Background: Ciliary dyneins monitor and respond to the mechanical state or curvature of the microtubular axoneme.
Results: NMR chemical shift mapping and binding assays define functional subdomains within a key component (LC1) of this system.
Conclusion: LC1 provides a direct tether between a dynein motor unit and the microtubule that modulates motor function.
Significance: This study provides insight into the mechanism by which dyneins are coordinated during ciliary beating.
Keywords: Cilia, Dynein, Microtubules, Nuclear Magnetic Resonance, Protein Structure
Abstract
Dynein light chain 1 (LC1/DNAL1) is one of the most highly conserved components of ciliary axonemal outer arm dyneins, and it associates with both a heavy chain motor unit and tubulin located within the A-tubule of the axonemal outer doublet microtubules. In a variety of model systems, lack of LC1 or expression of mutant forms leads to profound defects in ciliary motility, including the failure of the hydrodynamic coupling needed for ciliary metachronal synchrony, random stalling during the power/recovery stroke transition, an aberrant response to imposed viscous load, and in some cases partial failure of motor assembly. These phenotypes have led to the proposal that LC1 acts as part of a mechanical switch to control motor function in response to alterations in axonemal curvature. Here we have used NMR chemical shift mapping to define the regions perturbed by a series of mutations in the C-terminal domain that yield a range of phenotypic effects on motility. In addition, we have identified the subdomain of LC1 involved in binding microtubules and characterized the consequences of an Asn → Ser alteration within the terminal leucine-rich repeat that in humans causes primary ciliary dyskinesia. Together, these data define a series of functional subdomains within LC1 and allow us to propose a structural model for the organization of the dynein heavy chain-LC1-microtubule ternary complex that is required for the coordinated activity of dynein motors in cilia.
Introduction
The beating of cilia and flagella is driven by the inner and outer rows of dynein arms associated with the axonemal doublet microtubules. This motile behavior requires that a wave of dynein motor activity be propagated along the structure, usually from base to tip, and theoretical models suggest that one or more mechanosensitive switch systems might act to monitor axonemal curvature (1–3). In Chlamydomonas, evidence for the existence of such a switch mechanism came from experiments where paralyzed flagella of mutant strains lacking various axonemal substructures were induced to beat in a rhythmic oscillatory manner for one or more cycles following mechanical activation (4). Based on this analysis, it was further proposed that the flagellum contains two mechanosensory systems that monitor and respond to the mechanical state (i.e. curvature) of the axoneme. The first of these involves the central pair microtubule complex and radial spokes and controls the activity of the inner dynein arm system, whereas the second appears to be distinct at the molecular level and impinges on the outer arms.
Chlamydomonas outer arm dynein with its associated docking structures is a highly complex motor consisting of at least 20 distinct molecular components with a total mass in excess of 2 MDa (for a review, see Ref. 5). This dynein is built around three heavy chains (HCs2; α, β, and γ), which exhibit different ATPase and motor properties (6–8). The γ HC (encoded at DHC15 (9)) is also distinct in that it is the only axonemal dynein HC known to have additional components bound to the motor unit: specifically, cross-linking and vanadate-mediated photocleavage experiments suggested that one copy of light chain 1 (LC1) is bound to the ATP hydrolytic subdomain AAA1, and a second copy is bound to either AAA3 or AAA4 (10).3 Further evidence that LC1 is associated with the motor unit came from analysis of the oda2-t mutant that expresses a truncated 150-kDa form of the γ HC retaining only the N-terminal region; LC1 is not present in oda2-t outer arm dynein, whereas all other components are (11). LC1 also binds to tubulin located within the A-tubule of the outer doublets, and thus, the γ HC is tethered to the A-tubule via both its N-terminal domain that associates with the intermediate chain-LC complex and its motor unit (12). Furthermore, a third distinct tether comes into play under high Ca2+ conditions when the LC4 calmodulin homologue (encoded at DLE1 (9)) releases its Ca2+-dependent binding site on the γ HC N-terminal domain (it remains bound to this γ HC region through a second Ca2+-insensitive site) and instead binds to intermediate chain 1 within the intermediate chain-LC complex (13, 14).
LC1 (encoded at DLU1 (9)) is a 22-kDa leucine-rich repeat (LRR) protein that is one of the most conserved components within outer arm dynein. In humans, a homozygous mutation within the last LRR of the LC1 orthologue DNAL1 has recently been demonstrated to cause primary ciliary dyskinesia (15). This LC consists of an N-terminal helix followed by six ββα motifs derived from the LRRs and a C-terminal helical domain that protrudes from the main protein axis defined by the LRR barrel (16). The orientation of the C-terminal helical region is controlled by two residues (Met182 and Asp185) that exhibit high backbone dynamics; the remainder of the protein is relatively rigid with the exception of both termini and a flexible leucine spine within one interaction surface (17, 18). The C-terminal region is also highly charged: in mammals, the α9 helix is very acidic, whereas in Chlamydomonas and Trypanosoma, there are two conserved basic residues (Arg189 and Arg196 in Chlamydomonas) in addition to several Asp/Glu residues.
Previously we found that expression of mutant forms of LC1 in which these Arg residues or the Met/Asp exhibiting high dynamics were altered yielded dominant negative phenotypes in a wild-type background (12). These Chlamydomonas cells swam slowly as they were unable to coordinate their flagella movement due to random stalling during the beat cycle and especially at the power/recovery stroke transition. In addition, these strains had an aberrant response to viscous load. Under low viscosity conditions, they generated the low propulsive force characteristic of outer armless mutants (19). However, as load increased (and beat frequency decreased), the force output approached that of wild-type cells, a response that essentially phenocopied the oda2-t mutant. Introduction of different mutant forms of LC1 produced graded effects: for example, mutation of Met182 to Ala was much less disruptive than alterations to either Gly or Pro, which were predicted to change the dynamic behavior. Mutation of the equivalent residues in Trypanosoma brucei LC1 also caused major motility defects (20).
Some of the key issues for understanding this dynein control system are to identify the functional subdomains within LC1 and to determine their orientation with respect to the motor unit and microtubule. Here we have used chemical shift mapping to define the perturbed regions within five mutant forms of LC1 that exhibited a range of phenotypic effects in vivo. To gain further insight into the mechanisms involved, we have also used targeted mutagenesis to delineate the segment of this molecule involved in associating the dynein motor unit with the outer doublet A-tubule and have characterized the structural consequences of the PCD-causing mutation within the terminal LRR. Together, these data allow functional subdomains within LC1 and the overall geometry to be defined. The results implicate the LC1 C-terminal domain in controlling motor function through direct interactions within the HC AAA domains.
EXPERIMENTAL PROCEDURES
Molecular Biology and Mutagenesis
A cDNA containing the entire LC1 coding region in pBluescript II SK− was obtained previously (10). The M182A, D185G, D185P, R189E, and R196A mutations used for in vivo experiments (12), and the additional K3E, K8E, R12E, R17E, K18E, K39E, K62E, R79E, K83E, K84E, K126E, K135E, and N150S alterations were generated by oligonucleotide-directed mutagenesis using standard methods. The mutated coding regions were then obtained by PCR, subcloned into the pET16B vector (Novagen), and transformed into Escherichia coli BL21(DE3). When expressed, these proteins contain an N-terminal His10 tag and linker that may be cleaved from the target molecule by Factor Xa digestion leaving a single additional His residue at the N terminus (designated residue −1).
Preparation of LC1 Proteins
For 15N labeling, cells were grown at 37 °C in 1 liter of M9 minimal medium containing [15N]NH4Cl to an optical density at 600 nm of ∼0.6. The culture was then supplemented with isopropyl β-d-1-thiogalactopyranoside to a final concentration of 1 mm and grown for an additional 3 h to allow for protein expression. Double labeled samples of D185G and R189E LC1 were prepared in a similar manner from cells grown in the presence of [U-13C]glucose and [15N]NH4Cl. For unlabeled samples, cells were grown in LB complete medium, and protein expression was induced for 2 h.
