Background: The rhomboid protease GlpG is a prototype of the serine intramembrane protease.
Results: The crystal structure of GlpG in complex with diisopropyl fluorophosphonate was solved at 2.3 Å resolution.
Conclusion: The crystal structure provides a model for the tetrahedral transitional state and reveals conformational changes within the active site.
Significance: Learning how GlpG conducts catalysis is crucial for understanding the mechanism of intramembrane proteolysis by rhomboid proteases.
Keywords: Crystallography, Enzyme Catalysis, Intramembrane Proteolysis, Protease Inhibitor, Rhomboid Protease, Catalytic Mechanism
Abstract
Rhomboid proteases have many important biological functions. Unlike soluble serine proteases such as chymotrypsin, the active site of rhomboid protease, which contains a Ser-His catalytic dyad, is submerged in the membrane and surrounded by membrane-spanning helices. Previous crystallographic analyses of GlpG, a bacterial rhomboid protease, and its complex with isocoumarin have provided insights into the mechanism of the membrane protease. Here, we studied the interaction of GlpG with 3,4-dichloroisocoumarin and diisopropyl fluorophosphonate, both mechanism-based inhibitors for the serine protease, and describe the crystal structure of the covalent adduct between GlpG and diisopropyl fluorophosphonate, which mimics the oxyanion-containing tetrahedral intermediate of the hydrolytic reaction. The crystal structure confirms that the oxyanion is stabilized by the main chain amide of Ser-201 and by the side chains of His-150 and Asn-154. The phosphorylation of the catalytic Ser-201 weakens its interaction with His-254, causing the catalytic histidine to rotate away from the serine. The rotation of His-254 is accompanied by further rearrangement of the side chains of Tyr-205 and Trp-236 within the substrate-binding groove. The formation of the tetrahedral adduct is also accompanied by opening of the L5 cap and movement of transmembrane helix S5 toward S6 in a direction different from that predicted by the lateral gating model. Combining the new structural data with those on the isocoumarin complex sheds further light on the plasticity of the active site of rhomboid membrane protease.
Introduction
Rhomboid protease was first discovered in Drosophila genetics as a key member of the EGF receptor signaling pathway in flies (1, 2). The requirement of rhomboid in the pathway is due to its proteolytic activity against the membrane-tethered growth factors, which controls the release of the growth factors from signal-sending cells (3, 4). Sequence analysis has since shown that Drosophila rhomboid belongs to a conserved and widespread protein family (5, 6), with a characteristic core structure of six transmembrane (TM)2 domains (7). The biological functions of many rhomboid proteases are known (for reviews, see Refs. 8–10). Besides the role of Drosophila rhomboid in growth factor signaling, rhomboid proteases have been found to be involved in quorum sensing in Providencia (11), in the functions of mitochondria such as membrane remodeling and apoptosis (12–14), and in the life cycle of apicomplexan parasites (15–17) by cleaving various membrane protein substrates.
Rhomboid protease uses a Ser-His dyad to catalyze peptide hydrolysis (4, 18). The crystal structures of bacterial rhomboid GlpG have been characterized by us (7) and by others (19–22). The crystal structure confirmed the earlier prediction that the active site of the protease is embedded in the membrane and surrounded by multiple TM helices. Like site-2 protease and presenilin/γ-secretase, many rhomboid proteases are capable of cleaving substrates within their TM domains (23, 24). A previous study found that rhomboid protease is insensitive to most serine protease inhibitor classes (25), although the reason behind this phenomenon has never been clear. 3,4-Dichloroisocoumarin (DCI), a mechanism-based and broad-spectrum serine protease inhibitor (26), is the only compound that has demonstrated a consistent inhibitory effect for multiple rhomboid proteases (4, 18, 25).
Rhomboid protease must change conformation to allow substrate access to its internal active site, but how this occurs has been debated. The observations that the L5 cap, a surface loop that sits above the Ser-His catalytic dyad, is intrinsically flexible (27, 28) and that the lipid bilayer appears to be constricted around the protease (29, 30) have led to the hypothesis that a portion of the substrate's TM helix may partition in solution and bend into the active site through the opening created by the displaced L5 cap (23, 29). This model is easily compatible with the protease's dual capability of cleaving substrates not only within the TM domain but also in the juxtamembrane region (31–33). The second hypothesis is based on the observation that TM segment S5 is heavily tilted in one crystal form of GlpG (19). The unusual structure was proposed to represent the open conformation because the separation of S5 from the main body of the protease created a large lateral opening inside the membrane, and TM substrate was thought to go through the opening to reach the active site (19). Mutations have been found in S5 and an opposing helix (S2) that appear to enhance the activity of the protease (34–36).
The crystal structure of a complex between GlpG and 7-amino-4-chloro-3-methoxyisocoumarin was reported recently (37). This represents a major breakthrough because it confirms structurally the acyl-enzyme mechanism for rhomboid protease. The isocoumarin is bound to the protease through two covalent bonds, one with the catalytic Ser-201 and the other with the catalytic His-254. The bond with His-254 does not occur with natural peptide substrate and may have the unwanted effect of altering the conformation of the histidine or the inhibitor. Here, we have identified diisopropyl fluorophosphonate (DFP) as a potent inhibitor of GlpG and describe the crystal structure of its complex with the protease, which is stabilized by a single covalent bond with Ser-201. The structure of the phosphorylated protease also for the first time provides a model for the tetrahedral transition state of the hydrolytic reaction. On the basis of the crystal structure of the two protease-inhibitor complexes, we discuss conformational changes that may occur at the active site of rhomboid protease during catalysis.
