Abstract
Mycoplasma mycoides subsp. mycoidesSC (MmymySC)is the etiological agent of contagious bovine pleuropneumonia (CBPP), a highly contagious respiratory disease in cattle. The genome of Mmymy SC type strain PG1T has been sequenced to map all the genes and to facilitate further studies regarding the cell function of the organism and CBPP. The genome is characterized by a single circular chromosome of 1,211,703 bp with the lowest G+C content (24 mole%)and the highest density of insertion sequences (13% of the genome size)of all sequenced bacterial genomes. The genome contains 985 putative genes, of which 72 are part of insertion sequences and encode transposases. Anomalies in the GC-skew pattern and the presence of large repetitive sequences indicate a high genomic plasticity. A variety of potential virulence factors was identified, including genes encoding putative variable surface proteins and enzymes and transport proteins responsible for the production of hydrogen peroxide and the capsule, which is believed to have toxic effects on the animal.
Contagious bovine pleuropneumonia (CBPP) is the infectious disease that kills the largest number of cattle in Africa each year. It is a highly contagious respiratory disease, which is caused by Mycoplasma mycoides subsp. mycoides biotype small colony (MmymySC). CBPP is the only bacterial disease included in the A-list of the World Organization for Animal Health (http://www.oie.int) of prioritized communicable animal diseases, together with fourteen viral diseases. Thus, from a global socioeconomic perspective, it is the most important bacterial epizootic. CBPP also affects buffalo and can appear in different forms, ranging from hyperacute and acute variants with high mortality (up to 70%), to subacute and chronic forms with high risk of transmitting the infectious agent from symptomless carriers. The clinical symptoms of acute CBPP involve respiratory distress, cough, cessation of rumination, anorexia, and severe pleuritic pain. CBPP is mainly present in Africa south of Sahara, and it is also assumed to be prevalent in Asia. During the 1980s and 1990s, there have also been several outbreaks of CBPP in southern Europe.
MmymySC is a member of the class Mollicutes (trivial name, mollicutes), which has evolved from the Gram-positive bacteria that possess genomes with low G+C content (Phylum Firmicutes), and belongs to the genus Mycoplasma (trivial name, mycoplasmas). Mollicutes lack cell wall and are known as the smallest self-replicating organisms. According to phylogenetic studies of the 16S rRNA gene, MmymySC belongs to the M. mycoides cluster of the Spiroplasma group (Weisburg et al. 1989; Pettersson et al. 1996; Johansson et al. 1998). Five sequenced genomes of mollicutes have been published to date. M. genitalium (Fraser et al. 1995), M. pneumoniae (Himmelreich et al. 1996), Ureaplasma parvum (formerly Ureaplasma urealyticum; Glass et al. 2000; Robertson et al. 2002), and M. penetrans (Sasaki et al. 2002), which all belong to the pneumoniae group, as well as M. pulmonis (Chambaud et al. 2001), which belongs to the hominis group (Weisburg et al. 1989; Johansson et al. 1998). The relationship between MmymySC (and other members of the Spiroplasma group) and the other sequenced mollicutes is rather distant, that is, 75%–80% similarity as judged from 16S rRNA sequences. We have sequenced the genome of MmymySC type strain PG1T to get a better knowledge of the biology and pathogenicity of MmymySC and to promote efforts to develop recombinant vaccines and diagnostic tools for CBPP.
RESULTS AND DISCUSSION
General Genome Features
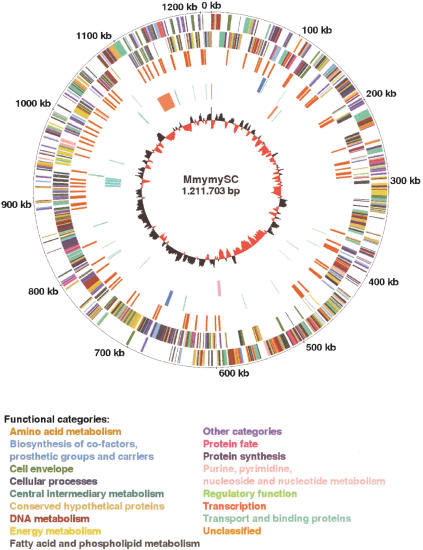
The general features of the genome of MmymySC type strain PG1T are shown in Figure 1 and Table 1. The genome consists of a single circular chromosome with a size of 1,211,703 bp and a G+C-content of 24.0 mole%, which is the lowest G+C-content among all genomes sequenced thus far. It possesses 985 putative genes (Supplemental Fig. 1; Supplemental Table 1 available online at www.genome.org), including 72 transposase genes located within insertion sequences (ISs). In addition, 83 truncated genes were found, including 52 transposase genes. Putative biological functions were assigned for 59% of the genes, whereas a further 14% were similar to genes with unknown function in other species. Interestingly, as much as 27% are unassigned genes that are unique for MmymySC, even though five additional genomes of mollicutes have previously been sequenced.
Figure 1.