Cells were harvested by centrifugation; resuspended in 50 ml of 20 mm Tris·Cl, pH 8.0, 50 mm NaCl; and frozen at −20 °C. Samples were subsequently defrosted, and cells were lysed by sonication. Following centrifugation to remove insoluble material, total soluble protein was applied to a Ni2+ resin column and washed with 20 mm Tris·Cl, pH 8.0, 500 mm NaCl, 20 mm imidazole. His-tagged protein was subsequently eluted with 20 mm Tris·Cl, pH 8.0, 500 mm NaCl, 250 mm imidazole. Peak fractions were pooled and dialyzed extensively against 20 mm Tris·Cl, pH 8.0, 50 mm NaCl, 10 mm EDTA to remove any paramagnetic Ni2+. Following dialysis, the His10 tag was cleaved from LC1 proteins by addition of 100 units of Factor Xa and incubation at room temperature overnight; samples were subsequently concentrated using Amicon Ultra-4 ultrafiltration units (10,000 molecular weight cutoff). Proteins were subjected to a final purification step and buffer exchange by gel filtration in a Superdex 200 10/300 GL column attached to an ÄKTApurifier chromatography work station and equilibrated with 2.5 mm Tris·Cl, pH 6.8, 50 mm NaCl; this buffer was used previously for chemical shift assignments and collection of other NMR data necessary for structure determination (21). All NMR experiments and binding assays utilized only monomeric LC1.
NMR Spectroscopy
Purified 15N-labeled LC1 samples were concentrated to a final volume of 600 μl and a concentration of ∼1 mm (∼22 mg/ml) or greater using Amicon Ultra-4 ultrafiltration units. Thirty-five microliters of D2O was then added, and two-dimensional 1H-15N HSQC spectra were collected using a Varian 800-MHz NMR spectrometer equipped with a cryogenic probe. The NMR spectra and chemical shift alterations were analyzed with NMRViewJ. To obtain the complete backbone assignments for the D185G form of LC1, the 1H-15N HSQC, HNCA, HNCO, and HBHA(CO)NH spectra were obtained from a 13C,15N-labeled sample using a Varian 500-MHz NMR spectrometer equipped with a cryogenic probe; the HNCACB experiment did not yield a usable spectrum due to the large size of LC1. Peak assignments were made with XEASY, and the backbone torsion angles were predicted from the chemical shifts using TALOS+ (22). The 1H-15N HSQC spectrum from 13C,15N-labeled R189E LC1 was sufficiently poorly resolved that it was not feasible to reassign the entire backbone for this mutant form.
Circular Dichroism Spectroscopy
The concentrations of purified fully denatured wild-type, D185G, and N150S LC1 proteins in 2.5 mm Tris·Cl, pH 6.8, 50 mm NaCl were determined by absorbance at 280 nm using an extinction coefficient (ϵ) of 17,810 liters·mol−1·cm−1. To determine the melting temperature of these LC1 proteins, the ellipticity signals at 222 nm (θ222) of 3 μm samples were measured in the temperature range 4–86 °C (65 °C for N150S) using a Jasco J-715 spectropolarimeter as described (23).
Axoneme Binding Assay
The Chlamydomonas reinhardtii wild-type cc125 and mutant oda6 that lacks outer dynein arms were grown in R medium as described previously (24). Cells were deflagellated with dibucaine, and flagellar axonemes were purified by standard methods. Wild-type axonemes were depleted of dynein arms by extraction with 0.6 m NaCl. Axonemes were resuspended in 20 mm Tris·Cl, pH 8.0, 5 mm MgSO4, 0.5 mm EDTA, 50 mm NaCl prior to addition of LC1 proteins. Following incubation at room temperature for 60 min, axonemes were pelleted in a microcentrifuge, washed with buffer, and prepared for electrophoresis. Samples were separated in 12.5% polyacrylamide gels containing SDS and stained with Coomassie Blue. Stained gels were imaged using an ImageQuant LAS4000 digital imaging system (GE Healthcare), and band intensities were quantified using ImageQuant TL software.
Other Computational Methods
The alignment of LC1 orthologues was performed using ClustalW, and the output was processed with BOXSHADE. The level of sequence conservation was titrated onto the LC1 structure with VENN (25). Spectral overlays were prepared using Adobe Photoshop CS4. The structural model of D185P mutant LC1 was generated with SWISS-MODEL (26).
RESULTS
Structural Orthology of LC1 Proteins
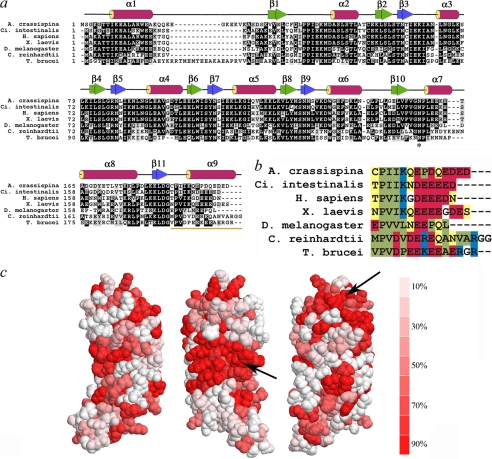
LC1 is one of the most highly conserved components of axonemal outer arm dyneins (Fig. 1, a and b); for example, the Chlamydomonas and human orthologues share 55% identity (70% similarity). Mapping the degree of sequence identity onto the Chlamydomonas LC1 structure revealed two major areas of surface conservation (Fig. 1c, arrows). The first is located within the midregion of the molecule centered on Trp99 (this exposed hydrophobic region may represent part of the γ HC binding surface), whereas the second is on the opposite face near the N terminus and includes a series of charged residues.
FIGURE 1.
LC1 structural organization and conservation of C-terminal domain. a, ClustalW sequence alignment of LC1 orthologues from Anthocidaris crassispina (GenBank no. BAA24184), Ciona intestinalis (GenBank no. Q8T888), Homo sapiens (GenBank no. Q4LDG9), Xenopus laevis (GenBank no. Q28G94), Drosophila melanogaster (GenBank no. NP610483), C. reinhardtii (GenBank no. AAD41040), and T. brucei (GenBank no. XP828643). The secondary structure derived from the Chlamydomonas protein structure (Protein Data Bank code 1M9L) is indicated above the alignment. The asterisk indicates the conserved Asn residue that is mutated to Ser in the PCD patients. b, color-coded alignment of the C-terminal helix: polar, yellow; hydrophobic, green; acidic, red; basic, blue; glycine, white. c, the cross-species conservation is displayed on the LC1 surface: the graded colors represent the degree of conservation (identity) from red (≥90%) to white (≤10%). The arrows indicate major highly conserved regions of the molecular surface.