EXPERIMENTAL PROCEDURES
Reagents
DFP and DCI were purchased from EMD Chemicals and Sigma, respectively. The detergents were purchased from Anatrace.
Hydrolysis of DCI
DCI was dissolved in Me2SO to prepare a 100 mm stock solution. All hydrolytic reactions were conducted in 100 μl of assay buffer (50 mm Tris (pH 8), 0.1 m NaCl, and 0.5% n-nonyl-β-d-glucopyranoside), with the exception of hydrolysis under alkaline conditions, where the reaction was conducted in 0.1 m NaOH. To investigate spontaneous hydrolysis at room temperature, the DCI stock solution was diluted to 500 μm in assay buffer and sampled at various time points. To examine the effect of GlpG on DCI hydrolysis, 40 μm GlpG was included in the reaction system. The products of hydrolysis were separated and analyzed by LC-MS using an Agilent 1200 infinity series LC system coupled to an Agilent 6120 quadrupole LC-MS system. (The HPLC running buffer was the same as the assay buffer, except that for hydrolysis in NaOH, the running buffer contained 100 mm Tris (pH 8).)
GlpG Activity Assay
The assay conditions were adapted from the literature (7, 31). In brief, 10 μg of BODIPY FL casein (Invitrogen) was mixed at 37 °C with 20 μm GlpG in 0.2 ml of assay buffer. The proteolysis of casein causes an increase in fluorescence intensity. Fluorescence emission at 513 nm was measured continuously at 37 °C with an excitation wavelength of 503 nm using a Tecan Safire multidetection microplate reader. Me2SO stock solutions of DCI and DFP (both 100 mm) were added to the enzyme solution to reach different final concentrations, and the mixtures were assayed for proteolytic activity using BODIPY FL casein. To examine the stability of the protease-inhibitor complex, GlpG was mixed with the inhibitor and incubated at room temperature for different periods of time before adding casein.
Crystal Structure Determination
Recombinant GlpG was prepared, purified, and crystallized as described previously (7). Before soaking, GlpG crystals were transferred to a fresh drop of reservoir solution (10 μl) containing 100 mm BisTris propane (pH 7.0) and 3 m NaCl. To compensate for the spontaneous hydrolysis of DFP in water, we tried either soaking at high inhibitor concentrations (∼500 mm) or repeated soaking at low concentrations (10∼20 mm). Repeated soaking was detrimental to x-ray diffraction, but the protein crystal seemed to tolerate well soaking once with high concentrations of inhibitor. To generate the protease-inhibitor complex, 1 μl of liquid DFP (5.7 m) was directly added to the 10-μl drop with the protein crystals. After sitting for 5 days at room temperature, the crystals were cryoprotected with 25% glycerol and flash-cooled in liquid nitrogen. Diffraction data were collected using an in-house system (Rigaku MicroMax x-ray generator with an R-AXIS IV detector). The diffraction images were processed by HKL2000 (46). The apostructure (Protein Data Bank code 2IC8), with water and detergent molecules removed, was used as a starting model for refinement and map inspection. After several rounds of model building using Coot (47) and refinement with REFMAC5 (48), DFP was modeled into the structure, which lowered both R and Rfree by ∼1%. Further positional and B-factor refinement was carried out by PHENIX (49). The final model has an Rfree of 23.8% and good geometry (see Table 1). Direct co-crystallization of GlpG with DFP has so far failed to yield crystals.
TABLE 1.
Crystallographic statistics
GlpG was crystallized in space group H32. r.m.s.d., root mean square deviation.
| GlpG-DFP | |
|---|---|
| Data collection | |
| Cell dimensions (Å) | a = b = 109.7, c = 125.1 |
| Wavelength (Å) | 1.54 |
| Resolution (Å)a | 40.0–2.3 (2.38–2.30) |
| Observed reflections | 130,991 |
| Unique reflections | 12,996 |
| Redundancy | 10.1 |
| Completeness (%)a | 99.2 (95.6) |
| 〈I/σ〉a | 15.7 (2.8) |
| Rmergea,b | 0.063 (0.502) |
| Refinement | |
| Resolution (Å) | 40.0–2.3 |
| Rwork/Rfreec | 0.219/0.238 |
| No. of atoms | |
| Protein | 1373 |
| DFP | 10 |
| Water | 43 |
| B-factors | |
| Protein | 60 |
| DFP | 59 |
| Water | 65 |
| r.m.s.d. | |
| Bond length (Å) | 0.007 |
| Bond angle | 0.985o |
a The highest resolution shell is shown in parentheses.
b Rmerge = Σ|Ii − 〈I〉|/ΣIi.
c Rwork = Σ|Fo − Fc|/ΣFo. Rfree is the cross-validation R-factor for the test set of reflections (5% of the total) omitted in model refinement.