Circular representation of the MmymySC genome. Outer concentric circle: genomic positions in bases, where position one is the first base of the dnaA gene. Second concentric circle: the predicted genes on the positive strand. Third concentric circle: the predicted genes on the negative strand. The genes are shown in bars of different colors representing different functional categories. Fourth concentric circle: IS elements. Fifth concentric circle: tRNA and rRNAs genes in green and blue bars, respectively. Sixth concentric circle: the capsule biosynthesis clusters shown in orange bars, the hydrogen peroxide biosynthesis cluster in a pink bar, and the genes encoding variable surface proteins in green bars. Seventh concentric circle: the GC-skew diagram; where the red color indicates that the leading strand contains more Gs than Cs, and the black color indicates the opposite case.
Table 1.
Genome Features
|
IS elements
|
||
|---|---|---|
| Data included | Data excluded | |
| Length (bp) | 1,211,703 | 1,050,226 |
| G + C content (mole%) | 24.0 | 23.7 |
| Putative protein CDSs | 985 | 913 |
| Average length of protein CDS (bp) | 982 | 957 |
| Coding regions (including stable RNA) in proportion to the genome length (%) | 80 | 83 |
| Functionally assigned protein CDSs (%) | 59 | 55 |
| Conserved hypothetical protein CDSs (%) | 14 | 16 |
| MmymySC-specific protein CDSs (%) | 27 | 29 |
| Ribosomal RNA operons | 2 | 2 |
| Transfer RNA genes | 30 | 30 |
| IS elements in proportion to the genome length | 13.3% | — |
| Total length of repetitive sequences (bp) | 346,843 | 185,366 |
CDS indicates coding sequence.
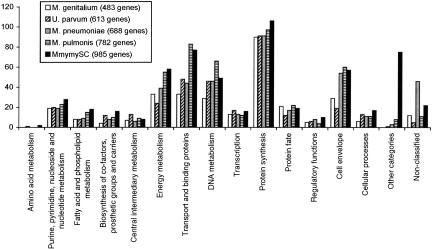
The number of genes that belong to the different functional categories in MmymySC is approximately the same as for the other sequenced mollicutes (Fig. 2). The large number of transport proteins in MmymySC, compared with the other species M. pulmonis, may result in MmymySC being better equipped to persist different tissue environments, reflecting its capability to form more or less systemic infections (Gourlay 1964; Masiga et al. 1972; Scanziani et al. 1997; Stradaioli et al. 1999; Grieco et al. 2001). The high number of MmymySC genes within “other categories” is due to the large amount of transposase genes located within the IS elements.
Figure 2.
The number of genes with assigned function divided into different functional categories for M. genitalium, U. parvum, M. pneumoniae, M. pulmonis, and MmymySC.
Repetitive Sequences
Intragenomic sequence comparisons show that MmymySC has a high degree of long repetitive sequences compared with other bacterial genomes (Supplemental Fig. 2). In total, the repetitive sequences in MmymySC constitute 29% of the genome. The largest repeats are 24, 13, and 12 kb. They are flanked by ISelements, which are known to cause genomic rearrangements and have been duplicated once in tandem. In many cases, the paralogous genes generated in these processes have most likely been subjects to negative evolutionary pressure, which have led to the truncation of one of the two duplicated genes.
More than 13% of the MmymySC genome consists of three kinds of ISelements, and it is therefore the most IS-dense bacterial genome that has been sequenced to date. ISMmy1 (Westberg et al. 2002), which is 1670 bp long, is present in eight full-length and one truncated copy. ISMmy1-like sequences were also found in the bovine pathogen M. bovis, whereas mycoplasmas that are phylogenetically closer to MmymySC lack ISMmy1 (Westberg et al. 2002). This observation indicates horizontal transfer of ISMmy1 between MmymySC and M. bovis. Southern blotting of 15 MmymySC strains with an ISMmy1 probe showed a unique hybridization pattern for the vaccine strain T1Sr49, which makes ISMmy1 a potential marker to distinguish the vaccine strain from naturally occurring field strains. The other two ISelements are IS1634 (Vilei et al. 1999), which is 1872 bp long, and IS1296 (Frey et al. 1995), which has a size of 1485 bp. IS1634 exists in 60 copies, in which two copies are split by other ISelements and one is truncated. IS1296 is present in 28 copies, including four that are interrupted by ISelements and seven truncated copies.
The IS elements are evenly distributed across the genome except for three larger IS-free regions, which are located at positions 285,937 to 363,559, 471,574 to 592,871, and 828,541 to 881,279 (Fig. 1). There is no obvious explanation for the absence of ISs in these regions except that they partly constitute several conserved regions of the mollicutes, such as the operons of the ribosomal protein and the ATP synthase genes, and the pyruvate dehydrogenase gene cluster.