Perturbations Caused by Dominant Negative Mutations in LC1 C-terminal Domain
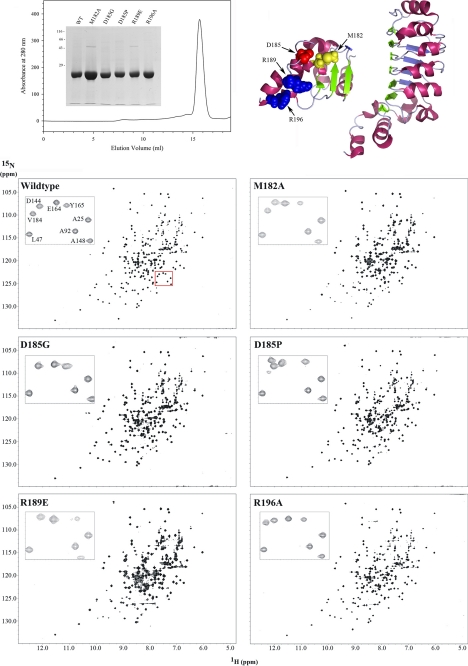
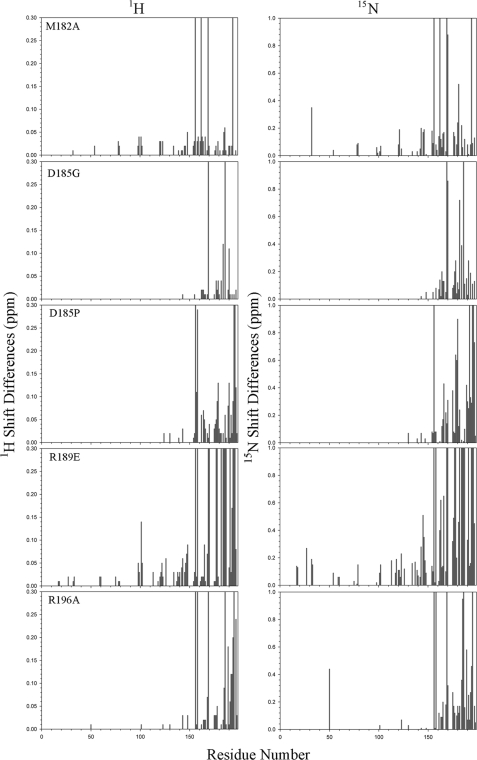
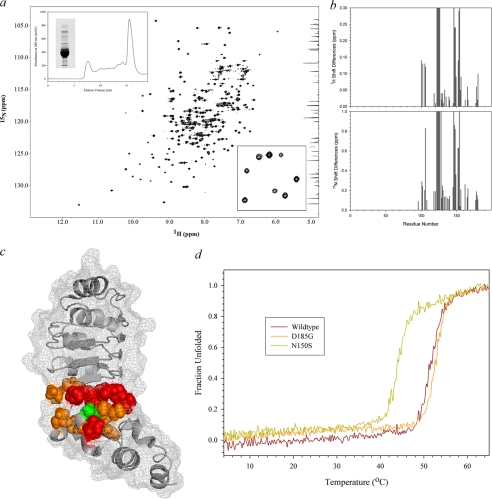
Previous single residue mutagenesis experiments (Ref. 12 and see Table 1) suggested that the C-terminal domain might represent a key functional subdomain of LC1, although intriguingly there are significant sequence differences observed within this region when assessed across a broad phylogenetic spectrum (Fig. 1, a–c). As long range perturbations could disrupt other segments of the molecule, we set out to determine the extent to which the introduced mutations affected the structure of LC1 and thus define the region responsible for the observed phenotypic effects. The chemical shifts (δ) for individual atoms are exquisitely sensitive to their local environment, and so NMR-based chemical shift mapping was used to identify those regions of the mutant proteins that undergo alterations in their chemical environment compared with wild type (Fig. 2). To achieve this, we prepared 15N-labeled wild-type and M182A, D185G, D185P, R189E, and R196A mutant forms of LC1 and collected two-dimensional 1H-15N HSQC spectra using an 800-MHz NMR spectrometer equipped with a cryogenic probe (Fig. 2). These particular mutations were chosen as they represent all residues altered previously and generated a graded series of phenotypic effects in vivo (12). All six molecules yielded well resolved peaks (Fig. 2, lower panel, insets), and the spectra exhibit large chemical shift dispersion in both 1H and 15N dimensions indicative of well folded proteins (Fig. 2, lower panels).
TABLE 1.
Functional consequences of C-terminal LC1 mutations
| Mutation | WT residue conserved in |
Swimming velocitya | LC1 in monomeric formb | Microtubule binding activityc | |
|---|---|---|---|---|---|
| H. sapiens | T. brucei | ||||
| μm/s | % | % WT | |||
| None | 115.1 ± 34.1 | 97.4 | 100 | ||
| M182A | No | No | 81.8 ± 19.3 | 95.9 | 142 |
| M182G | No | No | 69.2 ± 18.1 | NDd | ND |
| M182P | No | No | 55.7 ± 15.7 | ND | ND |
| D185G | No | Yes | 31.5 ± 12.2 | 95.7 | 122 |
| D185P | No | Yes | 38.2 ± 9.8 | 91.3 | 88 |
| R189A | No | Basic | 64.5 ± 20.9 | ND | ND |
| R189E | No | Basic | 32.0 ± 15.1 | 93.9e | 127 |
| R196A | No | Yes | 45.6 ± 13.4 | 99.3 | 122 |
| R196D | No | Yes | 46.7 ± 10.2 | ND | ND |
a These data are for cwd arg7-8 cells expressing mutant forms of LC1. From Ref. 12.
b Determined by gel filtration chromatography from the relative amounts of LC1 eluting from a Superdex 200 10/300 GL column in monomeric form versus larger oligomers. Only monomeric LC1 was used for binding assays.
c The values quoted were determined by quantitative densitometric analysis of the Coomassie Blue-stained gels shown in Fig. 9 using ImageQuant TL. Background subtraction utilized the rolling ball method.
d ND, not determined.
e Monomeric R189E LC1 was very stable, and only 0.47% was found in oligomeric form following >6 months at 4 °C.
FIGURE 2.
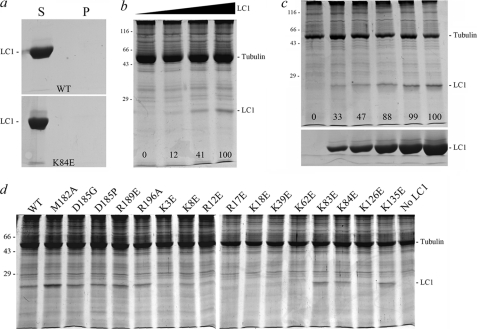
1H-15N HSQC spectra for wild-type and C-terminal mutant forms of LC1. The ribbon structure of LC1 and location of the four mutated residues within the C-terminal domain are shown at the top right. The final gel filtration profile of a representative 15N-labeled LC1 protein sample (R196A) is at the top left; inset, electrophoretic analysis of 15N-labeled LC1 proteins (12.5% SDS-polyacrylamide gel; Coomassie Blue stain). The two-dimensional 1H-15N HSQC spectra for wild-type and the M182A, D185G, D185P, R189E, and R196A mutant forms of LC1 are shown in the lower panels. Each peak represents an amide resonance derived from the peptide backbone, Asn/Gln side chains, or the Trp indole ring. All spectra reveal a similarly high degree of peak dispersion in both 1H and 15N dimensions, suggesting that the mutant proteins are well folded as is wild-type LC1. The insets show an enlargement of a small region (red box in wild-type spectrum) to illustrate the high resolution achieved for all LC1 proteins using the 800-MHz spectrometer.
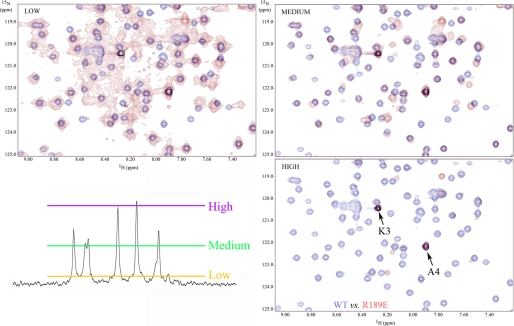
Superficially the spectra for mutant forms of LC1 appear similar to wild type. However, on careful examination, a significant number of chemical shift alterations (Δδ) were identified. In addition, the observed signals for many peaks from the R189E mutant (and to a lesser extent M182A) showed evidence of considerable line broadening and a concomitant decrease in peak height that is usually indicative of transitions between structural states occurring on the NMR time scale (i.e. in the μs to ms range); this line broadening was not due to aggregation as the sample remained completely monomeric even after many months. The broadening is illustrated in Fig. 3, which compares the R189E spectrum at different contour levels with wild type. The line broadening is especially evident in the low contour map where many R189E peaks were merged together. At a high contour, many R189E LC1 peaks were missing with the exception of those near the termini, which are highly dynamic (17).
