Background: The regulation of multi-functional DYNLL1 is poorly understood.
Results: ASCIZ activates Dynll1 gene expression and is inhibited by DYNLL1 binding to its transcription activation domain.
Conclusion: ASCIZ plays a key role in the auto-regulation of DYNLL1 levels.
Significance: This is the first case where a gene product directly inhibits its main transcriptional activator while bound at its own promoter.
Keywords: Cell Biology, DNA Damage, Dynein, Gene Expression, siRNA, ASCIZ, DYNLL1, TQT Motif
Abstract
The highly conserved DYNLL1 (LC8) protein was originally discovered as a light chain of the dynein motor complex, but is increasingly emerging as a sequence-specific regulator of protein dimerization with hundreds of targets and wide-ranging cellular functions. Despite its important roles, DYNLL1's own regulation remains poorly understood. Here we identify ASCIZ (ATMIN/ZNF822), an essential Zn2+ finger protein with dual roles in the DNA base damage response and as a developmental transcription factor, as a conserved regulator of Dynll1 gene expression. DYNLL1 levels are reduced by ∼10-fold in the absence of ASCIZ in human, mouse and chicken cells. ASCIZ binds directly to the Dynll1 promoter and regulates its activity in a Zn2+ finger-dependent manner. DYNLL1 protein in turn interacts with ten binding sites in the ASCIZ transcription activation domain, and high DYNLL1 levels inhibit the transcriptional activity of ASCIZ. In addition, DYNLL1 was also required for DNA damage-induced ASCIZ focus formation. The dual ability of ASCIZ to activate Dynll1 gene expression and to sense free DYNLL1 protein levels enables a simple dynamic feedback loop to adjust DYNLL1 levels to cellular needs. The ASCIZ-DYNLL1 feedback loop represents a novel mechanism for auto-regulation of gene expression, where the gene product directly inhibits the transcriptional activator while bound at its own promoter.
Introduction
Dynein light chain 1 (DYNLL1,3 also known as LC8) was originally identified as an associated light chain of the dynein motor complex (1), where it is believed to be required for proper folding and dimerization of the cargo-binding dynein intermediate chain (2–4). However, it is now clear that DYNLL1 has crucial roles beyond dynein assembly as a multifunctional regulator of protein dimerization with hundreds of likely targets (5–7). DYNLL1 itself forms a stable homodimer, where the dimer interface gives rise to two symmetric ligand-binding grooves that preferentially bind intrinsically disordered 7–10 residue target sequences anchored by a TQT motif (5, 7). DYNLL1 targets include transcription factors, DNA damage response proteins, as well as regulators of apoptosis, synaptic transmission, cell migration, nitric oxide signaling, and several other processes (5–9). Consistent with its wide-ranging functions, DYNLL1 is highly conserved throughout evolution, with just five amino acid substitutions over 89 residues between the human and Drosophila proteins, and its absence leads to embryonic lethality in Drosophila (10, 11), the only organism so far where a loss-of-function allele has been analyzed. Despite its important cellular functions, it remains largely unclear how DYNLL1 itself is regulated.
The ATM substrate Chk2-interacting Zn2+ finger protein (ASCIZ; also known as ATMIN and ZNF822) was recently identified as a protein with dual functions in the DNA damage response and embryonic development. ASCIZ forms DNA damage-induced nuclear foci, and is required for cell survival, specifically in response to lesions repaired by the base excision repair pathway (12–15), and in this context may function as a tumor suppressor of peripheral B cell lymphomas (16, 17). However, complete absence of ASCIZ causes late-embryonic lethality in mice (14, 15) with a range of organ development defects, most notably complete absence of lungs (14), that are most likely due to DNA damage-independent functions as a Zn2+ finger (ZnF) transcription factor (14, 18).
Here, we have sought to better understand the transcriptional roles of ASCIZ and found that it functions as a phylogenetically conserved transcriptional activator of Dynll1 gene expression as well as a sensor of DYNLL1 protein levels in a novel feedback mechanism for auto-regulated gene expression.
EXPERIMENTAL PROCEDURES
Animals and Cells
Embryonic tissues and MEFs (14), human U2OS and GFP-ASCIZ-667-overexpressing U2OS cells (12), and chicken DT40 cells (13) were prepared and cultured as described. MEFs and human cells were transfected with the indicated plasmids using FuGENE 6 (Promega). For retroviral complementation, the indicated constructs were cloned into MigR1 (19), virus was produced using the Phoenix packaging cell line, and virus-containing medium was used to infect early passage MEFs with centrifugation at 1100 × g for 90 min at room temperature.