RESULTS
Unlike DCI, DFP Causes Irreversible Inhibition of Rhomboid Protease GlpG
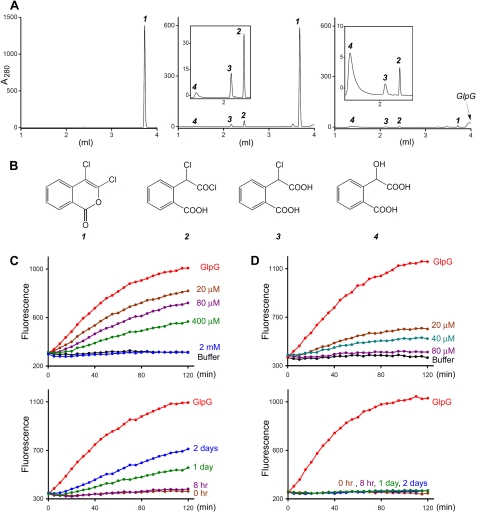
An earlier attempt to react crystalline GlpG with DCI had failed to generate a stable protease-inhibitor complex for crystallographic observation (27). The result was puzzling because DCI had been shown to prevent GlpG-catalyzed proteolysis in detergent solutions (18), and in the crystal lattice, the catalytic serine (Ser-201) of the protease is in an exposed position to react with the inhibitor (27). To better understand the inhibitory effect of DCI, we studied the reaction of the inhibitor with GlpG by LC-MS. Under our assay conditions, the inhibitor (1) underwent spontaneous hydrolysis to yield the acid chloride (2), the free carboxylic acid (3), and the fully dechlorinated α-hydroxy acid (4) (Fig. 1, A and B). Upon mixing with GlpG, the peak that corresponds to the intact inhibitor became greatly reduced. The amount of reduction was greater than that of the protease, indicating that the loss is not solely due to formation of the protease-inhibitor complex. Of the three hydrolytic products, only the amount of the α-hydroxy acid (4) increased (supplemental Fig. S1) (38). Taken together, these results suggest that a significant portion of the inhibitor bound to GlpG is enzymatically turned over (supplemental Fig. S2) and does not remain stably attached to the protease. Consistent with this possibility, we found that even at a DCI concentration of 2 mm, at which GlpG was fully inhibited initially, the proteolytic activity gradually returned when the inhibitor became hydrolyzed (Fig. 1C).
FIGURE 1.
Inhibition of GlpG by DCI and DFP. A, HPLC elution profiles for freshly prepared DCI (left panel), DCI after 5 h of incubation (middle panel), and DCI after 5 h of incubation with GlpG (right panel). Note that the scale for the y axis is larger for the freshly prepared sample. The insets show the details of the three product peaks. The extinction coefficients for the three hydrolytic products are much smaller than that for the double-ringed DCI. B, chemical structures for intact DCI (1), acid chloride (2), carboxylic acid (3), and α-hydroxy acid (4). C, the proteolysis of BODIPY FL casein by GlpG could be fully inhibited by 2 mm DCI (upper panel). With prolonged incubation, however, the inhibitory effect of DCI was partially lost (the initial concentration of the inhibitor was 2 mm) (lower panel). D, the inhibition of GlpG by DFP was irreversible (80 μm DFP) (lower panel).
Phosphorylating agents such as DFP are classic mechanism-based inhibitors for the serine protease. Under our assay conditions, DFP was a more potent inhibitor of GlpG than isocoumarin, achieving complete protease inactivation at a concentration of ∼80 μm (Fig. 1D). More important, the DFP-inhibited protease did not regain activity upon prolonged incubation, suggesting that the covalent bond between the inhibitor and GlpG is stable, which was confirmed by the crystallographic analysis described below. Our result contradicts the conclusion of a previous study stating that the organophophonate class of serine protease inhibitors is ineffective against rhomboid activity (25).
Crystal Structure of GlpG-DFP Complex
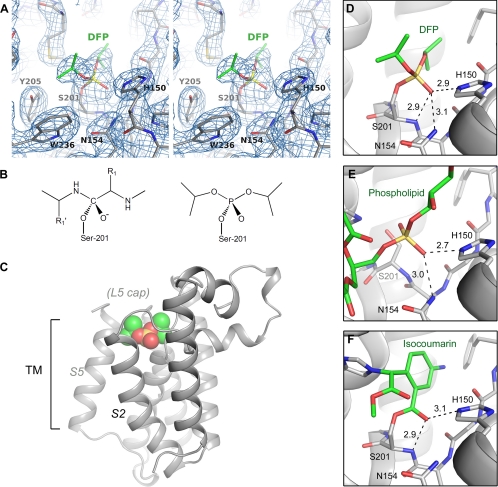
The protease-inhibitor complex was generated by directly soaking DFP in preformed GlpG crystals. A complete x-ray diffraction data set was collected to 2.3 Å resolution from a single DFP-treated crystal, and difference Fourier analysis immediately revealed several prominent changes in the active site of the membrane protease (supplemental Fig. S3). Near the catalytic serine (Ser-201), a strong positive peak indicates the presence of the covalently bound inhibitor; a long stretch of negative density is observed around the L5 loop, suggesting that the cap has become disordered; and pairs of positive and negative density peaks adjacent to Tyr-205, Trp-236, and His-254 indicate that their side chains have rotated into new positions. Fig. 2A shows the final 2Fo − Fc electron density map surrounding the inhibitor after model building and refinement (Table 1).
FIGURE 2.
Structure of DFP complex. A, stereo image showing the final 2Fo − Fc electron density map (contoured at 1.2σ levels) around the inhibitor. B, the chemical structure of the phosphorylated enzyme (right) mimics that of the oxyanion-containing tetrahedral intermediate (left). C, overall structure of the complex. The inhibitor is shown as a space-filling model. D, the phosphonyl oxygen of DFP occupies the oxyanion-binding site. Hydrogen bonds are shown as dashed lines, and the numbers are inter-atomic distances in angstroms. E, binding of a phospholipid to the active site of GlpG (Protein Data Bank code 2IRV) (20). F, structure of the isocoumarin complex (Protein Data Bank code 2XOW) (37). Note that the inhibitor is covalently bonded to both Ser-201 and His-254.