Six other transposase-like ORFs were found. One of them (MSC_0603) resembles transposases of the IS30 family, and one (MSC_0699) is similar to transposases of the IS3 family. The remaining four are possible remnant transposases of ISMmy1 (MSC_0120 and MSC_0125) and IS1296 (MSC_0213 and MSC_0836). However, no additional characteristic features of an ISelement have been found for these putative transposases.
Codon Usage
Genomes with low G+C content are particularly rich in As and Ts in the third position of their genetic codons. In MmymySC, 91.4 mole% of the nucleotides in the third position are A or T. Strikingly, the genome only possesses 10 CGG codons (Supplemental Fig. 3). This is in agreement with the fact that MmymySC only possesses a single tRNA (tRNAArg[ACG]) for decoding the CGN codons (where N is A, C, G, or T), whereas the other five sequenced mollicutes have two tRNAs for this purpose. It has been experimentally shown that translations of synthetic genes in M. capricolum, a close relative to MmymySC (>99% similarity between their 16S rRNA genes), are terminated at the CGG codons, indicating that the CGG codon is a nonsense codon in M. capricolum (Oba et al. 1991). The possession of only one tRNAArg(NCG) gene and the rare occurrence of CGG codons of the MmymySC genome indicate that the CGG codon is a nonsense codon also in MmymySC. The universal stop codon UGA is coding for tryptophan in most of the mollicutes. Interestingly, the UGA codon is 24 times as frequent in the MmymySC genome as is the synonymous codon UGG. A plausible explanation for the large amount of UGA codons is the evolutionary pressure toward a lower G+C content of the genome.
Virulence Factors
Despite large efforts, the mechanisms behind the ability of MmymySC to cause disease are virtually unknown. However, there are some theories that have been experimentally tested. Already in 1976, it was shown that intravenous injection of the capsule from MmymySC in calves evoked pulmonary edema as in natural lesions of CBPP, indicating that the capsule has a direct toxic effect (Buttery et al. 1976). There are also some indications that increased capsular content associates with reduced phagocytosis by host cells (Marshall et al. 1995). The MmymySC genome contains two clusters of genes involved in the synthesis of the capsule (Fig. 1). The first one is located between positions 127,251 and 130,842 and comprises three genes encoding two putative glycosyltransferases and a UTP-glucose-1-phosphate uridylyltransferase. The second one is located between positions 1,108,435 and 1,133,176 and consists of a gene coding for UTP-glucose-1-phosphate uridylyltransferase and a region that exists in three tandem copies. Each copy of that region contains genes encoding two putative glycosyltransferases, a UDP-glucose 4-epimerase, a UDP-galactopyranose mutase, and the ATP-binding component of an oligopeptide-specific ABC transporter. The large cluster is also intergenically interspersed with four IS1634 copies. The redundancy of capsule biosynthesis genes may enable MmymySC to produce a relatively high amount of capsule and thereby increase the virulence of the organism and reduce the risk for phagocytosis by the host cells. It may also be a way of varying the composition of the capsule in order to escape the immune system of the host.
Production of active oxygen-containing molecules have been suggested as potential virulence factors of mycoplasmas. Compared to the European strains, African and Australian strains of MmymySC form higher amounts of hydrogen peroxide (H2O2) by oxidizing glycerol (Vilei and Frey 2001). Because the European strains are less virulent than are the African and Australian strains (Nicholas et al. 1996), the formation of H2O2 is believed to be a factor of pathogenicity in MmymySC. The strain of the present study, MmymySC PG1T with an origin that is not known, contains two clusters of genes that are involved in glycerol transport and production of hydrogen peroxide. The first cluster contains the genes (glpO, glpK, and glpF) coding for glycerol-3-phosphate oxidase (GlpO), glycerol kinase (GlpK), and a glycerol uptake facilitator protein (GlpF). Glycerol that is taken up by GlpF can be phosphorylated by GlpK and subsequently converted to dihydroxyacetone phosphate and H2O2 by GlpO. The genes of the second cluster (gtsA, gtsB, and gtsC) encode the ABC transporter proteins involved in glycerol transport (Vilei and Frey 2001). The lppB gene encoding a lipoprotein precursor is located immediately downstream of the second cluster. Presumably, it codes for the glycerol-binding subunit, because the gene encoding the substrate-binding component normally is located in the vicinity of the associated ABC transporter genes and has the structure of a prolipoprotein coding gene. Glycerol that is phosphorylated by the ABC transporter can be used as substrate by GlpO for the production of H2O2. All genes in the second glycerol uptake cluster are present in the African and Australian strains, but in the European strains gtsB is truncated and gtsC and lppB are absent (Vilei and Frey 2001).