FIGURE 3.
R189E mutation causes extensive peak broadening. The central region of the 1H-15N HSQC spectrum for wild-type LC1 (blue) was superimposed on the equivalent region for R189E LC1 (red) drawn at three different contour levels. The approximate locations of these contour levels are illustrated on a representative vertical slice through the R189E spectrum at the bottom left. Many peaks are not well resolved for the mutant protein at a low contour and are missing from the spectrum at higher contours. In contrast, wild-type peaks are well resolved. The two very strong peaks present in the high contour map of R189E LC1 represent residues Lys3 and Ala4 that are very close to the N terminus and exhibit high mobility (low order parameter) (17). These observations suggest that R189E LC1 transitions between structural states on the μs to ms time scale.
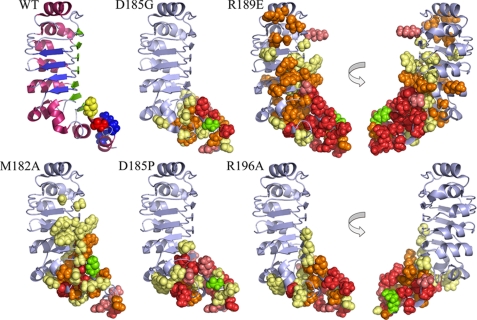
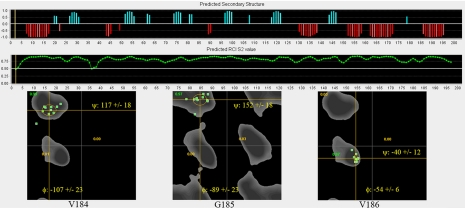
To assess the degree and extent of structural perturbations caused by these various mutations, the 1H and 15N Δδ values were clustered into four color-coded groups (≤0.1, ≤0.2, ≤0.3, and >0.3 ppm or peak missing), and the corresponding side chains are displayed on the LC1 structure (Fig. 4); the shift differences for every residue in both dimensions are shown in Fig. 5. For several mutations, e.g. D185G, significant changes occurred only in residues relatively close in three-dimensional space to the mutation site, and indeed all changes for this mutant were confined to the C-terminal domain. Even so, the number of residues exhibiting significant Δδ values suggests that the observed phenotypes cannot necessarily be ascribed to the mutation of a particular residue but rather may reflect a more global perturbation or reorientation of this domain. To address this directly, we reassigned the backbone for 13C,15N-labeled D185G LC1 from 1H-15N HSQC, HNCA, HNCO, and HBHA(CO)NH spectra and performed a TALOS+ backbone chemical shift analysis. This revealed that the overall helical structure of the C-terminal domain was not greatly perturbed but that the φ and ψ angles for residues surrounding the mutation site were significantly changed (Fig. 6).
FIGURE 4.
Chemical shift mapping of perturbations within mutant LC1 proteins. The wild-type LC1 structure and mutated residues are illustrated at the top left: yellow, Met182; red, Asp185; blue, Arg189 and Arg196. The locations of chemical shift alterations (Δδ) determined for the five mutant forms are shown in the other panels (two views of the R189E and R196A changes rotated by 180° around the y axis are illustrated). The side-chain atoms of residues that showed significant chemical shift (δ) perturbations compared with wild-type are represented as spheres. The color code is as follows: Δδ (in 1H and/or 15N dimension) ≤0.1 ppm, pale yellow; ≤0.2 ppm, orange; ≤0.3 ppm, deep salmon; >0.3 ppm or missing/unidentified, red; the mutated residue is indicated in green.
FIGURE 5.
Chemical shift differences for mutant LC1 proteins. The differences in chemical shifts for the mutant proteins compared with wild type in both 1H and 15N dimensions of the HSQC spectra are shown. Peaks that were missing/unidentified were assigned values of 0.3 and 1.0 ppm for the 1H and 15N dimensions, respectively.
FIGURE 6.
TALOS+ analysis of D185G LC1 structure. The backbone assignments for D185G LC1 were obtained from a 13C,15N-labeled sample, and the backbone torsion angles were predicted from the chemical shifts using TALOS+. The upper panels indicate the predicted secondary structure (blue, β sheet; red, α helix) and order parameter (S2). Ramachandran plots of φ/ψ angles for best matches in the database for the mutation site and immediately adjacent residues are shown in the lower panels. The results illustrate that this mutation does not dramatically alter the helical structure of the C-terminal domain. However, the φ/ψ angles for Gly185 (−89 ± 23° and 152 ± 18°) are very different from those for Asp185 in wild-type LC1 (48.2° and 4.7°), suggesting that the orientation of the terminal helix is altered.
Several mutations (R189E and to a lesser extent M182A and R196A) caused perturbations that were propagated along the two β sheet faces and intervening loop region of the ββα barrel toward the N terminus; in contrast, the α helical face of the barrel was essentially unaffected. The R189E transition was especially disruptive, and resonances for many residues within the C-terminal domain were greatly altered or missing. In addition, significant perturbations extended along the large β sheet face that is thought to provide the interaction surface with the γ HC. Thus, the dramatic phenotypic consequences observed for this mutation in vivo (Table 1) may reflect disruption of both the C-terminal domain and other LC1 regions.
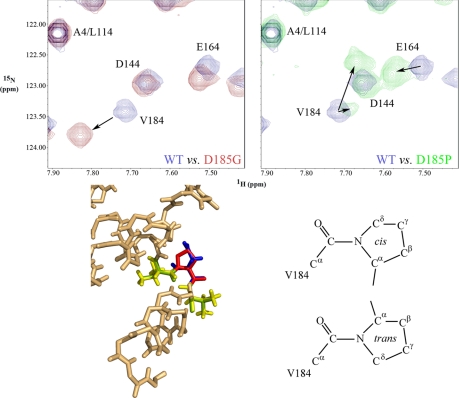
We previously altered both Met182 and Asp185 to Pro to reduce the high backbone dynamics at these positions that were predicted to control the orientation of the terminal α9 helix. Analysis of the D185P HSQC spectrum revealed that two amide peaks of unequal intensity were present for the preceding residue Val184; note that Pro residues do not contain a free amide and thus do not yield a peak in this NMR experiment. In contrast, alteration of Asp185 to Gly to increase backbone mobility resulted in a single Val184 peak that was shifted to a different location. Pro residues can exist in either cis or trans conformations (Fig. 7), and molecular modeling revealed that a cis-Pro residue is readily accommodated within the wild-type structure with little change in the location of Val184. Consequently, the less intense Val184 peak in the D185P spectrum that is very close to the wild-type position likely represents this isoform. This in turn suggests that the trans-Pro isomer, which would cause a significant alteration in the orientation of the C-terminal α9 helix, is the predominant form in solution.
FIGURE 7.