The hydroxyl oxygen of Ser-201 replaced the fluorine in DFP and is covalently bonded to the phosphorus atom. The phosphorylated enzyme resembles the tetrahedral intermediate of the proteolytic reaction (Fig. 2B). The two isopropyl alcohol groups of the inhibitor, which mimic peptide backbones on either side of the tetrahedral carbon, are fully extended and roughly parallel to the membrane plane (Fig. 2C). The phosphonyl oxygen of DFP is hydrogen-bonded to the backbone nitrogen of Ser-201 and to the side chains of His-150 and Asn-154 (Fig. 2D). The position of the oxygen probably corresponds to the binding site for the oxyanion developed during normal catalysis.
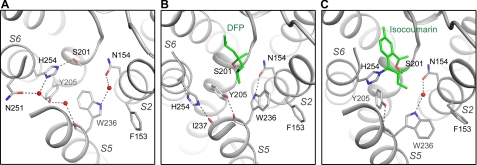
Conformational Changes within Active Site
The reaction with DFP weakened the interaction between Ser-201 and His-254, causing the latter to rotate away from the serine (Fig. 3, A and B). Movement of the catalytic histidine has been observed before in other serine proteases and is not uncommon especially in those that use a Ser-His dyad for catalysis, probably because the rotational freedom of the histidine is no longer restricted by an aspartate, as in the Ser-His-Asp catalytic triad (40–42).
FIGURE 3.
Conformational changes within active site. A, apostructure of GlpG (Protein Data Bank code 2IC8) (7). B, DFP complex. C, isocoumarin complex (Protein Data Bank code 2XOW) (37). The inhibitors are shown in green. Hydrogen bonds are shown as dashed lines. For clarity, the L5 cap is not shown in A and C where it has ordered structures.
The rotation of His-254 is coupled to the movements of Tyr-205 and Trp-236. In the apostructure, His-254 is hydrogen-bonded to Ser-201 and interacts with Asn-251 via a water molecule (7). The aromatic rings of His-254, Tyr-205, and Trp-236 are in van der Waals contact (Fig. 3A). In the DFP structure, the imidazole ring of His-254 rotated into a groove between two TM helices (S5 and S6) and became hydrogen-bonded to the backbone carbonyls of Ile-237 and His-254 (Fig. 3B). The side chain of Tyr-205 rotated slightly toward Trp-236, and a direct hydrogen bond between its phenol oxygen and the backbone oxygen of Trp-236 replaced the water-mediated hydrogen bond in the apostructure. The rotation of Tyr-205 caused the side chain of Trp-236 to flip around the Cβ–Cγ bond, so the benzene end of the indole ring now points toward the outside of the membrane protein and interacts with Phe-153. The indole nitrogen of Trp-236 is hydrogen-bonded to Asn-154, which replaced the water-mediated interaction observed in the apostructure.
In the isocoumarin structure, His-254 is covalently bonded to the methyl acetate group of the inhibitor (37). Although the downward rotation of His-254 is prohibited by the covalent bond, Tyr-205 and Trp-236 show similar movements as their counterparts in the DFP complex (Fig. 3C): the phenol ring of Tyr-205 rotated toward the tryptophan and is hydrogen-bonded to its backbone oxygen, and the indole ring of the tryptophan flipped outwards to make contact with Phe-153. Given the difference between the chemical structures of DFP and isocoumarin, these similarities are striking and raise the possibility that similar side chain rearrangement may also occur in this region when natural peptide substrate binds. Although subtle, the movement of Tyr-205 and Trp-236 could be important to the enzymatic activity of GlpG because Tyr-205 interacts with the catalytic histidine, whereas Trp-236 lies at the mouth of the substrate-binding groove. The Y205A mutation greatly reduced the proteolytic activity of GlpG, which confirms the importance of the tyrosine residue (34). It is interesting that some mutations introduced into the interface between TM helices S2 and S5 (e.g. F153A/W236A) enhanced the proteolytic activity of GlpG (34, 36). The proximity of the mutations to a region that we now know may undergo side chain rearrangement suggests that their effects on activity do not have to be related to the hypothesized lateral movement of an entire helix (34). In the isocoumarin structure, Trp-236 is not directly hydrogen-bonded to Asn-154 (Fig. 3C). The flipping of the indole ring created a cavity between Trp-236 and Asn-154, which was proposed to constitute the binding subsite for the P2′ side chain in the peptide substrate (37). In the DFP structure, this cavity is not present, and insertion of a hydrophobic side chain would have to disrupt the hydrogen bond between Trp-236 and Asn-154.
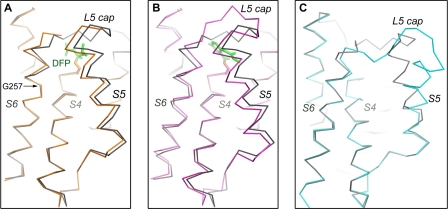
Global Conformational Changes
The experimental strategy (soaking) used to generate the protease-inhibitor complex precludes the possibility that large changes in the protein structure can be observed. Nevertheless, a comparison between the DFP structure and the apostructure reveals differences that are suggestive of the type of conformational changes that may occur globally in the membrane protein during catalysis.