RNA Expression Array Analysis
MEFs were prepared from three E12.5 embryos per genotype, incubated for 48 h in a 10-cm dish of Dulbecco's modified Eagle's medium with 10% fetal bovine serum with a medium change after 24 h, and then transferred to a 15-cm dish for 72 h. Total RNA was extracted using an RNeasy mini kit (Qiagen). Expression profiling using standard conditions and Illumina Mouse WG-6 v2 arrays was performed by the Australian Genome Research Facility (Melbourne). The microarray data have been deposited in NCBI Gene Expression Omnibus (accession number GSE30417).
Blots, Immunoprecipitations, cDNAs, Probes, and Antibodies
Northern blots using total RNA and immunoblots of the indicated protein extracts were performed as described (14). Probes were amplified by PCR from mouse genomic DNA, cloned into pGEM-T and agarose gel-purified before labeling. Northern blot signals were quantified using ImageQuant software. cDNAs were cloned in the indicated vectors, mutagenized using standard procedures, and validated by DNA sequence analysis before use in experiments. For immunoprecipitation of endogenous proteins, confluent U2OS cells from 2 × 15 cm dishes were lysed in ∼2.4 ml of IP buffer (150 mm NaCl, 50 mm Tris pH 7.4, 0.1% Triton X-100), soluble extracts precleared using protein A-Sepharose (GE Healthcare) and 2 μg of normal rabbit IgG (DAKO), and equal aliquots were incubated with 3 μg of anti-ASCIZ, or 3 μg of normal rabbit IgG and 50 μl of protein A-Sepharose for 2 h at 4 °C, before elution by boiling into 60 μl of SDS-loading buffer for 10 min. 10 μl of lysate and 25 μl of eluted material were analyzed by SDS-PAGE and immunoblotting, using protein A-HRP for detection. For immunoprecipitations from overexpressing cells, cells from a 10-cm dish were lysed in ∼600 μl of IP buffer (150 mm NaCl, 50 mm Tris pH 7.4, 0.1% Triton X-100), soluble extracts pre-cleared using protein-G-Sepharose (GE Healthcare) and 2 μg of normal rabbit IgG (DAKO), and equal aliquots were incubated with 2 μg of anti-ASCIZ (12) or anti-FLAG M2 (F 1804, Sigma) and 30 μl of protein G-Sepharose for 2 h at 4 °C, before elution by boiling into 50 μl of SDS-loading buffer for 10 min. Routinely, 10 μl of lysate and 20 μl of eluted material were analyzed by SDS-PAGE and immunoblotting. The following antibodies were used for immunoblots and immunoprecipitations: Actin (MAB1501, Millipore; 1/4000), ASCIZ (12) (available from Millipore; 1/4000), DYNLL1/2 (sc-13969, Santa Cruz Biotechnology; 1/1000), GFP (sc-8334, Santa Cruz Biotechnology; 1/1000); normal rabbit IgG (DAKO).
Transcription Reporter Assays
To measure TAD activity, ASCIZ fragments were cloned into the pCDNA3-Gal4 DNA binding domain (DBD) vector (20), and human U2OS cells were co-transfected with the pFR-Luc plasmid (Stratagene) for expression of the firely luciferase reporter gene under control of five GAL4 binding elements. To measure effects of ASCIZ on the Dynll1 promoter, ∼2 kbp of the human Dynll1 gene (upstream of the transcription start site (21)) were cloned into pGL3 (Promega) upstream of the firefly luciferase gene for transfection of primary MEFs. Cells were also co-transfected with pRL-CMV containing Renilla luciferase under control of the constitutively active CMV promoter for normalization of firefly/Renilla luciferase ratios. 24 h after transfection, cells were transferred to Nunclon Δ Surface 96-well plates and incubated overnight before determining reporter gene activities using the dual-luciferase reporter assay kit (Promega) and a Polarstar Optima (BMG Labtechnologies) instrument. Statistical analyses were performed using the unpaired student's t test.
