considered to bind actin monomers, they can also have effects at the filament barbed end. WH2 domains of N-WASP are able to tether barbed ends to surfaces during Arp2/3 complex-mediated motility (25). Similarly, Spire contains four WH2 repeats and is capable of preventing both profilin-actin addition to filaments and barbed end depolymerization with nanomolar potency (20). Although the localization of Spire on actin filaments is still debated, an electron microscopy study suggests barbed end binding (26).
In this paper we investigate the activity of FMNL3 on actin, as this formin is a potent filopodial generator (27). We show that the C terminus of FMNL3 dramatically increases the polymerization activity of the FH2 domain. The C terminus contains an actin binding motif similar to a WH2 but with important differences. In addition, the actions of the FMNL3 C terminus on actin are very different from those of the INF2 or mDia1 C termini. In monomeric form, the FMNL3 C terminus binds monomers and also slows barbed end elongation. In dimeric form, the potency of the barbed end elongation effect increases dramatically, suggesting that the effect is due to barbed end binding. The dimeric C terminus also accelerates actin polymerization from monomers. In contrast, the C termini of mDia1 and INF2 do not display barbed end binding abilities.
EXPERIMENTAL PROCEDURES
DNA Constructs
Constructs of FMNL3 (see Fig. 1B) were generated by amplifying cDNA from mouse 300.19 pre-B lymphoma cells and cloned into pGEX-KT as previously described (27). Point mutations were created using QuikChange (Stratagene). C-terminal constructs of mDia1 (mouse, amino acids 1148–1255) and INF2 (human, 994–1249) were produced by PCR amplification of the relevant regions from previously described clones (8, 14) or from purchased clones (INF2, Origene) and cloned into pGEX-KT. All clones were verified by DNA sequencing.
Buffers
The following buffers were used frequently: G-buffer (2 mm Tris, pH 8, 0.5 mm DTT, 0.2 mm ATP, 0.1 mm CaCl2, and 0.01% NaN3), G-Mg buffer (same as G-buffer but with 0.1 mm MgCl2 instead of CaCl2), 10× K50MEI (500 mm KCl, 10 mm MgCl2, 10 mm EGTA, and 100 mm imidazole, pH 7.0), 10× Na50MEI (same as 10× K50MEI but with 500 mm NaCl instead of KCl), and polymerization buffer (G-Mg buffer plus either 1× K50MEI or 1× Na50MEI and 0.5 mm thesit (the common name for the detergent nonaethylene glycol monododecyl ether (Sigma, P-9641), which was included to minimize protein adhesion to the tube/well). Polymerization buffer with 1× Na50MEI was used for pelleting assays because dodecyl sulfate precipitates as the potassium salt. Protein storage buffer was composed of 10 mm PIPES, pH 6.5, 500 mm NaCl, 1 mm EDTA, and 1 mm DTT.
Protein Preparation and Purification
We expressed constructs of FMNL3 (see Fig. 1B) as glutathione S-transferase (GST) fusion proteins in Escherichia coli using methods similar to those described previously (28). Briefly, Rosetta 2, non-DE3 cells (Novagen 71402) containing expression constructs were grown to A600 of 1.0 in TB (12 g/liter Tryptone, 24 g/liter yeast extract, 4.5 ml/liter glycerol, 14 g/liter dibasic potassium phosphate, and 2.6 g/liter mono-basic potassium phosphate) with 100 μg/ml ampicillin and 34 μg/ml chloramphenicol at 37 °C. After reduction to 16 °C, 0.5 mm isopropyl-1-thio-β-d-galactopyranoside was added, and the cultures were grown overnight. All subsequent purification steps were performed at 4 °C or on ice. Bacteria were pelleted and resuspended in Extraction Buffer (50 mm Tris-HCl, pH 8.0, 500 mm NaCl, 5 mm EDTA, 1 mm DTT, and 1 pill/50 ml Complete protease inhibitors (Roche Applied Science)) and lysed by sonication. After lysate ultracentrifugation, supernatants were loaded onto glutathione-Sepharose 4B (Amersham Biosciences), which was subsequently washed with Wash Buffer (Extraction Buffer without protease inhibitors and with 0.05% thesit). Thrombin (Sigma T-4265) was added to a 50% slurry of beads to 20 units/ml, and the suspension was mixed for 1 h. Cleaved protein was washed from the column with Wash Buffer, and thrombin was inactivated with 1 mm phenylmethylsulfonyl fluoride for 15 min, after which DTT was added to 10 mm. Constructs were further enriched by cation exchange chromatography as follows. All FMNL3 FH1-FH2-C constructs were loaded onto SourceS15 10/10 column (Amersham Biosciences) and eluted with a 30-column volume gradient from 150–500 mm NaCl in PIPES, pH 6.5 (with 1 mm EDTA and 1 mm DTT). Peak fractions were further concentrated by binding to SP-Sepharose Fast Flow beads followed by elution with storage buffer. All FH1-FH2 and Cterm constructs were cleaved from GST with thrombin and then enriched by step elution from SP-Sepharose Fast Flow. GST-Cterm fusion constructs were eluded from glutathione-Sepharose 4B with 50 mm reduced l-glutathione (Sigma, G4251) and further purified by gel filtration on Superdex200 (GE Biosciences). All FMNL3 constructs were separated into aliquots and stored at −80 °C, which did not affect the activities measured here.
FMNL3 Cterm, containing a single cysteine engineered into its N terminus, was labeled with fluorescein-maleimide (Amersham Biosciences PA23031) as follows. The protein was diluted to 20 μm in labeling buffer (5 mm Tris, pH 8, 50 mm NaCl, 0.5 mm EDTA, 0.25 mm Tris(2-carboxyethyl)phosphine) and then incubated with a 5-fold molar excess of fluorescein-maleimide for 1 h at 4 °C. Labeled protein was enriched with SP-Sepharose Fast Flow (Amersham Biosciences) and exchanged with storage buffer by overnight dialysis at 4 °C. Labeling was confirmed by MALDI spectroscopy.
Rabbit skeletal muscle actin was purified from acetone powder (29) and labeled with pyrenyliodoacetamide (30). Both unlabeled and labeled actin were gel-filtered on Superdex 75 (GE Biosciences) (31) and stored in G-buffer under constant dialysis at 4 °C (refreshed every 3–4 days).
Actin Polymerization by Fluorescence Spectroscopy
Unlabeled and pyrene-labeled actin were mixed in G-buffer to produce a 5% pyrene-actin stock. This stock was converted to Mg2+ salt by 2 min of incubation at 23 °C in 1 mm EGTA, 0.1 mm MgCl2 immediately before polymerization. Polymerization was induced by the addition of 10× K50MEI to a concentration of 1×, with the remaining volume made up by G-Mg. Additional proteins were mixed together for 1 min before their rapid addition to actin to start the assay. Pyrene fluorescence (excitation 365 nm, emission 410 nm) was monitored in a 96-well fluorescence plate reader (Tecan Infinite M1000, Mannedorf Switzerland). The time between mixing of final components and start of fluorometer data collection was measured for each assay and ranged between 15 and 20 s.