The most visible difference is that the L5 loop in the DFP complex has become partially disordered (supplemental Fig. S3). The “opening” of the cap is necessary for the exposure of the catalytic serine to react with the inhibitor (27). The two TM helices that are connected to the cap also shifted position slightly (Fig. 4A). Helix S5 became more tilted toward S6, and the N-terminal end of S6 pivoted slightly outwards at a conserved glycine residue (Gly-257). Similar structural changes were observed in the complex with isocoumarin (Fig. 4B), although the relevance of the S6 movement in the enzyme mechanism was less certain initially due to the concern that the movement might result from the covalent linkage between the isocoumarin and His-254, which comes from S6 (37). Isocoumarin binding induced a more pronounced change in S6, which visibly shifted the whole helix, whereas the binding of the smaller DFP left the C-terminal half of the helix unaffected. It is noteworthy that, in both complexes, TM helix S5 shifted slightly toward S6 to adapt to the side chain movements within the active site. In an earlier apo crystal structure (Protein Data Bank code 2NRF), which was hypothesized to represent the open conformation of the membrane protein (19, 34), TM helix S5 tilted in a different direction away from S2 and S6 (Fig. 4C). The heavily tilted S5 in 2NRF is involved in crystal packing and does not appear to be modeled correctly.
FIGURE 4.
Global conformational change. A, the DFP structure (orange) is superimposed onto the apostructure (gray). Only the Cα traces are shown. The inhibitor (green) and Ser-201 (orange) are shown as stick models. The L5 cap is disordered in the DFP structure. B, superposition of the isocoumarin structure (magenta) and the apostructure (gray). C, the “open conformation” of GlpG (Protein Data Bank code 2NRF) (19), shown in blue, is compared with the closed apostructure (gray). TM helix S5, proposed to function as a lateral gate (19, 34), is tilted in a different direction from those observed in the two inhibitor complexes.
DISCUSSION
The composition of the proposed oxyanion-binding site has been suggested by previous crystallographic analyses (20, 27, 37) and is consistent with the observation that both His-150 and Asn-154 are highly conserved (5, 6). In the open-cap structure (27), a water molecule is bound at a similar but not identical site between Ser-201 and Asn-154. (The side chain of His-150 has rotated away from the site.) In another apostructure, a phospholipid is bound inside the active site (20). One of the phosphate oxygens is attracted to the oxyanion-binding site and forms hydrogen bonds with His-150 and Asn-154 (Fig. 2E). The backbone amide of Ser-201 is not hydrogen-bonded to the oxygen because the serine cannot react with the bound phospholipid, as it does with the fluorophosphonate, and is pushed back significantly by the phosphate group. In the isocoumarin complex (37), the carbonyl oxygen of the inhibitor is hydrogen-bonded to the backbone amide of Ser-201 and to the side chain of His-150 (Fig. 2F) but appears to be a little too far from the side chain of Asn-154 (3.4 Å) to form a strong hydrogen bond.
The inhibition of soluble serine protease by DCI and other isocoumarin compounds was extensively studied by Powers and Kam (43). Although initial evidence suggested that DCI could form a diacylated complex with the protease (26), this has yet to be confirmed by crystallographic analysis. In the only crystal structure of a protease-DCI complex that we are aware of (44), the inhibitor is bound through a single covalent bond with the catalytic serine and appears to have lost both chlorine atoms (similar to step 4 in supplemental Fig. S2). All DCI-inhibited proteases regain activity gradually through deacylation (26), and in the extreme case of subtilisin, the compound is turned over quickly and does not have inhibitory activity (43). In contrast to DCI, the substituted isocoumarins, e.g. 7-amino-3-(2-bromoethoxy)-4-chloroisocoumarin, can form a more stable complex with the enzyme due to their ability to alkylate the catalytic histidine, which involves a different mechanism (formation of a quinone-imine methide intermediate) and has been confirmed by crystal structures (39). The alkylated acyl-enzyme cannot be reactivated by deacylation.
For rhomboid protease, DCI has a special significance because it was the first mechanism-based inhibitor found to inhibit Drosophila rhomboid-1, providing the initial evidence that the protein is a serine protease (4). Later studies continued to show that, although most other serine protease inhibitors are ineffective (25), DCI has robust inhibitory activity against a wide range of rhomboid proteases (18, 25). Our study confirmed the DCI inhibitory activity against Escherichia coli rhomboid GlpG and suggests that the inhibitor works by competing for reaction with the catalytic serine residue. It is unclear whether a diacylated complex has formed, but the acyl-GlpG is clearly unstable and easily breaks down to release an α-hydroxy acid. Whether other rhomboid proteases can form stable complexes with DCI is unknown. In the complex with factor D (a soluble serine protease), the reactive carbonyl group of DCI has rotated out of the oxyanion-binding site, which renders it less susceptible to nucleophilic attack by the water molecule (44). Therefore, it is possible that even without the covalent bond with the catalytic histidine, the acyl-enzyme can achieve stability through other means. In parallel with our chemical study on DCI, the reported crystal structure of GlpG in complex with 7-amino-4-chloro-3-methoxyisocoumarin demonstrates that the inhibitory mechanism of the substituted isocoumarin for rhomboid protease is identical to that for soluble serine protease (37). Finally, we showed that E. coli GlpG is sensitive to inhibition by DFP, a classic inhibitor of serine protease. Under our experimental conditions, DFP was also more potent than DCI in inhibiting the proteolytic activity of GlpG. Taken together, these developments suggest that, in terms of inhibition by class-specific inhibitors, rhomboid protease is not as different from the soluble serine proteases as initially thought.