Variable Surface Proteins
Some mycoplasmas are able to alternate their composition of surface proteins, so-called antigenic variation, in order to enhance colonization and to adapt to the host tissue environment at various stages of infection (Rosengarten and Wise 1990). The only reported gene to be involved in antigenic variation in MmymySC is vmm (MSC_0390; Persson et al. 2002), which encodes a phase variable lipoprotein precursor. The expression of Vmm can be switched on and off in a population of MmymySC by alternating the number of TA repeats in the promoter spacer of the vmm gene. The molecular mechanism behind the hypermutations in the promoter spacer is unknown, but it is likely that the altered number of repeats is caused by polymerase slippage during replication. Interestingly, the genome sequence reveals that five additional genes (MSC_0117, MSC_0364, MSC_1005, MSC_1033, and MSC_1058) encoding prolipoproteins have promoters with five to 12 TA repeats in the promoter, including the first four nucleotides in the -10 region (Fig. 3). The DNA sequence assembly contains clones with different numbers of TA repeats in the promoters of MSC_0117 and MSC_1005, which show that dinucleotide insertions and deletions occur relatively frequently in the cultivated MmymySC population. Three clones contain 10 TA repeats and one clone contains 11 TA repeats within the promoter of MSC_0117, and seven clones contain 11 TA repeats and one clone contains 12 TA repeats within the promoter of MSC_1005.
Figure 3.
The upstream region of genes encoding putative variable surface proteins. The promoter spacer regions are underlined, and the -35 and -10 regions are shown in capital letters. The sequences end with the start codon of the corresponding gene.
Furthermore, there are homonucleotide regions consisting of 15 to 23 As, which are located in the putative promoter of nine MmymySC surface protein genes (MSC_0809, MSC_810, MSC_812, MSC_813, MSC_815, MSC_816, MSC_817, MSC_818, and MSC_847; Fig. 3). Again, these repetitive sequences may be involved in transcriptional control. There are also two surface protein genes, which contain a mononucleotide stretch of 10 and 14 Ts within the coding part of the gene. These repetitive sequences may lead to size variation of the resulting proteins due to frame-shift caused by misincorporation of the correct number of repeat residues. In addition, there are 34 prolipoprotein genes and 144 transmembrane protein genes that have no assigned function and whose products are potential virulence factors as they may be involved in adherence or host cell interactions.
It is noteworthy that seven ISMmy1 elements have been inserted into promoters with TA repeats (data not shown), thus abolishing the expression of putatively phase variable proteins. Three of these spliced promoters are located upstream of genes encoding membrane-associated proteins. The other four lack the corresponding genes, which probably have been eliminated from the genome.
Replication, Transcription, and Translation
The set of genes encoding proteins involved in replication, transcription, and translation resembles the repertoire of the other sequenced mollicutes. The ribosomal RNA genes are clustered in two rRNA operons with the gene order 16SrRNA-23S rRNA-5SrRNA, which are separated by 586 kbp (Fig.1). The MmymySC genome comprises 30 tRNA genes (Fig. 1), and their corresponding tRNAs have specificity for all amino acids. A reduced set of tRNAs is common in mollicutes, of which M. pulmonis has the smallest set of 29 tRNA genes (Chambaud et al. 2001).
For most bacterial genomes, the GC-skew, defined as [(G - C) / (G + C)], has two nodes that are located at the origin and the terminus of replication. The GC-skew of MmymySC reveals the putative position of the terminus of replication, but it does not follow a normal pattern at the expected oriC locus (Fig. 1). Opposite to the putative terminus of replication in MmymySC, the dnaA and dnaN genes are located, which indicates that oriC may be located in the vicinity of these genes. This hypothesis is supported by C. Lartigue, P.S. Pugnet, and A. Blanchard (unpubl.), who recently produced a DNA plasmid with the dnaA region, obtained from the genome sequence of MmymySC, and have shown that it can replicate in MmymySC. Thus, OriC seems to be present in the dnaA region. Anomalous patterns of the GC-skew have previously been shown for Yersinia pestis (Parkhill et al. 2001), in which ISs were found at the borders of the three regions with deviating patterns. Because the ISelements in MmymySC are distributed throughout the whole genome, it is difficult to determine their influence on the GC-skew pattern. Notably, the GC-skew for MmymySC follows the direction of transcription; that is, the transcripts are located on the strand with more Gs than Cs. A plausible explanation for a typical GC-skew pattern is that spontaneous deamination of cytosine to uracil or 5-methylcytosine to thymine is more frequent on the leading strand than on the lagging strand due to longer exposure of the leading strand as single-stranded DNA during replication (Grigoriev 1998; Lobry and Sueoka 2002). The fact that the GC-skew of MmymySC has an abnormal pattern may be due to recent rearrangements of the genome.