Chromatin Immunoprecipitations
Cells were chemically crosslinked with 1% formaldehyde for 10 min. The reaction was quenched with 125 mm glycine, and the cells were rinsed twice with PBS. 5 × 106 cells were resuspended in lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris pH 8.1, 1× protease inhibitors (Sigma)) and sonicated using a temperature cooled (4 °C) Diagenode UCD-200 Bioruptor on high setting for five 30-s pulses with 30-s intervals between each pulse to produce chromatin fragments ranging from 200 bp to 600 bp. The chromatin was cleared by centrifugation at 16,000 × g for 10 min. Equal amounts of chromatin (equivalent to ∼1 × 106 cells) were diluted 10-fold with dilution buffer (167 mm NaCl, 0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl pH 8.1, 1× protease inhibitors). After removing a control aliquot, chromatin was incubated with or without (no antibody control) antibodies overnight at 4 °C with rotation (1 μg anti-DYNLL1/2 (sc-13969, Santa Cruz Biotechnology), 5 μg of anti-ASCIZ (12), 1–5 μg of normal rabbit IgG (12–370, Millipore). Immune complexes were collected with 20 μl of protein A dynabeads (Invitrogen) with rotation at 4 °C for 4 h. The dynabeads were washed sequentially for 5 min with a low salt buffer (150 mm NaCl, 0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl pH 8.1), high salt buffer (500 mm NaCl, 0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl pH 8.1), LiCl buffer (250 mm LiCl, 1% IGEPAL, 1% deoxycholic acid, 1 mm EDTA, 10 mm Tris pH 8.1) and two washes with TE buffer. Immunoprecipitated DNA and control samples were eluted and crosslinks reversed with 150 μl elution buffer (20 mm Tris-HCl pH 7.5, 5 mm EDTA, 50 mm NaCl, 1% SDS) and 2.5 μl 20 mg/ml proteinase K with shaking at 65 °C overnight. The immunoprecipitated DNA samples and control DNA samples were purified with a MinElute Reaction Cleanup kit (Qiagen).
ChIP assays were evaluated by real-time PCR using Brilliant II SYBR Green (Agilent) in a MX3000P Stratagene detection system with MxPro software (Stratagene). 2–3 μl of undiluted or of 1:15 diluted immunoprecipitated and control DNA samples were assessed in duplicate reactions. PCR primer sequences were designed to amplify fragments less than 150 bp. The following primer pairs were used: Dynll1 (−175 bp), 5′-gagcccggtctcaccgtgga-3′ and 5′-gatgcgccacggcttcggta-3′; Dynll1 (−613 bp), 5′-cgggggccccactgaggta-3′ and 5′-gaaatgggcggcacctgggg-3′; Dynll1 (−1216 bp), 5′-tggggaaatggctctgctggtg-3′ and 5′-aaggcacaggccaccaaacct-3′; Dynll1 (−2247 bp), 5′-gccaggcagtggggacctca-3′ and 5′-tccgaggaagcagcctcacct-3′; Gapdh (−768 bp), 5′-cgaagaacaacgaggagaagatc-3′ and 5′-cgaacctctccccattattgaa-3′.
Relative enrichment was determined by the 2−ΔCT method using the control DNA to normalize. Average relative enrichment at specific regions was determined from 3–4 independent ChIP assays. Unpaired Student's t-tests were used for statistical evaluation.
siRNA Treatments and Pilot siRNA High-throughput Screen
Human U2OS cells were treated with non-targeting control siRNA (Dharmacon), Asciz siRNA (12) or Dynll1 SMARTpool and four individual siRNAs (Dharmacon siGenome) and Dharmafect 3 or Dharmafect-Duo according to the supplier's recommendations.
The siRNA screen of the SMARTpool drug targets sub-library (Dharmacon) was performed in a 384-well plate format at the Victorian Centre for Functional Genomics. 850 stably GFP-ASCIZ-667 expressing U2OS cells (12) per well were reverse transfected with 0.02 μl of Dharmafect 3 reagent. Each plate contained ∼320 SMARTpools plus multiple non-targeting as well as positive control siRNAs. 72 h after transfection, cells were treated with 0.025% MMS for 4 h, fixed and automatically imaged on a Cellomics ArrayScan VTi high-throughput microscope and scored by the “trained” Cellomics Spot Detector application. The pilot screen was performed on duplicate plates housed in a Liconics tower incubator to reduce edge effects. Fig. 3A shows an original Cellomics image of the screen for siDynll1 and negative control wells. Details of the whole genome siRNA screen will be described elsewhere upon its completion.
FIGURE 3.