Proline isomerization yields two forms of D185P mutant LC1. The upper panels show overlays of the Val184 region of 1H-15N HSQC spectra for wild-type LC1 and the D185G (red; left panel) and D185P (green; right panel) mutant forms. Whereas the D185G alteration causes a significant movement of the Val184 peak, D185P yields two peaks of unequal intensity. One peak has near wild-type 1H and 15N chemical shifts, and molecular modeling of a cis-Pro residue onto Asp185 required little change in the location of Val184, suggesting that this peak represents the cis isomer. A second shifted peak of considerably greater intensity was also present and likely represents the trans isomer. An overlay of the wild-type structure and the D185P LC1 model is shown in the lower left panel, and the cis- and trans-Pro isomeric forms are indicated at right. The color code for the wild-type structure is: yellow, Val184 and Val186; blue, Asp185. For the D185P model, the color code is: green, Val184 and Val186; red, Pro185.
Microtubule Interaction Surface
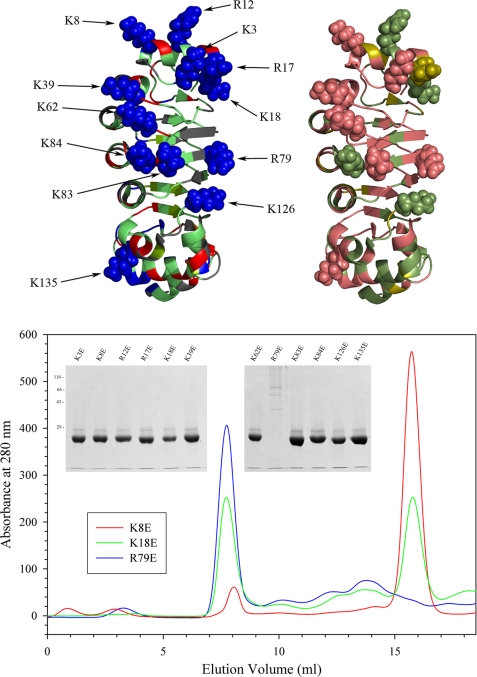
In addition to an interaction with the γ HC, LC1 also associates directly in situ with tubulin located in the A-tubule of the outer doublets (12). Defining the region of LC1 that mediates this association is important as it determines the geometry of the conformational switch and orientation of the LC1 C-terminal domain that has an impact on outer arm function. As the LC1-tubulin interaction is disrupted by high salt and readily detected using the zero-length cross-linking reagent 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide to generate an Mr 66,000 LC1-tubulin product (10), the interface must include both acidic and basic residues that interact directly. The exposed tubulin C-terminal domain is highly acidic, and therefore, we predicted that LC1 provides one or more basic residues to this interaction. To test this, we prepared 12 mutant forms of LC1 in which a single basic residue on the face opposite the presumptive γ HC interaction surface was changed to Glu (Fig. 8, top left panel). Interestingly, many of these basic residues are conserved between Chlamydomonas, trypanosomes, and humans and form a helical stripe that spans the short axis of the LC1 LRR barrel (Fig. 8, top right panel). With one exception, all these mutant proteins were soluble and could be purified in monomeric form; the R79E version of LC1 was obtained only as larger oligomers and could not be used further (Fig. 8, lower panel). Once purified, the mutant proteins were stable and remained almost completely monomeric even after many months at 4 °C (Table 2).
FIGURE 8.
Mutagenesis of basic residues within central LRR barrel. The 12 Arg/Lys residues mutated to Glu to assess their involvement in LC1-microtubule interactions are indicated in the top left panel. These residues form two clusters: a group near the N-terminal end of the central barrel and a helical stripe that extends along the barrel and crosses the protein short axis. The conservation of these residues between Chlamydomonas and humans is mapped onto the structure in the top right panel. The color code is as follows: identical, deep salmon; conservative substitution, olive; not conserved, green. Part of the N-terminal basic patch and much of the helical stripe is highly conserved. The lower panel shows the gel filtration profiles for K8E, K18E, and R79E forms of LC1 that exhibit distinct propensities for oligomerization: the peak eluting at ∼16 ml represents monomeric LC1, whereas oligomers elute earlier. The insets show the final purified mutant proteins separated in 12.5% SDS-polyacrylamide gels (Coomassie Blue stain).
TABLE 2.
Mutagenesis of basic residues in central LRR barrel
| Mutation | WT residue conserved in |
LC1 in monomeric forma | Monomer in aggregated form after prolonged storageb | Microtubule binding activityc | |
|---|---|---|---|---|---|
| H. sapiens | T. brucei | ||||
| % | % | % WT | |||
| None | 97.4 | <0.1 | 100 | ||
| K3E | Yes | No | 88.0 | 3.8 | 38 |
| K8E | Yes | Basic | 92.5 | 1.1 | 54 |
| R12E | No | No | 75.4 | <0.1 | 36 |
| R17E | Basic | No | 76.8 | <0.1 | 31 |
| K18E | No | No | 46.8 | 0.1 | 16 |
| K39E | Yes | Yes | 76.1 | <0.1 | 14 |
| K62E | Yes | Yes | 69.9 | <0.1 | 25 |
| R79E | Yes | Yes | ∼0d | NDe | ND |
| K83E | Yes | Yes | 82.5 | <0.1 | 144 |
| K84E | No | Yes | 63.7 | <0.1 | 93 |
| K126E | No | No | 71.1 | <0.1 | 31 |
| K135E | Yes | Basic | 93.1 | <0.1 | 118 |
a Determined by gel filtration chromatography from the relative amounts of LC1 eluting from a Superdex 200 10/300 GL column in monomeric form versus larger oligomers. Value quoted is the average of two separations. Only monomeric LC1 was used for binding assays.
b Purified monomeric LC1 proteins were stored at 4 °C for >4 months, and the percentage of aggregation occurring was determined by gel filtration in a Superdex 200 10/300 GL column.
c The values quoted were determined by quantitative densitometric analysis of the Coomassie Blue-stained gels shown in Fig. 9 using ImageQuant TL. Background subtraction utilized the rolling ball method.
d Essentially no monomeric R79E LC1 was evident in the chromatograms (see Fig. 6).
e ND, not determined.
Axonemal tubulins are highly modified by several processes, including polyglutamylation, polyglycylation, and acetylation; these modifications can have a major impact on dynein-axonemal microtubule interactions e.g. (27). Consequently, to assess the ability of mutant LC1 proteins to interact specifically with axonemal microtubules, we prepared axonemes from the oda6 mutant, which lacks outer dynein arms, or depleted wild-type axonemes of dynein arms by high salt extraction. Thus, the axoneme samples have the native LC1 interactions sites available. Monomeric LC1 does not sediment during centrifugation in the absence of axonemes (Fig. 9a), whereas wild-type LC1 bound oda6 axonemes in a dose-dependent manner (Fig. 9b). Furthermore, binding of LC1 to a fixed amount of dynein-depleted wild-type axonemes became saturated even in the presence of a large excess of LC1 (Fig. 9c). However, three mutants (K18E, K39E, and K62E) showed severe decreases (to <25% of wild type) in their ability to bind oda6 axonemes, and several others (K3E, K8E, R12E, R17E, and K126E) were significantly reduced from wild-type levels (Fig. 9d and Table 2). In contrast, the K83E, K84E, and K135E alterations and mutations in the C-terminal domain had little or no negative effect on microtubule binding activity (Tables 1 and 2).
FIGURE 9.