Transport and Biosynthesis
Mollicutes are known to have a restricted biosynthetic capacity. For instance, they lack a complete tricarboxylic acid cycle, have a scarce ability to synthesize amino acids, and are not able to synthesize purine and pyrimidine bases de novo. MmymySC has been shown to metabolize the exogenous sugars glucose, fructose, N-acetylglucosamine, glycerol, 2-oxobutyrate, and pyruvate at moderate concentrations and mannose and l-lactate at high concentrations (Abu-Groun et al. 1994). In contrast, it is not able to use maltose and trehalose. All genes of the phosphotransferase systems (PTSs) of glucose, fructose, and mannitol have been identified. The sugars transported into the cell by these systems are degraded by the enzymes of the Embden-Meyerhof-Parnas (EMP) pathway to pyruvate and subsequently to lactate and acetylcoenzyme A. The deoC gene is present, and it is encoding deoxyribose-5-phosphate aldolase, which connects the EMP pathway with the DNA metabolic pathway via 2-deoxyribose-5-phosphate and glyceraldehyde-3-phosphate. The oxidative branch of the pentose phosphate pathway is missing in MmymySC and in most other mollicutes except for Acholeplasma species (Pollack et al. 1997). In the nonoxidative branch, only transaldolase is missing, indicating an alternative route or enzyme for the conversion of sedoheptulose-7-phosphate and glyceraldehyde-3-phosphate to fructose-6-phosphate and erythrose-4-phosphate.
The biosynthesis of nucleotides in MmymySC follows either of two routes. First, adenine/hypoxanthine-guanine/uracil phosphoribosyltransferase catalyses the formation of nucleoside monophosphate from 5-phosphoribosyl-1-diphosphate (PRPP) and a nucleobase. However, because nucleobases are not synthesized de novo in mollicutes, exogenous nucleobases have to be transported into the cell. Second, nucleoside kinases generate nucleoside monophosphates by phosphorylation of nucleosides. In some mollicutes there is an interconversion of deoxynucleo-side monophosphates by nucleoside phosphotransferase, but it seems that MmymySC is lacking this actual gene. Experiments made by Wang et al. (2001) showed that MmymySC is capable of phosphorylation of all four deoxynucleosides by only two enzymes, thymidine kinase and an enzyme related to deoxyguanosine kinase.
In addition to the PTStransporters, eight complete ATP-binding cassette (ABC) transporters have been identified in MmymySC. The bioinformatics analysis indicates that these transporters are capable of transferring sugars, oligopeptides, spermidine and/or putrescine, phosphate, alkylphosphonate, glycerol, and a nonidentified solute across the plasma membrane. A unique feature of the spermidine/putrescine ABC-transporter system is that one of the permease components and the substrate-binding component are encoded by one gene (potCD) in MmymySC. These are normally encoded by two separate genes, potC and potD. The permease and substrate-binding domains of potCD are separated by ∼350 amino acids, and the signal peptide sequence of the potD genes is missing in the potD-like part of potCD. There are several additional PTSand ABC transport systems, although not all subunits have been identified. The missing components may be among the nonassigned hypothetical proteins.
Comparison to the Minimal Genome
A minimal gene set for cellular life has been postulated by comparing the genome sequences of M. genitalium and Haemophilus influenzae (Mushegian and Koonin 1996). Because these two species belong to different phyla, it was believed that their common genes would be essential for growth. A comparison of the gene set of the minimal genome to the MmymySC gene set showed that 11 out of 254 genes of the minimal genome are absent in MmymySC. Except for the genes encoding the heat-shock proteins GroEL and GroES, which are also missing in M. pulmonis and U. parvum, the genes coding for three hypothetical proteins (MG055, MG127, and MG143 in M. genitalium), the ribosomal protein S6 modification protein (MG012), the cytidine deaminase (MG052), the riboflavin kinase (MG145), the thymidylate synthase (MG227), the dihydrofolate reductase (MG228), and a histone-like protein (MG353) are absent.
In conclusion, MmymySC is the first bacterium that causes a severe disease in livestock and whose genome has been sequenced. Knowledge of the genome sequence of MmymySC will most likely facilitate the development of new vaccines, drugs, and diagnostic tools for CBPP. A plausible theory, as drawn from conclusions of the present work, is that the protein composition of the cell surface varies between different environmental conditions. Therefore, all combinations of the variable proteins are target candidates for vaccine development. In addition, substances that will inhibit the uptake of glycerol and production of the capsule are potential candidate drugs. Further analyses of the genome may reveal additional pathogenic mechanisms of MmymySC.
Because this is the first genome that has been sequenced in the Spiroplasma group of the mollicutes, it will serve as a good complement to the five previously published mollicute genomes for the study of the evolution of the mollicutes. The genome sequence reveals an ongoing process of large rearrangements of the genome, without any compulsions of preserving the direction of the transcripts.
METHODS
Construction of Random Libraries
The MmymySC type strain PG1T was grown in F medium (Bölske 1988). Genomic DNA was prepared and purified by proteinase K lysis and phenol/chloroform extraction. Five kinds of plasmid libraries were created. The A library was generated by nebulization, the B and C libraries by partial ApoI restriction, and the D and E libraries by partial Sau3AI restriction of genomic DNA. The size fractions were 0.8–.2 kbp for the A library, 2–4.5 kbp for the B and D libraries, and 4.5–9 kbp for the C and E libraries. The pUC18 plasmid was used as cloning vector for all five libraries. It was restricted with SmaI for the construction of the A library, EcoRI for the B and C libraries, and BamHI for the D and E libraries.