Barbed End Elongation Assays
Unlabeled actin (10 μm) was polymerized for 1 h at 23 °C followed by the addition of 20 μm phalloidin, then centrifuged at 100,000 rpm for 20 min in a TLA-120 rotor. The pellet was resuspended to 6 μm in 3× polymerization buffer (G-Mg with 3× KMEI) and then sheared by 5 passes through a 30-gauge needle. The resuspended polymerized actin was allowed to reanneal overnight at 23 °C. 30 μl of 1× polymerization buffer with or without formin protein was added to 30 μl filaments in a 96-well plate, shaken for 10 s, then centrifuged for 2 min at 1200 rpm. After 2 min at 23 °C, 60 μl of 2 μm monomers (5% pyrene, Mg2+-converted) were mixed with the filaments with a cut pipette tip. Fluorescence (365/410 nm) was recorded for 2700 s. Elongation velocity was obtained by linear fitting the initial 10% of elongation. Final concentrations in the assay were 1.5 μm phalloidin-stabilized polymerized actin and 0.5 μm monomer. Slopes of pyrene fluorescence from elongation time courses were taken at 10% completion using Kaleidagraph (Synergy Software, Reading, PA) and were converted to elongation rates (assuming 10 μm−1s−1 for actin alone (32)).
Calculating Filament Concentration
Slopes were measured at 50% polymerization and converted to filament concentration either under the assumption of unrestricted ATP-actin monomer addition to barbed ends with a rate constant (K+) of 10 μm−1s−1 (32) or with the observed elongation rates in the presence of the specific FMNL3 construct as measured above according to the equation [F] = S′/(K+ × m0.5), where [F] is the filament concentration (μm), S′ is the slope converted to polymerization rate (μm/s), and m0.5 is the monomer concentration (μm) at 50% polymerization. S′ is calculated by the equation S′= S × (Mt/(fmax − fmin), where S is raw slope in arbitrary units (a.u.)/s, Mt is the concentration of total polymerizable monomer (μm), and fmax and fmin are fluorescence (a.u.) of fully polymerized and unpolymerized actin, respectively.
Actin Filament Bundling Assays
Actin (6 μm) was polymerized 1 hr at 23 °C in polymerization buffer followed by addition of 6 μm phalloidin (Sigma P-2141). 100-μl filaments were pipetted into an Eppendorf 1.5 ml screwcap microcentrifuge tube using cut pipette tips to minimize shearing. 100 μl of polymerization buffer and 100 μl of formin were then mixed with filaments by gentle flicking and then incubated 5 min at 23 °C. Samples were centrifuged at 13,000 rpm 5 min at 4 °C in a microcentrifuge. 240 μl of supernatant was removed, lyophilized, and resuspended in 40 μl of SDS-PAGE sample buffer. The pellets were washed 2× by gently adding 300 μl of polymerization buffer and recentrifuged, and buffer was removed. Any residual buffer volume was eliminated by lyophilization. The dried pellet was resuspended in 50 μl of SDS-PAGE sample buffer. Supernatants and pellets were analyzed by Coomassie-stained SDS-PAGE. Protein bands were quantified by densitometry using ImageJ software (National Institutes of Health).
Fluorescence Anisotropy Measurements
Fluorescein-labeled FMNL3 Cterm was diluted to 20 nm in 1× K50MEI/G-Mg containing 0.02% thesit. A stock of actin monomers mixed with 1.5× mol eq of latrunculin B (Calbiochem) was prepared in polymerization buffer (1× K50MEI in G-Mg). To 5 μl of fluorescein-labeled Cterm, 35 μl of actin- latrunculin B solution was added, and fluorescence anisotropy was measured using a fluorescence plate reader with a 491-nm excitation wavelength and a 519-nm emission wavelength. For competition anisotropy experiments, actin monomers (with 1.5 mol eq of latrunculin B) were premixed with unlabeled Cterm or WH2 peptide. 20 μl of this mixture was then added to 20 μl fluorescein-labeled Cterm and measured as described above. Actin monomer concentration in competition assays was 1 μm.
Filament Severing Assay
Actin (4 μm) was polymerized for 1 h at 23 °C in polymerization buffer. Polymerized actin (2.5 μl) was incubated with 2.5 μl of polymerization buffer containing FMNL3 for 2 min at 23 °C. 10 μl of dilution buffer (25 mm imidazole pH 7.0, 25 mm KCl, 4 mm MgCl2, 1 mm EGTA, 0.5% methylcellulose) containing rhodamine-phalloidin (2 μm final concentration) was added, and samples were immediately diluted 150-fold with fluorescence buffer (250 mm NaCl, 100 mm DTT, 3 mg/ml glucose, 18 μg/ml catalase, and 100 μg/ml glucose oxidase in dilution buffer). Samples (2 μl) were adsorbed on 12-mm round glass coverslips coated with 0.01% poly-l-lysine. Cut pipette tips were used to minimize filament shearing. Filaments were visualized through a TRITC filter using a Nikon-Eclipse TE-2000 microscope with a 60 × 1.4 numerical aperture objective. Images were acquired using a Roper Cool-Snap camera. Filament length was measured using NIS-Elements software (Nikon Inc), and 200–300 filaments were measured from at least four different fields.
RESULTS
The C terminus of FMNL3 Accelerates Actin Polymerization by Its FH2 Domain
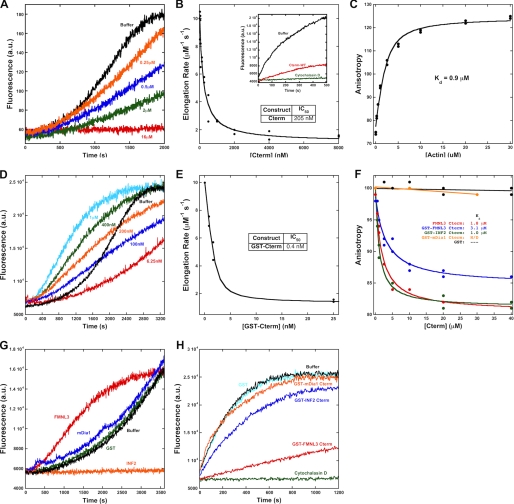
To study the effect of the FMNL3 C terminus on actin polymerization, we produced proteins containing the FH1-FH2 domains (abbreviated FF) and the FH1-FH2-C domains (FFC, Fig. 1A). The FF-WT and FFC-WT constructs have very different abilities to accelerate actin polymerization. In assays containing 2 μm actin monomers and 40 nm FMNL3 construct, FFC-WT accelerates polymerization potently with little lag, whereas FF-WT slightly slows polymerization rate (Fig. 2A). To analyze the polymerization potencies of these constructs in more detail, we plotted the time required to reach half-maximal polymerization (t½) as a function of formin concentration. The concentration dependence of FFC-WT on polymerization follows a relatively regular pattern down to low nm concentrations and becomes slower than actin alone below 1 nm (Fig. 2B and supplemental Fig. 1A). In contrast, FF-WT slows actin polymerization at almost all concentrations tested (Fig. 2B), possibly reflecting inhibitory effects on barbed end elongation. We tested barbed end elongation using pyrene-actin assays. Both constructs inhibit elongation by about 75%, with similar IC50 values (Fig. 2, C and D).