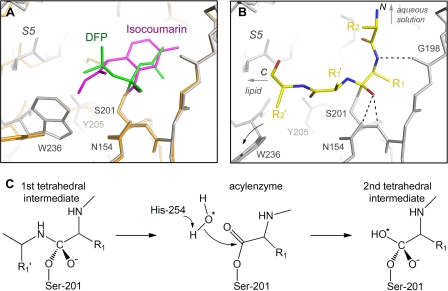
The DFP structure described in this work is only the second inhibitor complex structure for rhomboid protease. The first crystal structure, between GlpG and 7-amino-4-chloro-3-methoxyisocoumarin, contains a covalent bond between the inhibitor and the catalytic histidine. An initial concern was that this unnatural linkage might affect the conformation of the inhibitor or the protease (37), as has happened in an elastase-isocoumarin complex (39). Comparison with the DFP structure alleviated much of this concern. (i) After superposition, the benzoyl carbonyl oxygen of the isocoumarin is found to occupy a very similar position as the phosphonyl oxygen of DFP, despite the fact that the carbonyl has a planar geometry, whereas the phosphate has a tetrahedral geometry (Fig. 5A); (ii) within the active site, the side chains of Tyr-205 and Trp-236 show similar rotations; and (iii) the opening of the L5 cap and the slight tilting of S5 and S6 are observed in both crystal structures. Superposition of the two structures also reveals differences, most of which are concentrated on the lipid-facing side of the catalytic serine (left in Fig. 5A). Besides the freely rotating His-254 in the DFP structure, the bulkier methyl acetate group of the isocoumarin pushed the indole ring of Trp-236 farther out toward the lipid. The phenol groups of Tyr-205 also have slightly different angles. In the isocoumarin structure, TM helix S5 has moved farther away from Ser-201, probably generating room to accommodate the covalent bond between His-254 and the inhibitor. On the basis of the DFP structure, we constructed a model of peptide substrate bound at the GlpG active site after the nucleophilic attack of Ser-201 from the si-face of the scissile peptide bond (Fig. 5B) (27, 36). The model is similar to the one proposed previously (37) and contains a turn at the scissile bond, which projects the N terminus of the substrate up and toward the aqueous solution. The peptide segment on the C-terminal side of the scissile bond is roughly parallel to the membrane plane. The conformational flexibility suggested by the two inhibitor complexes is probably important for the membrane protease to accommodate different substrate sequences while maintaining a seal around the peptide (C-terminal to the scissile bond) before it encounters lipid.
FIGURE 5.
Comparison between DFP and isocoumarin structures. A, superposition of the two complex structures. DFP is shown in gray, and isocoumarin is shown in orange. The bound inhibitors are shown in green and magenta, respectively. B, hypothetical model of peptide substrate bound at the GlpG active site. The peptide substrate is shown in yellow. The transition state oxyanion is stabilized by hydrogen bonds with Ser-201, His-150 (not shown), and Asn-154. The side chain of the P1 residue (R1) is modeled based on one of the isopropyl groups of DFP, which points to a cavity suggested previously by mutagenesis to be the S1 subsite (37). Fixing the side chain in the S1 subsite projects the amide nitrogen and the N-terminal portion of the polypeptide up toward the aqueous solution. The position of the amide nitrogen may enable it to form a hydrogen bond with the backbone carbonyl of Gly-198 (37). Toward the C-terminal side of the scissile bond, the side chain of the P1′ residue is modeled to point up, away from the binding groove. This is based on the consideration that P1′ can accommodate polar residues such as aspartate (31). The side chain of Trp-236 is rotated to generate room for the P2′ residue. C, simplified mechanism of GlpG-catalyzed peptide hydrolysis to illustrate the relationship between the tetrahedral transition state and the acyl-enzyme.
The DFP structure provides for the first time a model for the tetrahedral transition state, which corresponds to an earlier time point on the reaction coordinate than the acyl-enzyme represented by the isocoumarin structure (Fig. 5C). Some of the movements observed between the two crystal structures could be related to this difference. One particularly interesting feature of the DFP structure is the rotation of the catalytic histidine. In the classic mechanism, the catalytic histidine not only enhances the nucleophilicity of the catalytic serine but also functions as a base to facilitate the attack of the acyl-enzyme by a water molecule (45). In the DFP structure, His-254 is wedged between two TM helices and is unable to perform the second task (Fig. 3B). Therefore, for rhomboid protease, either the hydrolysis of the acyl-enzyme requires no assistance from the histidine, or a second round of conformational change has to take place to restore His-254 to its original position next to Ser-201. The progression from the tetrahedral intermediate to the acyl-enzyme is accompanied by major changes in both substrate and enzyme: first, the tetrahedral carbon has to collapse back to the planar structure of a carbonyl; and second, the scissile peptide bond formally breaks to release the C-terminal peptide fragment, which contains the majority of the TM sequence. The dissociation of the C-terminal fragment from the complex will leave the active site exposed on the side to lipid, unless the membrane protease changes conformation again to reseal the gap. It is possible that these changes might trigger the rotation of His-254 back toward Ser-201 to participate in the hydrolysis of the acyl-enzyme.
Supplementary Material
Acknowledgments
We thank Professor Jonathan Ellman for sharing the LC-MS equipment and Dr. Somenath Chowdhury for helping us use the equipment.
This work was supported, in whole or in part, by National Institutes of Health Grant GM082839 (to Y. H.).

This article contains supplemental Figs. S1–S3.
The atomic coordinates and structure factors (code 3TXT) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- TM
- transmembrane
- DCI
- 3,4-dichloroisocoumarin
- DFP
- diisopropyl fluorophosphonate
- BisTris propane
- 1,3-bis(tris(hydroxymethyl)methylamino)propane.