DNA Sequencing
Initially, shotgun sequencing was performed on all five plasmid libraries. The plasmid clones of the A library were prepared for DNA sequencing by PCR, and the plasmid clones of the four other libraries were prepared by purification of the plasmids with a plasmid preparation kit from MilliPore. Both ends of the plasmid inserts were sequenced with BigDye Terminator Cycle Sequencing Ready Reaction Kit (Perkin Elmer) or DYEnamic ET terminator cycle sequencing premix kit (Amersham Biosciences), and the sequencing reaction products were loaded on ABI PRISM 3700 (PE Applied Biosystems) and MegaBACE 1000 DNA sequencers (Molecular Dynamics).
In the directed sequencing phase, nonrepetitive gap sequences and sequences of poor quality were sequenced by using genomic DNA as template (Heiner et al. 1998). The high copy-number of the ISelements, the large sizes of several long repeats, and the high sequence similarities between their copies caused problems with the genome assembly. To solve these problems, two kinds of strategies were performed. First, each IScopy was isolated within plasmid clones or PCR amplicons and subsequently sequenced by primer-walking. Because incompletely extended primers in the PCR could hybridize to incorrect IScopies and cause false positives, the PCR procedures were thoroughly optimized with long extension times and high annealing temperatures. Second, repeats larger than the individual clones were sequenced by primer-walking on a number of clones, which contained several positions of genome polymorphisms, covering different parts of the repetitive regions. The number of long tandem repeats was determined by pulse-field gel electrophoresis (PFGE) of restriction fragments containing the entire repetitive region.
Genome Restriction Map
The MmymySC genome was mapped by two-dimensional PFGE of MluI and SmaI restricted fragments and one-dimensional PFGE of SalI, AatII, AviII, PvuI, and NcoI restricted fragments. This map and a previously published genome map of MmymySC (Pyle et al. 1990) were used for determination of the accuracy of the genome assembly.
Assembly and Genome Analysis
Basecalling, vector sequence elimination and assembly of the sequences were performed with PHRED (Ewing et al. 1998) and PHRAP (P. Green, University of Washington; http://www.phrap.org/). The assembly was visualized and edited in the CONSED program (Gordon et al. 1998). The genome sequence was analyzed and annotated with the aid of GENDB (Meyer et al. 2003), a flexible open source genome annotation system for prokaryote genomes. Open reading frames (ORFs) were predicted by using GLIMMER 2.0 (Salzberg et al. 1998) and searched for homology with sequences of the public databases with BLASTN and BLASTP (Altschul et al. 1990). Protein motifs were searched for in the Pfam database (Bateman et al. 1999) by using HMMER (S.R. Eddy; http://hmmer.wustl.edu/). Signal peptide sequences were predicted by SIGNALP (Nielsen et al. 1997), and putative transmembrane proteins were identified by TMHMM 2.0 (Krogh et al. 2001).
The tRNA genes were identified with tRNAscan-SE (Lowe and Eddy 1997). Codon usage was calculated by codonW (J. Peden, University of Nottingham; http://molbiol.ox.ac.uk/cu/). Intragenomic sequence similarity searches were performed by the graphical dotplot program Dotter (Sonnhammer and Durbin 1995).
Acknowledgments
We are grateful to Anna-Lena Andersson, Ulla Wrethagen, Sara Jungberg, Helena Rönning, Anna Westring, and Marianne Persson for valuable technical assistance; the Center for Genome Research at Bielefeld University for its bioinformatics support; and the research groups of Joachim Frey and Alain Blanchard for kindly providing us with unpublished data. This work has been supported financially by grants from the Swedish Foundation for Strategic Research and the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1673304.
Footnotes
[Supplemental material is available online at www.genome.org. The genome sequence data from this study have been submitted to EMBL under accession number BX293980. The home page of the genome of Mycoplasma mycoides subsp. mycoides SC can be found at http://gendb.genetik.uni-bielefeld.de/projects/mmymysc.html. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: J. Frey and A. Blanchard.]