FIGURE 2.
Addition of the Cterm accelerates actin polymerization by FMNL3 FF. A, pyrene-actin polymerization assays containing 2 μm actin monomers (5% pyrene) with 40 nm concentrations of the indicated construct of FMNL3. Additional concentrations are shown in supplemental Fig. 1. B, time to 50% completion (t½) in polymerization assays from curves in Fig. 2A and supplemental Fig. 1. The inset is an expanded view of the lowest concentrations tested. The table gives minimum t½ for each construct as well as the EC50 to reach that minimum t½. C, elongation of 0.5 μm actin monomers (25% pyrene) from phalloidin-stabilized filament seeds (3.1 μm) in the presence of 400 nm of the indicated FMNL3 construct or 50 nm cytochalasin D. D, inhibition of filament elongation by FMNL3. Slopes from the initial 10% of the time course were converted to elongation rates (assuming 10 μm−1s−1 for actin alone (32)). E, filament production at 50% polymerization (from data such as that in Fig. 2A and supplemental Fig. 1) in the presence of the indicated FMNL3 constructs. Calculation of filaments produced takes into account the altered barbed end elongation rates for the FMNL3 constructs (Fig. 2C; see “Experimental Procedures”). F, Filament bundling by FMNL3 constructs was determined by low speed pelleting assay. Phalloidin-stabilized actin filaments (2 μm) were incubated with increasing concentrations of the indicated FMNL3 constructs, centrifuged at 16,000 × g, and resolved on SDS-PAGE followed by Coomassie stain. Pelleted actin bands were quantified by densitometry. See supplemental Fig. 2 for more information. Similar results were obtained in three separate experiments.
Using these elongation rates, we calculated the number of filaments produced by FF-WT and FFC-WT as a function of formin concentration (Fig. 2E). Both constructs cause a net increase in filament number compared with actin alone. The FFC-WT construct, however, is about 2.5-fold more efficient at producing filaments at 50 nm and produces filaments at lower concentrations (EC50 of 2.6 nm) than does FF-WT (EC50 of 4.9 nm). Combined with the t½ data, these results show that the C terminus of FMNL3 greatly increases the speed and efficiency of filament production.
Addition of C Terminus Increases Bundling/Severing Efficiency of FMNL3
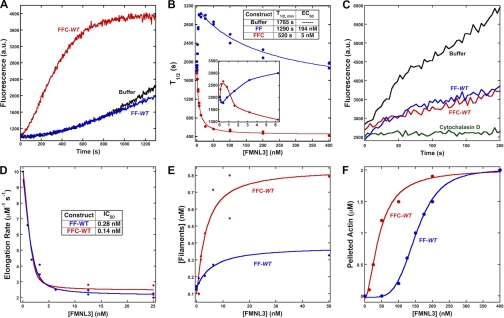
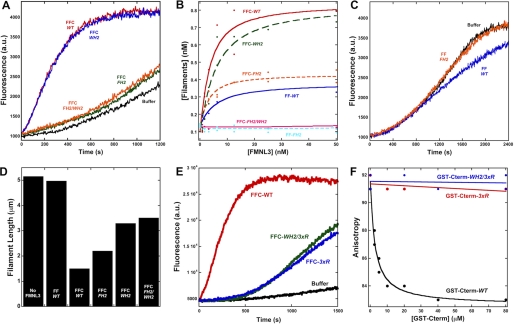
To determine the ability of FF-WT and FFC-WT to bundle actin filaments, we measured their abilities to pellet phalloidin-stabilized polymerized actin upon low speed centrifugation. Both FF-WT and FFC-WT pellet 2 μm actin in a concentration-dependent manner (Fig. 2F and supplemental Fig. 2). The FFC-WT is a more efficient bundler, with an EC50 of 44 nm, whereas FF-WT has an EC50 of 152 nm. In addition, FF-WT displays a decidedly sigmoidal bundling curve in this assay.
Previously, we demonstrated that an FFC construct of FMNL1 severed actin filaments (28). Here, we performed single time point severing assays (2-min incubation) with FF-WT and FFC-WT. FFC-WT generates shorter filaments compared with buffer or to FF-WT (Fig. 3, A–C and E), suggestive of severing activity. Additionally, this effect requires the Cterm be in cis with the FH1 and FH2 domains, as a combination of Cterm-WT and FF-WT has no effect on filament lengths compared with FF-WT alone (Fig. 3, D and E). The ability of FFC-WT to sever filaments is sensitive to ionic strength (supplemental Fig. 3). However, FMNL3-mediated severing is not sensitive to phosphate (supplemental Fig. 3), unlike the severing activity of INF2 (14), suggesting that FMNL3 can sever filaments regardless of whether they are bound to ADP-Pi or ADP alone. Additionally, phalloidin blocks severing by FFC-WT (supplemental Fig. 3).
FIGURE 3.
Addition of the C terminus of FMNL3 enhances filament severing. A–E, 2 μm polymerized actin was incubated with buffer (A) or 400 nm of FFC-WT (B), FF-WT (C), or FF-WT and 400 nm Cterm-WT in trans (D) for 2 min, then stabilized with 10 μm rhodamine-phalloidin and immediately diluted 100-fold with fluorescence buffer. After dilution, samples were adsorbed to poly-l-lysine-coated glass coverslips and viewed by fluorescence microscopy. Scale bar = 5 μm. E, quantification of filaments lengths from these assays is shown. The n value of filaments counted for each condition was >400; bar = S.E.
FMNL3 C Terminus Alone Inhibits Barbed End Elongation and Binds Actin Monomers
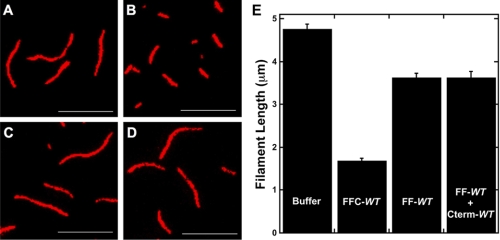
The ability of formins to bind barbed ends depends on a conserved isoleucine residue within the FH2 domain (33). Using pyrene-actin elongation assays, we tested whether mutating this residue in FMNL3 (Ile-649) would abolish its ability to inhibit barbed end elongation. Surprisingly, although the I649A mutation is sufficient to abolish elongation inhibition in the FF construct (FF-FH2), the mutation does not block the effect of the FFC-FH2 construct (Fig. 4).
FIGURE 4.