Axoneme binding activity of mutant LC1 proteins. a, the supernatants (S) and pellets (P) for wild-type and K84E mutant LC1 spun in a microcentrifuge in the absence of axonemes. This panel demonstrates that monomeric LC1 does not sediment during centrifugation. b illustrates the dose-dependent binding of LC1 to axonemes from the outer armless mutant oda6. The numbers on the image indicate the relative amounts of LC1 bound to the axoneme sample (0.83 mg/ml) as the LC1 concentration was increased from 0 to 0.18, 0.94, and 1.88 mg/ml. c, increasing amounts of D185P LC1 (0, 0.07, 0.14, 0.28, 0.42, and 0.7 mg/ml final concentration) were added to dynein-depleted wild-type axonemes (0.7 mg/ml). The pellets from the binding assay are shown in the top panel, and equivalent loadings of the corresponding supernatants are shown in the lower panel. The numbers on the upper panel indicate the relative amounts of LC1 bound to a fixed amount of axonemes and demonstrate that the binding becomes saturated. d illustrates the effects of individual point mutations on LC1-axoneme binding; only pellets from the binding assay are shown. All assays contained the same concentration of both axonemal (0.9 mg/ml) and LC1 (0.56 mg/ml) proteins except for M182A (0.81 mg/ml). Several of the Lys → Glu mutations severely affected the ability of LC1 to bind axonemes, whereas the more C-terminal mutations did not: these data are quantified in Tables 1 and 2. All samples were separated in 12.5% SDS-polyacrylamide gels and stained with Coomassie Blue.
Identification of the K126E mutation as affecting axoneme binding was unexpected in that it is located far from other alterations that yielded a similar effect. K126E LC1 was stable; the purified protein did not aggregate and indeed remained completely monomeric even after several months at 4 °C (data not shown). However, this residue is located close to Arg79, which caused severe disruption and essentially complete aggregation when altered to Glu, suggesting that changes propagated through the structure might be responsible for the decrease in binding activity.
Consequences of the N150S PCD Mutation
Asn150 is part of the terminal LRR consensus sequence, and the side chain of this residue packs within the protein core (16). In humans, the homozygous N150S mutation causes PCD and has been predicted to convert the α7 helix to a loop (15). To directly assess the consequences of this disease-causing alteration, we determined the extent of the perturbation by chemical shift mapping and thermal titration circular dichroism spectroscopy. N150S LC1 could be purified in monomeric form and yielded a dispersed, well resolved 1H-15N HSQC spectrum (Fig. 10, a and insets) even though significant precipitation occurred during data collection at 25 °C. Resonances for Ile105, Ala106, Met122–Lys126, Thr128, Leu146–Gly149, and Leu152–Asn154 either could not be identified or were shifted by >0.2 ppm (Fig. 10b). These residues map to the β7, β8/β9, and β10 strands of the fourth, fifth and sixth LRRs, respectively, and the α7 helix that is immediately adjacent to the mutation site (Fig. 10c). Although some of these residues are exposed, the side chains of Ile105, Met122, Asn125, Leu147, and Leu152 are located within the core, and thus, this mutation disrupts the internal packing. Consistent with this, we observed that the melting temperature as assessed by circular dichroism spectroscopy decreased from 51.5 °C for wild-type LC1 to 44 °C for the N150S mutant (Fig. 10d). In contrast, the Tm for D185G LC1 was slightly increased over wild type. The unfolding profile for LC1 appears to be two-state.
FIGURE 10.
Structural analysis of N150S PCD mutation. a, the entire 1H-15N HSQC spectrum of N150S LC1 is shown. The upper inset depicts the gel filtration and electrophoretic profiles of the mutant protein, and the lower inset shows an enlarged region of the spectrum illustrating that it is well resolved. Plots of the chemical shift changes for each residue in both dimensions are shown at right; missing/unidentified peaks were assigned values of 0.3 and 1.0 ppm for the 1H and 15N dimensions, respectively. b, the N150S mutation (green) and residues with chemical shifts that are >0.2 ppm different from wild type (orange) or missing/unidentified (red) are illustrated on the LC1 ribbon structure with surface mesh. Significant changes surround the mutation site and are propagated into the fourth and fifth LRRs. c, the thermal stability of wild-type, N150S, and D185G LC1 was assessed by circular dichroism spectroscopy in the range 4–65 °C. N150S LC1 was considerably destabilized and exhibited a Tm of 44 °C as compared with 51.5 and 52.5 °C for wild-type and D185G mutant LC1, respectively.
DISCUSSION
LC1 is one of the most highly conserved components of axonemal outer arm dyneins, and multiple lines of evidence have now revealed that it plays a key role in ciliary motility. In Chlamydomonas, dominant negative mutations in the C-terminal domain caused flagellar stalling and an abnormal response to viscous load (12). Similar mutations in Trypanosoma also lead to defective motility (20), whereas RNAi-driven knockdowns caused failure of outer arm dynein assembly (28). In the context of a ciliated epithelium, reduction of LC1 levels in the planarian Schmidtea mediterranea resulted in lower beat frequency and disruption of the hydrodynamic coupling necessary for generating metachronal synchrony but did not appear to affect dynein assembly (29). Furthermore, a single residue alteration within the terminal LRR of the human orthologue DNAL1 has been demonstrated to cause PCD (15), and lack of a LC1/DNAL1 orthologue has also been reported to yield non-flagellate zoospores in the pathogenic fungus Phytophthora nicotianae (30).
Subdomain Organization of LC1 and Orientation of Conformational Switch
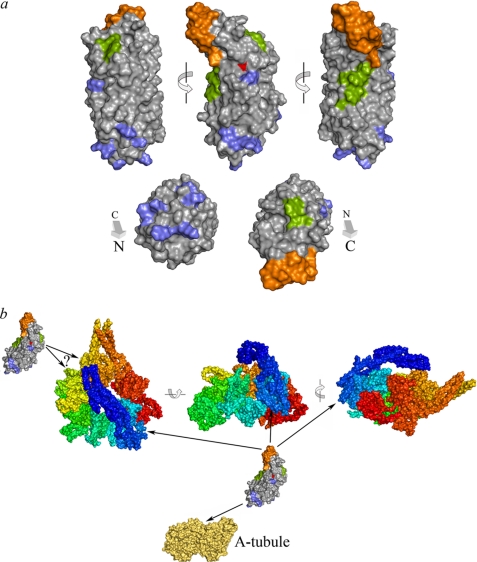
LC1 interacts with both the γ HC motor domain and tubulin located within the outer doublet A-tubule. Chemical shift mapping indicates that perturbation of the C-terminal helical domain accounts for the dominant negative effects observed in vivo. Indeed, for one of the most disruptive alterations (D185G), all identified chemical shift changes were within the C-terminal region. Furthermore, the interaction with the γ HC is hydrophobic and so likely involves one or both of the exposed Trp residues. The mutagenesis experiments described here also allow the region responsible for binding tubulin to be localized to one face of the N-terminal region of the LRR barrel. These data are mapped onto the LC1 structure in Fig. 11a and together define discrete functional subdomains.
FIGURE 11.
Functional subdomains within LC1 and architecture of dynein conformational switch. a, three views of the LC1 long axis and views from either terminus of the molecular surface are shown at the top. The tubulin-binding region is defined by the basic residues (blue) that disrupt this activity, whereas the C-terminal domain (orange) represents the alterations present in the D185G mutation that exhibits a strong phenotype in vivo with the least structural perturbation. These two sections are at opposite ends of the molecule, strongly suggesting that the C-terminal region indeed interacts directly with the γ HC AAA domains. This latter association is known to be hydrophobic, and two potential regions, a hydrophobic patch centered on Trp99 and a smaller region involving Trp130 and Tyr156, which may individually or together mediate this interaction, are indicated in green. The PCD-associated mutation site is shown in red. b, a model for the overall geometry of LC1-mediated association of the γ HC motor domain with the outer doublet A-tubule is illustrated as van der Waals surface representations of the dynein motor unit (rainbow color from the N-terminal linker in blue to AAA6 in red; Protein Data Bank code 3QMZ) (38), LC1 (from a), and the tubulin dimer (yellow-orange; Protein Data Bank code 3IZO) (39); the structures are all illustrated at the same scale. One copy of LC1 is predicted to be bound to AAA1 through the hydrophobic patch(es) with the C-terminal domain interacting with the AAA ring. The second copy of LC1 appears to associate with AAA3 or AAA4. The precise relative orientations of the components remain uncertain.