References
- Abu-Groun, E.A., Taylor, R.R., Varsani, H., Wadher, B.J., Leach, R.H., and Miles, R.J. 1994. Biochemical diversity within the “Mycoplasma mycoides” cluster. Microbiology 140: 2033-2042. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. [DOI] [PubMed] [Google Scholar]
- Bateman, A., Birney, E., Durbin, R., Eddy, S.R., Finn, R.D., and Sonnhammer, E.L. 1999. Pfam 3.1: 1313 multiple alignments and profile HMMs match the majority of proteins. Nucleic Acids Res. 27: 260-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bölske, G. 1988. Survey of Mycoplasma infections in cell cultures and a comparison of detection methods. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 269: 331-340. [DOI] [PubMed] [Google Scholar]
- Buttery, S.H., Lloyd, L.C., and Titchen, D.A. 1976. Acute respiratory, circulatory and pathological changes in the calf after intravenous injections of the galactan from Mycoplasma mycoides subsp. mycoides. J. Med. Microbiol. 9: 379-391. [DOI] [PubMed] [Google Scholar]
- Chambaud, I., Heilig, R., Ferris, S., Barbe, V., Samson, D., Galisson, F., Moszer, I., Dybvig, K., Wroblewski, H., Viari, A., et al. 2001. The complete genome sequence of the murine respiratory pathogen Mycoplasma pulmonis. Nucleic Acids Res. 29: 2145-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using phred, I: Accuracy assessment. Genome Res. 8: 175-185. [DOI] [PubMed] [Google Scholar]
- Fraser, C.M., Gocayne, J.D., White, O., Adams, M.D., Clayton, R.A., Fleischmann, R.D., Bult, C.J., Kerlavage, A.R., Sutton, G., Kelley, J.M., et al. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270: 397-403. [DOI] [PubMed] [Google Scholar]
- Frey, J., Cheng, X., Kuhnert, P., and Nicolet, J. 1995. Identification and characterization of IS1296 in Mycoplasma mycoides subsp. mycoides SC and presence in related mycoplasmas. Gene 160: 95-100. [DOI] [PubMed] [Google Scholar]
- Glass, J.I., Lefkowitz, E.J., Glass, J.S., Heiner, C.R., Chen, E.Y., and Cassell, G.H. 2000. The complete sequence of the mucosal pathogen Ureaplasma urealyticum. Nature 407: 757-762. [DOI] [PubMed] [Google Scholar]
- Gordon, D., Abajian, C., and Green, P. 1998. Consed: A graphical tool for sequence finishing. Genome Res. 8: 195-202. [DOI] [PubMed] [Google Scholar]
- Gourlay, R.N. 1964. Antigenicity of Mycoplasma mycoides, I: Examination of body fluids from cases of contagious bovine pleuropneumonia. Res. Vet. Sci. 5: 473-482. [PubMed] [Google Scholar]
- Grieco, V., Boldini, M., Luini, M., Finazzi, M., Mandelli, G., and Scanziani, E. 2001. Pathological, immunohistochemical and bacteriological findings in kidneys of cattle with contagious bovine pleuropneumonia (CBPP). J. Comp. Pathol. 124: 95-101. [DOI] [PubMed] [Google Scholar]
- Grigoriev, A. 1998. Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 26: 2286-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiner, C.R., Hunkapiller, K.L., Chen, S.M., Glass, J.I., and Chen, E.Y. 1998. Sequencing multimegabase-template DNA with BigDye terminator chemistry. Genome Res. 8: 557-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelreich, R., Hilbert, H., Plagens, H., Pirkl, E., Li, B.C., and Herrmann, R. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 24: 4420-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson, K.-E., Heldtander, M.U., and Pettersson, B. 1998. Characterization of mycoplasmas by PCR and sequence analysis with universal 16S rDNA primers. Methods Mol. Biol. 104: 145-165. [DOI] [PubMed] [Google Scholar]
- Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E.L. 2001. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305: 567-580. [DOI] [PubMed] [Google Scholar]
- Lobry, J.R. and Sueoka, N. 2002. Asymmetric directional mutation pressures in bacteria. Genome Biol. 3: 0058.1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe, T.M. and Eddy, S.R. 1997. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, A.J., Miles, R.J., and Richards, L. 1995. The phagocytosis of mycoplasmas. J. Med. Microbiol. 43: 239-250. [DOI] [PubMed] [Google Scholar]
- Masiga, W.N., Windsor, R.S., and Read, W.C. 1972. A new mode of spread of contagious bovine pleuropneumonia? Vet. Rec. 90: 247-248. [DOI] [PubMed] [Google Scholar]
- Meyer, F., Goesmann, A., McHardy, A.C., Bartels, D., Bekel, T., Clausen, J., Kalinowski, J., Linke, B., Rupp, O., Giegerich, R., et al. 2003. GenDB: An open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 31: 2187-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushegian, A.R. and Koonin, E.V. 1996. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl. Acad. Sci. 93: 10268-10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas, R.A., Santini, F.G., Clark, K.M., Palmer, N.M., De Santis, P., and Bashiruddin, J.B. 1996. A comparison of serological tests and gross lung pathology for detecting contagious bovine pleuropneumonia in two groups of Italian cattle. Vet. Rec. 139: 89-93. [DOI] [PubMed] [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. 1997. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int. J. Neural Syst. 8: 581-599. [DOI] [PubMed] [Google Scholar]