A mutation that compromises FH2 barbed end binding does not affect barbed end elongation inhibition by the FFC construct. A, elongation of 0.5 μm actin monomers (25% pyrene) from phalloidin-stabilized filament seeds (3.1 μm) in the presence of 12.5 nm of the indicated FMNL3 construct or 50 nm cytochalasin D. B, concentration dependence of filament elongation inhibition by FH2 mutants. Slopes were taken at 10% completion of elongation reactions and were converted to elongation rates (assuming 10 μm−1s−1 for actin alone (32)).
We, therefore, tested whether the C terminus of FMNL3 could affect actin dynamics (Fig. 1B; Cterm-WT). We first tested Cterm-WT in monomeric form, cleaved from its N-terminal GST purification tag. In pyrene-actin assays, Cterm-WT inhibits actin polymerization from monomers in a concentration-dependent manner (Fig. 5A). In elongation assays, Cterm-WT inhibits barbed end elongation with an IC50 of 205 nm (Fig. 5B).
FIGURE 5.
The C terminus of FMNL3 is sufficient to alter actin dynamics, binds actin monomers, and slows barbed end elongation. A, pyrene-actin polymerization assays using 2 μm actin monomers (5% pyrene) and the indicated amounts of Cterm-WT. B, inhibition of filament elongation by Cterm-WT. Inset, raw pyrene-actin elongation assay using 1.5 μm polymerized filaments, 0.5 μm actin monomers (20% pyrene), and 2 μm Cterm-WT. C, direct binding assays using polarization anisotropy of 20 nm fluorescein-labeled Cterm-WT and increasing concentrations of latrunculin B-bound actin monomers in polymerization buffer. D, Pyrene-actin polymerization assays using 2 μm actin monomers (5% pyrene) and the indicated amounts of GST-Cterm-WT. E, inhibition of filament elongation by GST-Cterm-WT. F, competition binding assays using 20 nm fluorescein-labeled Cterm-WT, 1 μm latrunculin B-bound actin monomers, and increasing concentrations of unlabeled Cterm-WT or GST-Cterm-WT. G, pyrene-actin polymerization assays using 2 μm actin monomers (5% pyrene) and 1 μm concentrations of the indicated GST-Cterm construct. H, elongation of 1.5 μm polymerized actin in the presence of 0.5 μm actin monomers (20% pyrene) and 50 nm concentrations of the indicated GST-Cterm construct.
These assays suggest that Cterm-WT binds actin monomers and possibly binds filament barbed ends, as it inhibits barbed end elongation at concentrations significantly lower than the monomer concentration in these assays. To test actin monomer binding directly, we developed a polarization anisotropy assay in which Cterm-WT was labeled with fluorescein-maleimide on a cysteine engineered onto its N terminus to create FL-Cterm-WT. FL-Cterm-WT binds latrunculin B-bound actin monomers with a Kd of 0.9 μm (Fig. 5C).
We further tested whether Cterm-WT binds filament barbed ends using the dimeric GST-Cterm construct (GST-Cterm-WT). Our reasoning was that interaction with the barbed end (containing two actin subunits) would be enhanced in the dimeric state. Furthermore, the C terminus is presented as a dimer in full-length FMNL3 because it follows the dimeric FH2 domain. GST-Cterm-WT inhibits barbed end elongation with a significantly lower IC50 (0.4 nm) than the monomeric construct (Fig. 5E). Interestingly, the affinity of GST-Cterm-WT for actin monomers (Kd = 3.1 μm) is similar to that of the monomeric Cterm-WT (Kd = 1.8 μm), as measured by competition anisotropy assays with FL-Cterm-WT (Fig. 5F). Furthermore, submicromolar concentrations of GST-Cterm-WT cause an acceleration in the polymerization rate of actin from monomers (Fig. 5D), as opposed to the inhibition caused by the monomeric protein (Fig. 5A).
Past studies showed that the C termini of INF2 and mDia1 could bind actin monomers and strongly influence polymerization in combination with the FH2 domain (12, 14). We compared the FMNL3 C terminus to those of mDia1 and INF2 using the dimeric GST fusions. In elongation assays, neither GST-mDia1-Cterm nor GST-INF2-Cterm inhibits elongation at low (50 nm) concentration (Fig. 5H). At high concentration (10 μm), GST-mDia1-Cterm also has no effect on elongation (not shown). In contrast, 10 μm GST-INF2-Cterm causes complete elongation inhibition, consistent with the ability of the INF2 WH2 motif to bind actin monomers (not shown). In polymerization assays, GST-mDia1-Cterm does not increase actin polymerization rates at 1 μm, whereas GST-INF2-Cterm has an inhibitory effect, again presumably due to monomer binding (Fig. 5G). To test actin monomer binding, we used anisotropy assays in which we measured the abilities of the constructs to compete with FL-Cterm-WT. GST-mDia1-Cterm fails to compete with FL-Cterm-WT for actin monomer binding at concentrations up to 30 μm (Fig. 5F), suggesting either that it binds to a different region of the actin monomer or binds actin monomers with low affinity. In contrast, GST-INF2-Cterm competes with FL-Cterm-WT for actin monomer binding with a Kd of 1.0 μm (Fig. 5F).
The C Terminus of FMNL3 Contains a WH2-like Sequence Responsible for Actin Binding
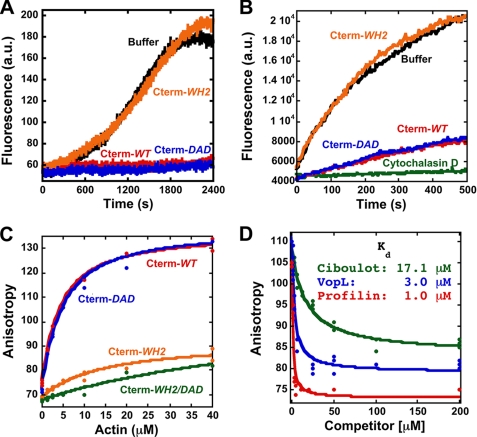
We were interested in the identity of the actin binding site in FMNL3 C terminus. Prior work suggested the existence of a WH2-like sequence upstream of the DAD in FMNL2 and FMNL3 (13). We mutated both the WH2-like sequence and the DAD in the FMNL3-Cterm construct and tested their effects on actin. In both cases we mutated adjacent aliphatic residues (Leu-970/Ile-971 in the WH2 and Ile-996/Ile-997 in DAD) because such residues are crucial for actin binding by WH2 domains (3). The WH2 mutation (Cterm-WH2) abolishes the effects on polymerization from monomers (Fig. 6A) and elongation (Fig. 6B), whereas the DAD mutation (Cterm-DAD) does not. In anisotropy binding assays using fluorescein-labeled proteins, Cterm-WH2 has much less affinity for actin monomers than wild type, whereas Cterm-DAD maintains full monomer binding affinity (Fig. 6C). In the dimeric GST fusion construct, the Cterm-WH2 mutant loses the ability to inhibit elongation and to stimulate polymerization from monomers (not shown). These results show that the WH2-like sequence is largely responsible for the Cterm effects on actin.