REFERENCES
- 1. Mayer U., Nüsslein-Volhard C. (1988) A group of genes required for pattern formation in the ventral ectoderm of the Drosophila embryo. Genes Dev. 2, 1496–1511 [DOI] [PubMed] [Google Scholar]
- 2. Wasserman J. D., Urban S., Freeman M. (2000) A family of rhomboid-like genes: Drosophila rhomboid-1 and roughoid/rhomboid-3 cooperate to activate EGF receptor signaling. Genes Dev. 14, 1651–1663 [PMC free article] [PubMed] [Google Scholar]
- 3. Lee J. R., Urban S., Garvey C. F., Freeman M. (2001) Regulated intracellular ligand transport and proteolysis control EGF signal activation in Drosophila. Cell 107, 161–171 [DOI] [PubMed] [Google Scholar]
- 4. Urban S., Lee J. R., Freeman M. (2001) Drosophila Rhomboid-1 defines a family of putative intramembrane serine proteases. Cell 107, 173–182 [DOI] [PubMed] [Google Scholar]
- 5. Koonin E. V., Makarova K. S., Rogozin I. B., Davidovic L., Letellier M. C., Pellegrini L. (2003) The rhomboids: a nearly ubiquitous family of intramembrane serine proteases that probably evolved by multiple ancient horizontal gene transfers. Genome Biol. 4, R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lemberg M. K., Freeman M. (2007) Functional and evolutionary implications of enhanced genomic analysis of rhomboid intramembrane proteases. Genome Res. 17, 1634–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang Y., Zhang Y., Ha Y. (2006) Crystal structure of a rhomboid family intramembrane protease. Nature 444, 179–180 [DOI] [PubMed] [Google Scholar]
- 8. Freeman M. (2008) Rhomboid proteases and their biological functions. Annu. Rev. Genet. 42, 191–210 [DOI] [PubMed] [Google Scholar]
- 9. Hill R. B., Pellegrini L. (2010) The PARL family of mitochondrial rhomboid proteases. Semin. Cell Dev. Biol. 21, 582–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Urban S. (2006) Rhomboid proteins: conserved membrane proteases with divergent biological functions. Genes Dev. 20, 3054–3068 [DOI] [PubMed] [Google Scholar]
- 11. Stevenson L. G., Strisovsky K., Clemmer K. M., Bhatt S., Freeman M., Rather P. N. (2007) Rhomboid protease AarA mediates quorum-sensing in Providencia stuartii by activating TatA of the twin-arginine translocase. Proc. Natl. Acad. Sci. U.S.A. 104, 1003–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McQuibban G. A., Saurya S., Freeman M. (2003) Mitochondrial membrane remodeling regulated by a conserved rhomboid protease. Nature 423, 537–541 [DOI] [PubMed] [Google Scholar]
- 13. Cipolat S., Rudka T., Hartmann D., Costa V., Serneels L., Craessaerts K., Metzger K., Frezza C., Annaert W., D'Adamio L., Derks C., Dejaegere T., Pellegrini L., D'Hooge R., Scorrano L., De Strooper B. (2006) Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell 126, 163–175 [DOI] [PubMed] [Google Scholar]
- 14. Chao J. R., Parganas E., Boyd K., Hong C. Y., Opferman J. T., Ihle J. N. (2008) Hax1-mediated processing of HtrA2 by Parl allows survival of lymphocytes and neurons. Nature 452, 98–102 [DOI] [PubMed] [Google Scholar]
- 15. Opitz C., Di Cristina M., Reiss M., Ruppert T., Crisanti A., Soldati D. (2002) Intramembrane cleavage of microneme proteins at the surface of the apicomplexan parasite Toxoplasma gondii. EMBO J. 21, 1577–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. O'Donnell R. A., Hackett F., Howell S. A., Treeck M., Struck N., Krnajski Z., Withers-Martinez C., Gilberger T. W., Blackman M. J. (2006) Intramembrane proteolysis mediates shedding of a key adhesin during erythrocyte invasion by the malaria parasite. J. Cell Biol. 174, 1023–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Santos J. M., Ferguson D. J., Blackman M. J., Soldati-Favre D. (2011) Intramembrane cleavage of AMA1 triggers Toxoplasma to switch from an invasive to a replicative mode. Science 331, 473–477 [DOI] [PubMed] [Google Scholar]
- 18. Lemberg M. K., Menendez J., Misik A., Garcia M., Koth C. M., Freeman M. (2005) Mechanism of intramembrane proteolysis investigated with purified rhomboid proteases. EMBO J. 24, 464–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu Z., Yan N., Feng L., Oberstein A., Yan H., Baker R. P., Gu L., Jeffrey P. D., Urban S., Shi Y. (2006) Structural analysis of a rhomboid family intramembrane protease reveals a gating mechanism for substrate entry. Nat. Struct. Mol. Biol. 13, 1084–1091 [DOI] [PubMed] [Google Scholar]
- 20. Ben-Shem A., Fass D., Bibi E. (2007) Structural basis for intramembrane proteolysis by rhomboid serine proteases. Proc. Natl. Acad. Sci. U.S.A. 104, 462–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lemieux M. J., Fischer S. J., Cherney M. M., Bateman K. S., James M. N. (2007) The crystal structure of the rhomboid peptidase from Haemophilus influenzae provides insight into intramembrane proteolysis. Proc. Natl. Acad. Sci. U.S.A. 104, 750–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vinothkumar K. R. (2011) Structure of rhomboid protease in a lipid environment. J. Mol. Biol. 407, 232–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ha Y. (2009) Structure and mechanism of intramembrane protease. Semin. Cell Dev. Biol. 20, 240–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Erez E., Fass D., Bibi E. (2009) How intramembrane proteases bury hydrolytic reactions in the membrane. Nature 459, 371–378 [DOI] [PubMed] [Google Scholar]