- Oba, T., Andachi, Y., Muto, A., and Osawa, S. 1991. CGG: An unassigned or nonsense codon in Mycoplasma capricolum. Proc. Natl. Acad. Sci. 88: 921-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill, J., Wren, B.W., Thomson, N.R., Titball, R.W., Holden, M.T., Prentice, M.B., Sebaihia, M., James, K.D., Churcher, C., Mungall, K.L., et al. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413: 523-527. [DOI] [PubMed] [Google Scholar]
- Persson, A., Jacobsson, K., Frykberg, L., Johansson, K.-E., and Poumarat, F. 2002. Variable surface protein Vmm of Mycoplasma mycoides subsp. mycoides small colony type. J. Bacteriol. 184: 3712-3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson, B., Leitner, T., Ronaghi, M., Bölske, G., Uhlén, M., and Johansson, K.-E. 1996. Phylogeny of the Mycoplasma mycoides cluster as determined by sequence analysis of the 16S rRNA genes from the two rRNA operons. J. Bacteriol. 178: 4131-4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack, J.D., Williams, M.V., and McElhaney, R.N. 1997. The comparative metabolism of the mollicutes (Mycoplasmas): The utility for taxonomic classification and the relationship of putative gene annotation and phylogeny to enzymatic function in the smallest free-living cells. Crit. Rev. Microbiol. 23: 269-354. [DOI] [PubMed] [Google Scholar]
- Pyle, L.E., Taylor, T., and Finch, L.R. 1990. Genomic maps of some strains within the Mycoplasma mycoides cluster. J. Bacteriol. 172: 7265-7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, J.A., Stemke, G.W., Davis Jr., J.W., Harasawa, R., Thirkell, D., Kong, F., Shepard, M.C., and Ford, D.K. 2002. Proposal of Ureaplasma parvum sp. nov. and emended description of Ureaplasma urealyticum (Shepard et al. 1974) Robertson et al. 2001. Int. J. Syst. Evol. Microbiol. 52: 587-597. [DOI] [PubMed] [Google Scholar]
- Rosengarten, R. and Wise, K.S. 1990. Phenotypic switching in mycoplasmas: Phase variation of diverse surface lipoproteins. Science 247: 315-318. [DOI] [PubMed] [Google Scholar]
- Salzberg, S.L., Delcher, A.L., Kasif, S., and White, O. 1998. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 26: 544-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, Y., Ishikawa, J., Yamashita, A., Oshima, K., Kenri, T., Furuya, K., Yoshino, C., Horino, A., Shiba, T., Sasaki, T., et al. 2002. The complete genomic sequence of Mycoplasma penetrans, an intracellular bacterial pathogen in humans. Nucleic Acids Res. 30: 5293-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanziani, E., Paltrinieri, S., Boldini, M., Grieco, V., Monaci, C., Giusti, A.M., and Mandelli, G. 1997. Histological and immunohistochemical findings in thoracic lymph nodes of cattle with contagious bovine pleuropneumonia. J. Comp. Pathol. 117: 127-136. [DOI] [PubMed] [Google Scholar]
- Sonnhammer, E.L. and Durbin, R. 1995. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167: GC1-10. [DOI] [PubMed] [Google Scholar]
- Stradaioli, G., Sylla, L., Mazzarelli, F., Zelli, R., Rawadi, G., and Monaci, M. 1999. Mycoplasma mycoides subsp. mycoides SC identification by PCR in sperm of seminal vesiculitis-affected bulls. Vet. Res. 30: 457-466. [PubMed] [Google Scholar]
- Vilei, E.M. and Frey, J. 2001. Genetic and biochemical characterization of glycerol uptake in Mycoplasma mycoides subsp. mycoides SC: Its impact on H2O2 production and virulence. Clin. Diagn. Lab. Immunol. 8: 85-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilei, E.M., Nicolet, J., and Frey, J. 1999. IS1634, a novel insertion element creating long, variable-length direct repeats which is specific for Mycoplasma mycoides subsp. mycoides small-colony type. J. Bacteriol. 181: 1319-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Westberg, J., Bölske, G., and Eriksson, S. 2001. Novel deoxynucleoside-phosphorylating enzymes in mycoplasmas: Evidence for efficient utilization of deoxynucleosides. Mol. Microbiol. 42: 1065-1073. [DOI] [PubMed] [Google Scholar]
- Weisburg, W.G., Tully, J.G., Rose, D.L., Petzel, J.P., Oyaizu, H., Yang, D., Mandelco, L., Sechrest, J., Lawrence, T.G., Van Etten, J., et al. 1989. A phylogenetic analysis of the mycoplasmas: Basis for their classification. J. Bacteriol. 171: 6455-6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westberg, J., Persson, A., Pettersson, B., Uhlén, M., and Johansson, K.-E. 2002. ISMmy1, a novel insertion sequence of Mycoplasma mycoides subsp. mycoides small colony type. FEMS Microbiol. Lett. 208: 207-213. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.oie.int/; Office International des Epizooties (World Organization for Animal Health) home page.
- http://www.phrap.org/; P. Green, University of Washington.
- http://hmmer.wustl.edu/; S.R. Eddy (2001) HMMER: Profile hidden Markov models for biological sequence analysis.
- http://molbiol.ox.ac.uk/cu/; J. Peden, University of Nottingham.
- http://gendb.genetik.uni-bielefeld.de/projects/mmymysc.html; Mycoplasma mycoides subsp. mycoides SC genome home page.