FIGURE 6.
The C terminus of FMNL3 contains a WH2-like sequence whose mutation disrupts monomer binding and inhibition of filament elongation. A, pyrene-actin polymerization assays using 2 μm actin monomers (5% pyrene) and 16 μm concentrations of the indicated Cterm constructs. B, filament elongation assays using 3.1 μm polymerized actin, 1 μm actin monomers (25% pyrene), and 8 μm concentrations of the indicated Cterm constructs. C, direct binding assays using polarization anisotropy of 20 nm concentrations of the indicated fluorescein-labeled Cterm constructs and increasing concentrations of latrunculin B-bound actin monomers in polymerization buffer. D, a competition binding assay with FMNL3 and increasing concentrations of Profilin or WH2 peptides Ciboulot or VopL. Assays were performed with 5 μm latrunculin B-bound actin monomers and 20 nm FL-Cterm-WT in polymerization buffer.
Although the WH2-like sequence possesses some attributes of other WH2s, it lacks a leucine residue that is highly conserved in most WH2s (Fig. 1C). To characterize this binding site further, we tested the abilities of two WH2 motifs (from Ciboulot and VopL) as well as profilin to compete with FL-Cterm-WT for actin monomer binding. All three proteins compete with FL-Cterm-WT (Fig. 6D). These results suggest that the C terminus of FMNL3 binds to a similar interface of actin monomers as these other proteins (15, 34–36).
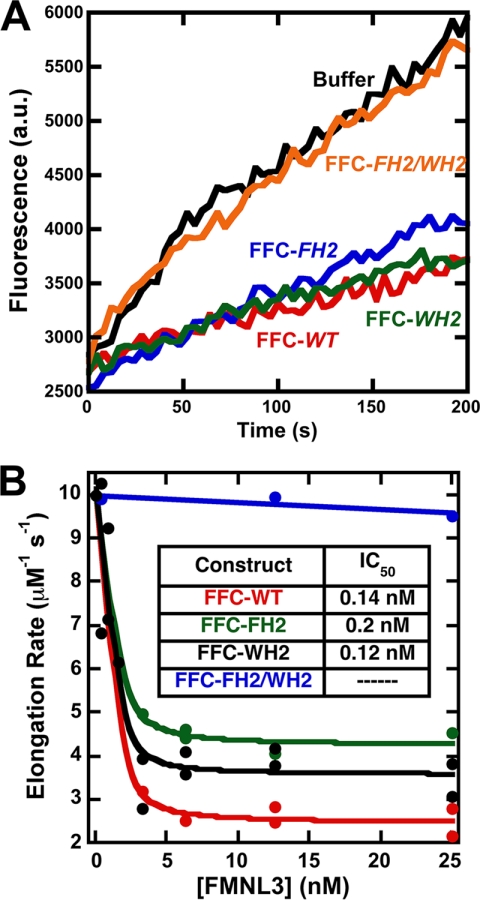
We then asked whether filament elongation inhibition by the WH2-like sequence accounted for the fact that the FFC-FH2 mutant maintained elongation inhibition, whereas the FF-FH2 mutant did not (Fig. 4A). In pyrene-actin elongation assays, the double mutant FFC-FH2/WH2 does not inhibit elongation at any concentration tested, whereas individual mutations of the WH2 or FH2 alone maintain potent elongation inhibition (Fig. 7). Together, these results show that the WH2-like sequence within the C terminus of FMNL3 can bind both actin monomers and filament barbed ends. When combined with the FH2 domain, the WH2 motif constitutes a second barbed end binding sequence that inhibits elongation.
FIGURE 7.
Mutation in the WH2-like sequence FH2-independent elongation inhibition. A, raw curves from filament elongation assays using 3.1 μm polymerized actin, 1 μm actin monomers (25% pyrene), and 6.25 nm concentrations of the indicated constructs. B, inhibition of filament elongation by the indicated FFC mutant constructs.
The C Terminus Stimulates Actin Polymerization Activity of FH2 Domain in Absence of High Affinity Actin Binding
We then tested whether the WH2-like sequence was important for accelerating polymerization in the FFC construct. Surprisingly, the WH2 mutation (FFC-WH2) has very little effect on the ability of the FFC construct to accelerate polymerization (Fig. 8A; supplemental Fig. 1E). In contrast, the FH2 mutation alone (FFC-FH2) drastically reduces FMNL3 polymerization activity, although it still accelerates polymerization at higher concentrations (Fig. 8, A and B). The residual polymerization ability of FFC-FH2 is almost completely eliminated in the FFC-FH2/WH2 double mutant (Fig. 8A and supplemental Fig. 1F). The FH2 mutation completely eliminates polymerization effects of the FF construct (Fig. 8, B and C). In severing assays, the severing activity of the FFC construct is more strongly compromised by the WH2 mutation than by the FH2 mutation, and the double FH2/WH2 mutant does not appreciably change severing activity from that of the WH2 mutation alone (Fig. 8D).
FIGURE 8.
Differential effects of C-terminal mutants on actin polymerization by the FFC construct. A, pyrene-actin polymerization assays containing 2 μm actin monomers (5% pyrene) with 40 nm concentrations of the indicated FFC constructs. B, filament production at 50% polymerization by the indicated FMNL3 constructs. C, pyrene-actin polymerization assays containing 2 μm actin monomers (5% pyrene) with 40 nm concentrations of the indicated FF constructs. D, quantification of filament lengths in a severing assay using 2 μm polymerized actin and 400 nm concentrations of the indicated FMNL3 construct. E, pyrene-actin polymerization assays containing 2 μm actin monomers (5% pyrene) with 25 nm FFC-WT (red), FFC-3xR (blue), or FFC-WH2/3xR (green). F, competition binding assays using 20 nm FL-Cterm-WT, 1 μm latrunculin B-bound actin monomers, and increasing concentrations of GST-Cterm-WT, GST-Cterm-3xR, or GST-Cterm-WH2/3xR.
A previous study showed that basic residues in the C terminus of mDia1 were important for accelerating actin polymerization by mDia1-FFC (12). FMNL3 contains three successive arginines at the C terminus of the WH2-like sequence (Fig. 1C). We mutated these residues to alanines (FFC-3xR) to test whether they played a role in C-terminal acceleration of actin polymerization by FFC. The FFC-3xR construct appreciably extends the lag phase of actin polymerization (Fig. 8E and supplemental Fig. 1G). Combining the WH2 and 3xR mutations does not result in a further reduction in polymerization ability. Similar to the Cterm-WH2 mutant, the Cterm-3xR mutant displays no apparent affinity for actin monomers, as measured by competition anisotropy assay (Fig. 8F).