- 25. Urban S., Wolfe M. S. (2005) Reconstitution of intramembrane proteolysis in vitro reveals that pure rhomboid is sufficient for catalysis and specificity. Proc. Natl. Acad. Sci. U.S.A. 102, 1883–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harper J. W., Hemmi K., Powers J. C. (1985) Reaction of serine proteases with substituted isocoumarins: discovery of 3,4-dichloroisocoumarin, a new general mechanism-based serine protease inhibitor. Biochemistry 24, 1831–1841 [DOI] [PubMed] [Google Scholar]
- 27. Wang Y., Ha Y. (2007) Open-cap conformation of intramembrane protease GlpG. Proc. Natl. Acad. Sci. U.S.A. 104, 2098–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maegawa S., Koide K., Ito K., Akiyama Y. (2007) The intramembrane active site of GlpG, an E. coli rhomboid protease, is accessible to water and hydrolyzes an extramembrane peptide bond of substrates. Mol. Microbiol. 64, 435–447 [DOI] [PubMed] [Google Scholar]
- 29. Wang Y., Maegawa S., Akiyama Y., Ha Y. (2007) The role of L1 loop in the mechanism of rhomboid intramembrane protease GlpG. J. Mol. Biol. 374, 1104–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bondar A. N., del Val C., White S. H. (2009) Rhomboid protease dynamics and lipid interactions. Structure 17, 395–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maegawa S., Ito K., Akiyama Y. (2005) Proteolytic action of GlpG, a rhomboid protease in the Escherichia coli cytoplasmic membrane. Biochemistry 44, 13543–13552 [DOI] [PubMed] [Google Scholar]
- 32. Erez E., Bibi E. (2009) Cleavage of a multispanning membrane protein by an intramembrane serine protease. Biochemistry 48, 12314–12322 [DOI] [PubMed] [Google Scholar]
- 33. Adrain C., Strisovsky K., Zettl M., Hu L., Lemberg M. K., Freeman M. (2011) Mammalian EGF receptor activation by the rhomboid protease RHBDL2. EMBO Rep. 12, 421–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Baker R. P., Young K., Feng L., Shi Y., Urban S. (2007) Enzymatic analysis of a rhomboid intramembrane protease implicates transmembrane helix 5 as the lateral substrate gate. Proc. Natl. Acad. Sci. U.S.A. 104, 8257–8262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Urban S., Baker R. P. (2008) In vivo analysis reveals substrate-gating mutants of a rhomboid intramembrane protease display increased activity in living cells. Biol. Chem. 389, 1107–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brooks C. L., Lazareno-Saez C., Lamoureux J. S., Mak M. W., Lemieux M. J. (2011) Insights into substrate gating in H. influenzae rhomboid. J. Mol. Biol. 407, 687–697 [DOI] [PubMed] [Google Scholar]
- 37. Vinothkumar K. R., Strisovsky K., Andreeva A., Christova Y., Verhelst S., Freeman M. (2010) The structural basis for catalysis and substrate specificity of a rhomboid protease. EMBO J. 29, 3797–3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Benoit F., Holmes J. L., Isaacs N. S. (1969) The mass spectra of carboxylic acids. I. Fragmentation mechanisms in maleic and fumaric acids and related compounds. Org. Mass Spectrom. 2, 591–601 [Google Scholar]
- 39. Vijayalakshmi J., Meyer E. F., Jr., Kam C. M., Powers J. C. (1991) Structural study of porcine pancreatic elastase complexed with 7-amino-3-(2-bromoethoxy)-4-chloroisocoumarin as a nonreactivatable doubly covalent enzyme-inhibitor complex. Biochemistry 30, 2175–2183 [DOI] [PubMed] [Google Scholar]
- 40. Radisky E. S., Lee J. M., Lu C. J., Koshland D. E., Jr. (2006) Insights into the serine protease mechanism from atomic resolution structures of trypsin reaction intermediates. Proc. Natl. Acad. Sci. U.S.A. 103, 6835–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bone R., Sampson N. S., Bartlett P. A., Agard D. A. (1991) Crystal structures of α-lytic protease complexes with irreversibly bound phosphonate esters. Biochemistry 30, 2263–2272 [DOI] [PubMed] [Google Scholar]
- 42. Larsen N. A., Lin H., Wei R., Fischbach M. A., Walsh C. T. (2006) Structural characterization of enterobactin hydrolase IroE. Biochemistry 45, 10184–10190 [DOI] [PubMed] [Google Scholar]
- 43. Powers J. C., Kam C. M. (1994) Isocoumarin inhibitors of serine peptidases. Methods Enzymol. 244, 442–457 [DOI] [PubMed] [Google Scholar]
- 44. Cole L. B., Kilpatrick J. M., Chu N., Babu Y. S. (1998) Structure of 3,4-dichloroisocoumarin-inhibited factor D. Acta Crystallogr. Sect. D 547, 11–17 [DOI] [PubMed] [Google Scholar]
- 45. Fersht A. (1999) Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding, 1st Ed., W. H. Freeman Publishers [Google Scholar]
- 46. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 47. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Winn M. D., Murshudov G. N., Papiz M. Z. (2003) Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 374, 300–321 [DOI] [PubMed] [Google Scholar]
- 49. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.