As at least one of the two copies of LC1 appears to span the ∼6-nm gap between the γ HC AAA ring domain and the A-tubule (12, 31), it must be aligned approximately along its long axis of 69.8 Å; the two short axes are only 28.4 and 30.0 Å (17). Thus, taken together, the geometric considerations and functional data suggest that one copy of LC1 is arranged with the N-terminal domain binding the A-tubule and the hydrophobic region(s) interacting with the AAA1 domain (Fig. 11b). This in turn requires that the LC1 C-terminal region is also in direct contact with the HC motor unit; it might either be directly inserted into the cleft between AAA domains or aligned along an exposed face. One key question is now to identify the residues on the γ HC with which this LC1 subdomain interacts as this will provide additional clues as to how LC1 is involved in the control of outer arm function.
Cross-linking and vanadate-mediated photocleavage data support association of an LC1 monomer with AAA1 with a second copy bound to either AAA3 or AAA4 (10)3; the ambiguity is derived from assignment of the V2a and V2b cleavage sites (32) to specific AAA domains. Based on the structural data that are now available for both the HC and LC1, it is uncertain whether both copies of LC1 could be bound to the A-tubule at the same time, although they might interact sequentially during the mechanochemical cycle, or indeed how LC1 if bound to AAA4 could even interact with the A-tubule at all if the γ HC motor unit is arrayed in the same orientation as others within the outer arm (33, 34). Thus, the possibility remains that this second copy of LC1 is associated with the HC in a different manner and/or performs a function distinct from LC1 bound to AAA1.
Microtubule-binding Subdomain
Mutation of single Lys residues within the LRR barrel had a pronounced effect on the ability of LC1 to interact with axonemal microtubules. With one exception (discussed further below), all these residues form a patch near the N terminus (Fig. 11a). Identification of multiple Lys residues involved in binding the A-tubule also provides an explanation for the very high yield Mr 66,000 product containing LC1 and tubulin that is generated by treatment of wild-type axonemes with 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (10). In effect, multiple cross-links between Lys residues on LC1 and their interacting acidic residues on tubulin are possible, any one of which generates the same dimeric product as assessed by gel electrophoresis.
We also observed that alteration of Lys126 led to decreased microtubule binding even though this residue is located far from others that yielded a similar phenotype. This residue is adjacent to Arg79, which caused protein aggregation when mutated to Glu. Although K126E LC1 was stable, a possible explanation is that altering the Arg79/Lys126 region causes widespread disruption to a large portion of the LC1 surface that in the case of R79E yields aggregation but for K126E results only in decreased microtubule binding affinity.
Function of LC1 C-terminal Domain
In metazoans, the C-terminal helical domain is highly charged, containing five or more acidic residues, whereas in protists (i.e. Chlamydomonas and Trypanosoma), there are also two conserved basic residues (Arg189 and Arg196 in Chlamydomonas) (Fig. 1b). Structural analyses indicate that the orientation of this C-terminal helix is controlled by two residues (Met182 and Asp185 in Chlamydomonas) that show high backbone dynamics (17). Binding of LC1 to the γ HC requires hydrophobic interactions, and most single residue alterations within the C-terminal domain did not affect LC1-γ HC associations. The one exception was D185G, which resulted in a slight increase in detergent solubility of LC1 (12). Thus, it is unlikely that this LC1 region is merely acting as part of a tether attaching the motor unit to the A-tubule. Rather, given the strong phenotypic consequences observed from minor alterations, it more likely makes key contacts within the AAA domain that lead to the modulation of motor activity. This suggestion does not negate the possibility that LC1 also plays an important role in stabilizing the motor domain with respect to the A-tubule. This might ensure that the AAA ring remains a constant distance from the A-tubule within a bent region of the flagellum when the interdoublet gap increases (35) and/or prevent or modulate the ability of the AAA ring to move with respect to the axonemal long axis (36, 37); this latter feature might convert the γ HC motor into an ATP-dependent brake that limits sliding.
Why Does N150S LC1 Cause Primary Ciliary Dyskinesia?
Asn150 is part of the LRR consensus, and in wild type, this side chain is packed within the protein core. In humans, altering this conserved residue to Ser resulted in a partial outer arm assembly defect. Molecular modeling suggested that the N150S alteration might convert the α7 helix to a loop and disturb the terminal LRR fold (15). Our chemical shift mapping revealed that the perturbation is much more pervasive and extends through the last three LRRs, disrupting the core. Consequently, N150S LC1 is much less stable than wild type or any of the other mutant forms we have made. Indeed, significant precipitation occurred within the few hours at 25 °C necessary to collect the 1H-15N HSQC spectrum, and this protein had a melting temperature that was 8 °C lower than wild type. Thus, at physiological temperature in humans, it is likely that a significant fraction of this PCD-causing mutant form does not fold correctly and/or even assemble within the dynein complex. As the lack of LC1 would lead to exposure of a hydrophobic region on one HC, this mutation might therefore result in the partial failure of outer arm assembly due to indirect effects mediated through aggregation of other component(s) that are no longer stable in the absence of LC1. This scenario would be consistent with the observed reduction of outer arms in trypanosomes following LC1 RNAi (28), although similar knockdowns in planaria did not appear to affect arm assembly (29).
In conclusion, we have defined a series of functional subdomains within the LC1 component of outer arm dynein. These data provide insight into the mechanism by which this highly conserved protein modulates dynein motor function within the ciliary axoneme.
Acknowledgments
We thank Dr. Mark Maciejewski (facility manager, Gregory P. Mullen NMR Structural Biology Facility, University of Connecticut Health Center) for collecting and processing the NMR data and for many helpful discussions, Rekha King for analyzing the N150S LC1 spectrum, and Li Luo (Biophysics Core Facility, University of Connecticut Health Center) for assistance with circular dichroism spectroscopy.
This work was supported, in whole or in part, by National Institutes of Health Grant GM051293 (to S. M. K.).
S. E. Benashski and S. M. King, unpublished results.
- HC
- heavy chain
- HSQC
- heteronuclear single quantum coherence
- LC
- light chain
- LRR
- leucine-rich repeat
- PCD
- primary ciliary dyskinesia
- AAA
- ATPase associated with cellular activities.