DISCUSSION
In this study we find that the addition of the C terminus of FMNL3 dramatically accelerates actin polymerization by the FH2 domain as well as increases its efficiency of filament bundling and severing. We characterize an actin binding motif within the C terminus of FMNL3 that directly interacts with actin monomers and filament barbed ends. Profilin and WH2 domains disrupt binding of the C terminus to actin, suggesting a common binding interface. Point mutagenesis of adjacent aliphatic residues within this motif strongly inhibit the actin binding ability of the C terminus. Curiously, however, these mutations do not influence the ability of the FFC construct to accelerate actin polymerization. This study extends work by others, suggesting that the C termini of FMNL2 and FMNL3 influence interactions with actin (13). In addition, our work reveals that the C termini of three different formins (FMNL3, mDia1, INF2) adopt three different strategies in influencing actin polymerization.
The C Terminus of FMNL3 Contains a WH2-like Sequence
The C-terminal actin binding motif we identify corresponds to a previously predicted WH2-like sequence, based on alignments with other known WH2-containing proteins (13). The WH2 domain is present in a wide variety of actin-binding proteins (3, 15, 16, 37) and consists of an amphiphilic helix that binds between subdomains 1 and 3 of actin followed by an extended region that makes a crucial contact with the lateral face of actin (15). The binding interface for the WH2 helix corresponds roughly the barbed end of an actin monomer (38) and partially overlaps with the profilin binding interface (36). Three aliphatic residues, including two adjacent residues in the helix and a third residue in the extended region, make crucial hydrophobic contacts in the WH2/actin interaction (Fig. 1C). This third aliphatic residue is a leucine in almost all cases and is often followed by one or two basic residues (3, 15).
The FMNL3 actin binding motif is roughly similar to a WH2 domain, with one potentially important difference. FMNL3 contains two adjacent aliphatic residues within the predicted α-helix but does not have a leucine as the third aliphatic residue, although an alanine might functionally substitute to some degree (Fig. 1C). Two findings suggest that this motif is similar to a WH2; 1) three WH2 domains (VopL, Ciboulot, and INF2) as well as profilin compete with Cterm-WT for binding to actin monomers, and 2) mutation of the two adjacent aliphatic residues (Leu-970 and Ile-971) strongly decreases binding affinity. We wonder, however, whether the binding interactions in the extended region are slightly different for FMNL3 as opposed to most WH2 domains.
A curious feature of FMNL3 C terminus is its ability to slow barbed end elongation in three different contexts: as the monomeric Cterm-WT construct, as the dimeric GST-Cterm-WT construct, and as the dimeric FFC-FH2 mutant construct (Fig. 9A). The WH2s from N-WASP and Spire exhibit barbed end binding ability, whereas those of Cobl do not (20, 21, 25, 26). Our results show that the barbed end binding affinity for the FMNL3 C terminus is dramatically increased when it is dimeric, approaching the affinity of FMNL3 FH2 domain, although its actin monomer binding affinity remains unchanged. This effect is consistent with the thermodynamic prediction that a dimeric ligand (the barbed end) will bind more tightly to a dimeric binding protein than to a monomer.
FIGURE 9.
Contributions of the FMNL3 C terminus to actin binding and actin filament severing. A, comparison of the observed effects of FMNL3 Cterm (1), GST-FMNL3 Cterm (2), and FMNL3 FH2-C (3) on barbed end elongation. FMNL3 FH2-C has two possible barbed end binding states, FH2-bound and Cterm-bound. B, comparison of actin monomer affinity of FMNL3-Cterm with the C termini of INF2 and mDia1. The proposed binding interface of FMNL3 and INF2 Cterm is positioned in the cleft at the barbed end of the actin monomer, located between subdomains 1 and 3. The binding site of mDia1-Cterm is unknown. C, proposed mechanism of filament severing by FMNL3-FFC. FH2 dimer dissociates (1), allowing filament side binding by the FH2 domain (2), which enables binding by the cis Cterm to subunits adjacent to those binding the FH2, destabilizing the filament (3), and ultimately resulting in severing (4). Barbed end affinity of the FH2 causes it to remain with the barbed end of the newly severed filament. D, comparison of the C termini from mouse FMNL3, INF2 (CAAX variant), and mDia1. Schematic alignments are based on the last 10 residues of their FH2 domains. The DAD boundaries represent the “core DAD” regions for FMNL3 and INF2 and extend to the basic residues in mDia1 (5). The arrow in FMNL3 denotes the site of splice variation after T999.
Another curious feature of FMNL3 C terminus is that, while inhibiting actin polymerization in monomeric form, it accelerates actin polymerization as the dimeric GST fusion protein. This property may be related to the nucleation activities of several COWs, including Spire, Cobl, and VopL (19, 22–24). One model for nucleation by these proteins is through positioning monomers in configurations that allow for subsequent actin monomer addition. In a similar fashion, the dimeric C-terminal construct might stabilize an actin dimer-like configuration, which allows recruitment of actin monomers in a polymerizable state. In COWs, at least one additional actin binding sequence is required for effective nucleation, such as a basic region just N-terminal to the three WH2s in Cobl (21), the L3 sequence in Spire (23), and the dimeric C-terminal domain in VopL (22, 24). It is unclear as to whether the FMNL3 C terminus possesses such a sequence.
Comparison of FMNL3 with Other Formins
The defining characteristic of the formin class of actin nucleating factors is the presence of the FH2 domain (5). The FH2 is a high affinity barbed end binding domain that processively moves with elongating barbed ends in all but one case (39). However, FH2 domains vary widely in their abilities to nucleate actin filaments, with FH2s from mDia1 and mDia2 being extremely potent nucleators, whereas DAAM1 FH2 possesses almost no nucleation activity (8, 12, 40). Previously, we have shown that FMNL1 FFC is a poor nucleator (7), and we show here that the FMNL3-FF construct has even lower nucleation activity.
Recently, the role of sequences C-terminal to the FH2 domain on actin polymerization has been appreciated for several formins. For INF2, the addition of the C terminus enhances actin polymerization from monomers in addition to introducing a filament severing activity on ADP-actin filaments and accelerating depolymerization (14). The addition of C-terminal sequences to several other formins, including mDia1, DAAM1, Bni1p, and Bnr1p, increases polymerization from monomers without introducing a severing or depolymerization activity (12). Here, we show that the C terminus of FMNL3 dramatically increases polymerization activity as well as increasing severing.
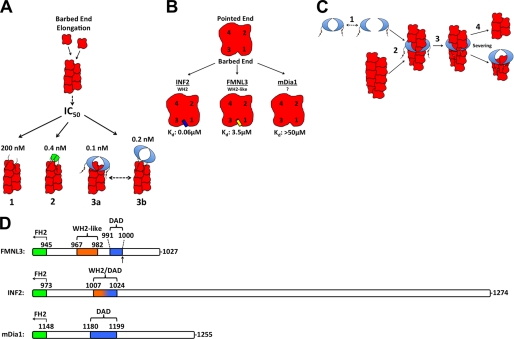
These works show that C-terminal sequences contribute strongly to the ability of formins to influence actin dynamics but do so in different ways. We discuss briefly the characteristics of the three best-characterized formin C termini: INF2, mDia1, and FMNL3 (Table 1 and Fig. 9, B and D). INF2 contains a clear WH2 C-terminal to the FH2. This WH2 binds actin monomers with high affinity (Kd of 60 nm (14)) but does not display apparent barbed end binding (this study). High affinity actin binding by the WH2 is crucial for INF2 depolymerization activity. Interestingly, INF2 WH2 overlaps almost completely with its DAD (Fig. 9D), and mutations in the crucial aliphatic residues abolish both actin binding and DID binding (41).
TABLE 1.
Properties of formin C-terminal regions
The effects of INF2, mDia1, and FMNL3 on actin polymerization, elongation, and monomer binding are presented.
For mDia1, the C terminus binds actin monomers with significantly lower affinity than for INF2, with an apparent Kd in the hundreds of μm (Fig. 9B) (12). The mDia1 C terminus accelerates actin polymerization from monomers at micromolar concentrations when used as the dimeric GST-Cterm fusion (12). Mutational analysis suggests that the relevant motif for actin binding might not be a canonical WH2 domain nor does it depend on key residues within the core DAD, as mutation of a leucine that would be important for WH2 function (Leu-1185) does not abolish actin polymerization effects (12). Rather, mutation of two adjacent basic residues C-terminal to the core DAD (Lys-1198, Arg-1199) compromises actin polymerization effects. Thus, the mDia1 C-terminal actin binding motif appears somewhat different from that of INF2 and contributes different properties (accelerated polymerization but not depolymerization).
For FMNL3, a third variation appears to exist. FMNL3 possesses an actin binding motif that binds actin monomers with intermediate affinity (Kd of 0.9 μm) between INF2 and mDia1. In addition, the FMNL3 C terminus binds barbed ends and slows actin filament elongation, which is not the case for INF2 or mDia1. Like INF2, the FMNL3 actin binding motif resembles a WH2 domain but is missing the third crucial leucine found in canonical WH2s. In addition, the FMNL3 actin binding motif does not overlap at all with its DAD, which has been mapped to residues Gly-991—Phe-1000 (Fig. 9D) (13).
Function of FMNL3 C Terminus in the Context of the FH2 Domain
It is interesting that FMNL3 contains two barbed end binding sequences (the FH2 and the C terminus) in close proximity. Both sequences slow barbed end elongation with high affinity in dimeric form. The presence of the C terminus in general is also crucial for potent acceleration of actin polymerization by the FH2 construct. Puzzlingly, mutations of two aliphatic residues in the WH2-like sequence, which dramatically reduce its actin binding ability, do not compromise actin polymerization acceleration in the FFC construct. This phenomenon has also been observed with a construct of N-WASP containing a single WH2 motif (25). Mutants of the WH2 in this N-WASP construct still stimulate nucleation through the Arp2/3 complex. These results provoke the question, What is the purpose of high affinity actin binding in these cases? In N-WASP, the WH2 motif appears to be important in keeping the resulting barbed end tethered to the membrane (25), which is important as Arp2/3 complex remains at the pointed end. In the case of FMNL3, the FH2 domain presumably remains at the barbed end after nucleation, so the function of the Cterm as a membrane tether seems superfluous.
Although mutation of the two aliphatic residues in the WH2-like sequence (WH2 mutation) does not affect polymerization by the FFC construct, mutation of three arginines (3xR mutation) only four residues C-terminal does have a significant effect. Both the WH2 and the 3xR mutations abolish actin monomer binding by the C terminus. We presume that these residues all contribute to one actin binding site. Therefore, it is extremely curious that the two mutants affect FFC-mediated polymerization differently. In this respect, there might be similarities between FMNL3 and mDia1 (12) in that adjacent C-terminal basic residues contributing little actin binding affinity alone can work together with the FH2 domain to enhance polymerization.
Mutation of the aliphatic residues in the WH2-like sequence do, however, decrease severing efficiency of FFC-WH2, which raises the question as to how FMNL3 might sever filaments. A closer look at the crystal structures of WH2 domains and the FH2 domain bound to actin may provide some clues. In the case of Bni1p complexed with TMR-actin, the αD helix, which contains the well conserved isoleucine (Ile-1431) crucial for FH2-mediated polymerization, binds the pocket formed by subdomains 1 and 3 of actin (42). As discussed above, the amphiphilic α-helix of the WH2 also binds within this same pocket, and this is likely the binding site for the FMNL3 C terminus. Because these sites are already occupied by the FH2 domain on the barbed end subunits, the FMNL3 C terminus might bind other subunits within the filament, destabilizing he filament and causing severing. This mechanism has been proposed for Cobl (21). For FMNL3, severing could happen at barbed ends or at filament sides, as FMNL3 is capable of side binding (Fig. 9C).
The actin filament bundling activity of FF-WT as well as many of the mutants tested displays sigmoidal concentration dependence, which could suggest a cooperative component of bundling for these constructs. However, the low speed pelleting assays used here, which simply measure changes in total pelletable actin, are inappropriate for detailed characterization of this potential effect. Followup studies using more detailed assays (rheometry, direct bundling studies by microscopy) are necessary in these investigations.
We suspect that the FMNL3 WH2-like motif contributes to more than increasing severing efficiency. Other possibilities include cooperation with profilin for supplying monomers to the FH2-bound barbed end or for synergistic interaction with other actin-binding proteins that might functionally associate with FMNL3 in cells. It will be interesting in this respect to test the effect of profilin on polymerization by these constructs. Finally, we point out that the FMNL3 C terminus is subject to splice variation, altering the sequence C-terminal to the DAD. Although this splice variation appears to be outside of the region containing the WH2-like sequence, it is unclear at this point what effect the splice variants would have on the ability of FMNL3 to accelerate actin polymerization.
Supplementary Material
Acknowledgments
We gratefully thank Roberto Dominguez for providing the VopL and Ciboulot WH2 peptides. We also thank Maddy Higgs for help with writing and Lance Timon-Yoprizati for tireless additions to a very dynamic process.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM069818 (to H. N. H.) and F31 GM089149 (Ruth L. Kirschstein Predoctoral National Research Service Award (to E. G. H.)). This work was also supported by the Hitchcock Foundation (to H. N. H.).

This article contains supplemental Figs. 1–3.
- FH2
- formin homology 2
- DAD
- diaphanous autoinhibitory domain
- WH2
- WASp homology 2
- TRITC
- tetramethylrhodamine isothiocyanate
- FF
- FH1-FH2
- FFC
- FH1-FH2-C domains
- a.u.
- arbitrary units
- Cterm
- C-terminus
- 3xR
- 3× arginine residues
- COWs
- compound WH2 domain proteins.