REFERENCES
- 1. Sugino K., Naitoh Y. (1982) Simulated cross-bridge patterns corresponding to ciliary beating in Paramecium. Nature 295, 609–611 [Google Scholar]
- 2. Lindemann C. B. (2002) Geometric Clutch model version 3: the role of the inner and outer arm dyneins in the ciliary beat. Cell Motil. Cytoskeleton 52, 242–254 [DOI] [PubMed] [Google Scholar]
- 3. Brokaw C. (2009) Thinking about flagellar oscillation. Cell Motil. Cytoskeleton 66, 425–436 [DOI] [PubMed] [Google Scholar]
- 4. Hayashibe K., Shingyoji C., Kamiya R. (1997) Induction of temporary beating in paralyzed flagella of Chlamydomonas mutants by application of external force. Cell Motil. Cytoskeleton 37, 232–239 [DOI] [PubMed] [Google Scholar]
- 5. King S. M., Kamiya R. (2009) in The Chlamydomonas Source Book: Cell Motility and Behavior (Witman G. B., ed) 2nd Ed., Vol. 3, pp. 131–208, Elsevier, San Diego, CA [Google Scholar]
- 6. Pfister K. K., Fay R. B., Witman G. B. (1982) Purification and polypeptide composition of dynein ATPases from Chlamydomonas flagella. Cell Motil. 2, 525–547 [DOI] [PubMed] [Google Scholar]
- 7. Pfister K. K., Witman G. B. (1984) Subfractionation of Chlamydomonas 18 S dynein into two unique subunits containing ATPase activity. J. Biol. Chem. 259, 12072–12080 [PubMed] [Google Scholar]
- 8. Sakakibara H., Nakayama H. (1998) Translocation of microtubules caused by the αβ, β and γ outer arm dynein subparticles of Chlamydomonas. J. Cell Sci. 111, 1155–1164 [DOI] [PubMed] [Google Scholar]
- 9. Hom E. F., Witman G. B., Harris E. H., Dutcher S. K., Kamiya R., Mitchell D. R., Pazour G. J., Porter M. E., Sale W. S., Wirschell M., Yagi T., King S. M. (2011) A unified taxonomy for ciliary dyneins. Cytoskeleton 68, 555–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Benashski S. E., Patel-King R. S., King S. M. (1999) Light chain 1 from the Chlamydomonas outer dynein arm is a leucine-rich repeat protein associated with the motor domain of the γ heavy chain. Biochemistry 38, 7253–7264 [DOI] [PubMed] [Google Scholar]
- 11. Liu Z., Takazaki H., Nakazawa Y., Sakato M., Yagi T., Yasunaga T., King S. M., Kamiya R. (2008) Partially functional outer-arm dynein in a novel Chlamydomonas mutant expressing a truncated γ heavy chain. Eukaryot. Cell 7, 1136–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Patel-King R. S., King S. M. (2009) An outer arm dynein light chain acts in a conformational switch for flagellar motility. J. Cell Biol. 186, 283–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sakato M., Sakakibara H., King S. M. (2007) Chlamydomonas outer arm dynein alters conformation in response to Ca2+. Mol. Biol. Cell 18, 3620–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. King S. M. (2010) Sensing the mechanical state of the axoneme and integration of Ca2+ signaling by outer arm dynein. Cytoskeleton 67, 207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mazor M., Alkrinawi S., Chalifa-Caspi V., Manor E., Sheffield V. C., Aviram M., Parvari R. (2011) Primary ciliary dyskinesia caused by homozygous mutation in DNAL1, encoding dynein light chain 1. Am. J. Hum. Genet. 88, 599–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu H., Maciejewski M. W., Marintchev A., Benashski S. E., Mullen G. P., King S. M. (2000) Solution structure of a dynein motor domain associated light chain. Nat. Struct. Biol. 7, 575–579 [DOI] [PubMed] [Google Scholar]
- 17. Wu H., Blackledge M., Maciejewski M. W., Mullen G. P., King S. M. (2003) Relaxation-based structure refinement and backbone molecular dynamics of the dynein motor domain-associated light chain. Biochemistry 42, 57–71 [DOI] [PubMed] [Google Scholar]
- 18. Wu H., King S. (2003) Backbone dynamics of dynein light chains. Cell Motil. Cytoskeleton 54, 267–273 [DOI] [PubMed] [Google Scholar]
- 19. Yagi T., Minoura I., Fujiwara A., Saito R., Yasunaga T., Hirono M., Kamiya R. (2005) An axonemal dynein particularly important for flagellar movement at high viscosity. Implications from a new Chlamydomonas mutant deficient in the dynein heavy chain gene DHC9. J. Biol. Chem. 280, 41412–41420 [DOI] [PubMed] [Google Scholar]
- 20. Ralston K. S., Kisalu N. K., Hill K. L. (2011) Structure-function analysis of dynein light chain 1 identifies viable motility mutants in bloodstream-form Trypanosoma brucei. Eukaryot. Cell 10, 884–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu H., Maciejewski M. W., Benashski S. E., Mullen G. P., King S. M. (1999) 1H, 15N and 13C resonance assignments for the 22 kDa LC1 light chain from Chlamydomonas outer arm dynein. J. Biomol. NMR 13, 309–310 [DOI] [PubMed] [Google Scholar]
- 22. Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mosavi L. K., Peng Z. Y. (2003) Structure-based substitutions for increased solubility of a designed protein. Protein Eng. 16, 739–745 [DOI] [PubMed] [Google Scholar]
- 24. Witman G. B. (1986) Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 134, 280–290 [DOI] [PubMed] [Google Scholar]
- 25. Vyas J., Gryk M. R., Schiller M. R. (2009) VENN, a tool for titrating sequence conservation onto protein structures. Nucleic Acids Res. 37, e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peitsch M. C. (1996) ProMod and Swiss-Model: internet-based tools for automated comparative protein modelling. Biochem. Soc. Trans. 24, 274–279 [DOI] [PubMed] [Google Scholar]
- 27. Kubo T., Yanagisawa H. A., Yagi T., Hirono M., Kamiya R. (2010) Tubulin polyglutamylation regulates axonemal motility by modulating activities of inner-arm dyneins. Curr. Biol. 20, 441–445 [DOI] [PubMed] [Google Scholar]
- 28. Baron D. M., Kabututu Z. P., Hill K. L. (2007) Stuck in reverse: loss of LC1 in Trypanosoma brucei disrupts outer dynein arms and leads to reverse flagellar beat and backward movement. J. Cell Sci. 120, 1513–1520 [DOI] [PubMed] [Google Scholar]
- 29. Rompolas P., Patel-King R. S., King S. M. (2010) An outer arm dynein conformational switch is required for metachronal synchrony of motile cilia in planaria. Mol. Biol. Cell 21, 3669–3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Narayan R. D., Blackman L. M., Shan W., Hardham A. R. (2010) Phytophthora nicotianae transformants lacking dynein light chain 1 produce non-flagellate zoospores. Fungal Genet. Biol. 47, 663–671 [DOI] [PubMed] [Google Scholar]
- 31. Oda T., Hirokawa N., Kikkawa M. (2007) Three-dimensional structures of the flagellar dynein-microtubule complex by cryoelectron microscopy. J. Cell Biol. 177, 243–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. King S. M., Witman G. B. (1988) Structure of the γ heavy chain of the outer arm dynein from Chlamydomonas flagella. J. Cell Biol. 107, 1799–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nicastro D., Schwartz C., Pierson J., Gaudette R., Porter M. E., McIntosh J. R. (2006) The molecular architecture of axonemes revealed by cryoelectron tomography. Science 313, 944–948 [DOI] [PubMed] [Google Scholar]
- 34. Ishikawa T., Sakakibara H., Oiwa K. (2007) The architecture of outer dynein arms in situ. J. Mol. Biol. 368, 1249–1258 [DOI] [PubMed] [Google Scholar]
- 35. Lindemann C. B., Mitchell D. R. (2007) Evidence for axonemal distortion during the flagellar beat of Chlamydomonas. Cell Motil. Cytoskeleton 64, 580–589 [DOI] [PubMed] [Google Scholar]
- 36. Movassagh T., Bui K. H., Sakakibara H., Oiwa K., Ishikawa T. (2010) Nucleotide-induced global conformational changes of flagellar dynein arms revealed by in situ analysis. Nat. Struct. Mol. Biol. 17, 761–767 [DOI] [PubMed] [Google Scholar]
- 37. Ueno H., Yasunaga T., Shingyoji C., Hirose K. (2008) Dynein pulls microtubules without rotating its stalk. Proc. Natl. Acad. Sci. U.S.A. 105, 19702–19707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Carter A. P., Cho C., Jin L., Vale R. (2011) Crystal structure of the dynein motor domain. Science 331, 1159–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Alushin G. M., Ramey V. H., Pasqualato S., Ball D. A., Grigorieff N., Musacchio A., Nogales E. (2010) The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature 467, 805–810 [DOI] [PMC free article] [PubMed] [Google Scholar]