REFERENCES
- 1. Chhabra E. S., Higgs H. N. (2007) Nat. Cell Biol. 9, 1110–1121 [DOI] [PubMed] [Google Scholar]
- 2. Campellone K. G., Welch M. D. (2010) Nat. Rev. Mol. Cell Biol. 11, 237–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dominguez R. (2009) Crit. Rev. Biochem. Mol. Biol. 44, 351–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Husson C., Cantrelle F. X., Roblin P., Didry D., Le K. H., Perez J., Guittet E., Van Heijenoort C., Renault L., Carlier M. F. (2010) Ann. N.Y. Acad. Sci. 1194, 44–52 [DOI] [PubMed] [Google Scholar]
- 5. Higgs H. N., Peterson K. J. (2005) Mol. Biol. Cell 16, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goode B. L., Eck M. J. (2007) Annu. Rev. Biochem. 76, 593–627 [DOI] [PubMed] [Google Scholar]
- 7. Harris E. S., Rouiller I., Hanein D., Higgs H. N. (2006) J. Biol. Chem. 281, 14383–14392 [DOI] [PubMed] [Google Scholar]
- 8. Li F., Higgs H. N. (2003) Curr. Biol. 13, 1335–1340 [DOI] [PubMed] [Google Scholar]
- 9. Lu J., Meng W., Poy F., Maiti S., Goode B. L., Eck M. J. (2007) J. Mol. Biol. 369, 1258–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yamashita M., Higashi T., Suetsugu S., Sato Y., Ikeda T., Shirakawa R., Kita T., Takenawa T., Horiuchi H., Fukai S., Nureki O. (2007) Genes Cells 12, 1255–1265 [DOI] [PubMed] [Google Scholar]
- 11. Kovar D. R., Harris E. S., Mahaffy R., Higgs H. N., Pollard T. D. (2006) Cell 124, 423–435 [DOI] [PubMed] [Google Scholar]
- 12. Gould C. J., Maiti S., Michelot A., Graziano B. R., Blanchoin L., Goode B. L. (2011) Curr. Biol. 21, 384–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vaillant D. C., Copeland S. J., Davis C., Thurston S. F., Abdennur N., Copeland J. W. (2008) J. Biol. Chem. 283, 33750–33762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chhabra E. S., Higgs H. N. (2006) J. Biol. Chem. 281, 26754–26767 [DOI] [PubMed] [Google Scholar]
- 15. Chereau D., Kerff F., Graceffa P., Grabarek Z., Langsetmo K., Dominguez R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16644–16649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hertzog M., van Heijenoort C., Didry D., Gaudier M., Coutant J., Gigant B., Didelot G., Préat T., Knossow M., Guittet E., Carlier M. F. (2004) Cell 117, 611–623 [DOI] [PubMed] [Google Scholar]
- 17. Higgs H. N., Blanchoin L., Pollard T. D. (1999) Biochemistry 38, 15212–15222 [DOI] [PubMed] [Google Scholar]
- 18. Higgs H. N., Pollard T. D. (2001) Annu. Rev. Biochem. 70, 649–676 [DOI] [PubMed] [Google Scholar]
- 19. Ahuja R., Pinyol R., Reichenbach N., Custer L., Klingensmith J., Kessels M. M., Qualmann B. (2007) Cell 131, 337–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bosch M., Le K. H., Bugyi B., Correia J. J., Renault L., Carlier M. F. (2007) Mol. Cell 28, 555–568 [DOI] [PubMed] [Google Scholar]
- 21. Husson C., Renault L., Didry D., Pantaloni D., Carlier M. F. (2011) Mol. Cell 43, 464–477 [DOI] [PubMed] [Google Scholar]
- 22. Namgoong S., Boczkowska M., Glista M. J., Winkelman J. D., Rebowski G., Kovar D. R., Dominguez R. (2011) Nat. Struct. Mol. Biol. 18, 1060–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Quinlan M. E., Heuser J. E., Kerkhoff E., Mullins R. D. (2005) Nature 433, 382–388 [DOI] [PubMed] [Google Scholar]
- 24. Yu B., Cheng H. C., Brautigam C. A., Tomchick D. R., Rosen M. K. (2011) Nat. Struct. Mol. Biol. 18, 1068–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Co C., Wong D. T., Gierke S., Chang V., Taunton J. (2007) Cell 128, 901–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ito T., Narita A., Hirayama T., Taki M., Iyoshi S., Yamamoto Y., Maéda Y., Oda T. (2011) J. Mol. Biol. 408, 18–25 [DOI] [PubMed] [Google Scholar]
- 27. Harris E. S., Gauvin T. J., Heimsath E. G., Higgs H. N. (2010) Cytoskeleton 67, 755–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Harris E. S., Li F., Higgs H. N. (2004) J. Biol. Chem. 279, 20076–20087 [DOI] [PubMed] [Google Scholar]
- 29. Spudich J. A., Watt S. (1971) J. Biol. Chem. 246, 4866–4871 [PubMed] [Google Scholar]
- 30. Pollard T. D., Cooper J. A. (1984) Biochemistry 23, 6631–6641 [DOI] [PubMed] [Google Scholar]
- 31. MacLean-Fletcher S., Pollard T. D. (1980) Cell 20, 329–341 [DOI] [PubMed] [Google Scholar]
- 32. Kuhn J. R., Pollard T. D. (2005) Biophys. J. 88, 1387–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xu Y., Moseley J. B., Sagot I., Poy F., Pellman D., Goode B. L., Eck M. J. (2004) Cell 116, 711–723 [DOI] [PubMed] [Google Scholar]
- 34. Lee S. H., Kerff F., Chereau D., Ferron F., Klug A., Dominguez R. (2007) Structure 15, 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liverman A. D., Cheng H. C., Trosky J. E., Leung D. W., Yarbrough M. L., Burdette D. L., Rosen M. K., Orth K. (2007) Proc. Natl. Acad. Sci. U. S. A. 104, 17117–17122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schutt C. E., Myslik J. C., Rozycki M. D., Goonesekere N. C., Lindberg U. (1993) Nature 365, 810–816 [DOI] [PubMed] [Google Scholar]
- 37. Paavilainen V. O., Bertling E., Falck S., Lappalainen P. (2004) Trends Cell Biol. 14, 386–394 [DOI] [PubMed] [Google Scholar]
- 38. Otterbein L. R., Graceffa P., Dominguez R. (2001) Science 293, 708–711 [DOI] [PubMed] [Google Scholar]
- 39. Michelot A., Derivery E., Paterski-Boujemaa R., Guérin C., Huang S., Parcy F., Staiger C. J., Blanchoin L. (2006) Curr. Biol. 16, 1924–1930 [DOI] [PubMed] [Google Scholar]
- 40. Li F., Higgs H. N. (2005) J. Biol. Chem. 280, 6986–6992 [DOI] [PubMed] [Google Scholar]
- 41. Chhabra E. S., Ramabhadran V., Gerber S. A., Higgs H. N. (2009) J. Cell Sci. 122, 1430–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Otomo T., Tomchick D. R., Otomo C., Panchal S. C., Machius M., Rosen M. K. (2005) Nature 433, 488–494 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.