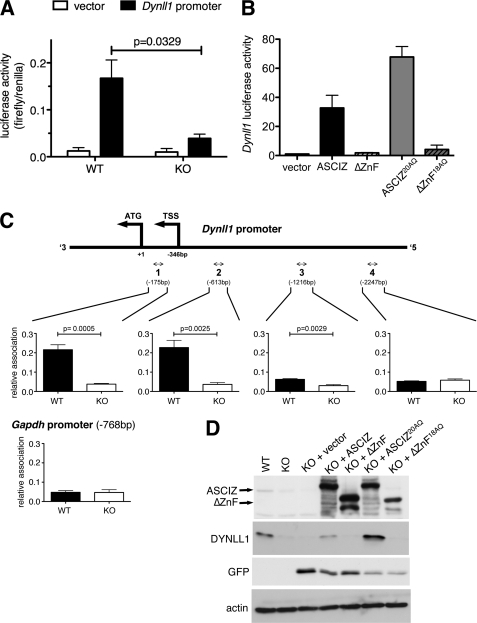
ASCIZ regulates Dynll1 as a ZnF transcription factor. A, firefly luciferase reporter activity of the human Dynll1 promoter (solid bars) or pGL3 empty vector (open bars) transiently transfected into early-passage primary WT and Asciz−/− MEFs (three independent embryos per genotype). Values are means ± S.E. relative to Renilla luciferase as transfection control. B, luciferase reporter assays as in A using an immortalized Asciz−/− MEF cell line transiently co-transfected with GFP-fusion constructs of the human 823-residue ASCIZ isoform, a ZnF-deleted ASCIZ fragment, 823-residue ASCIZ with all 20 SQ/TQ motifs mutated to AQ, or a ZnF-deleted and AQ-mutated fragment, as well as Dynll1-firefly luciferase and Renilla luciferase vectors. Values are means ± S.E. normalized to the empty GFP-only vector control (n = 3–4 independent experiments). C, ChIP assays using the ASCIZ antibody and real-time quantitative PCR analyses of the indicated regions (relative to the ATG start codon) of the Dynll1 promoter, and the corresponding Gapdh control from the same precipitations. Values are means ± S.E. of four different sets of WT and Asciz−/− littermate early-passage MEFs. TSS, transcription start site. D, immunoblot analysis of early-passage WT and Asciz−/− littermate primary MEFs, and the same Asciz−/− MEFs after retroviral complementation with the empty vector, the human 823-residue ASCIZ isoform, a ZnF-deleted ASCIZ fragment, 823-residue ASCIZ with all 20 SQ/TQ motifs mutated to AQ, or a ZnF-deleted and AQ-mutated fragment. Virus-encoded GFP serves as an infection control.
Microscopy
For manual fluorescence microscopy analyses, cells were fixed in 2% paraformaldehyde after treatment with 0.025% MMS for 4 h, washed in PBS, and directly imaged for GFP fluorescence (supplemental Fig. S2). For immunofluorescence (Fig. 4G), fixed cells were permeabilized with ice-cold methanol for 1 min, and stained with 1:250 anti-FLAG M2 for 1–2 h and 1:500 anti-mouse IgG-Alexa594 (Invitrogen) for 60 min, and counterstained with 1 μg/ml DAPI. Micrographs were taken on a Zeiss Axiovert-25 microscope with a Zeiss AxioCam MRc using a 40× objective, and analyzed using AxioVision software.
FIGURE 4.
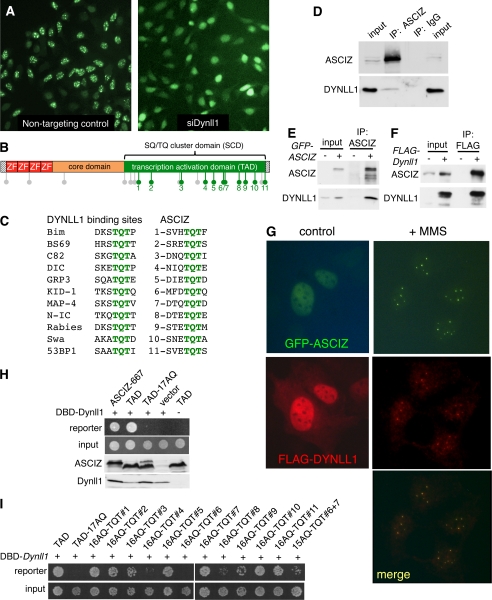
Direct interaction between ASCIZ and DYNLL1 proteins. A, fluorescence micrographs GFP-ASCIZ-667 localization in human U2OS cells treated with non-targeting control siRNA or Dynll1 siRNA following 4-h treatment with 0.025% MMS. Note that GFP-ASCIZ is retained in the nucleus following DYNLL1 depletion, but fails to form DNA damage-induced nuclear foci. B, schematic diagram of ASCIZ domain topology. ZF, Zn2+ finger; lollipops indicate SQ/TQ (gray) or TQT motifs (green). C, compilation of arbitrarily selected bona fide DYNLL1 binding sites (5–7) (left panel) and eleven ASCIZ TQT motifs numbered in sequential order (compare panel B). D, co-immunoprecipitation (IP) of endogenous ASCIZ and DYNLL1 with ASCIZ but not IgG control antibodies from human U2OS cells. Input represents ∼12.5% of lysate relative to loading of IP lanes. E, co-immunoprecipitation of endogenous DYNLL1 with the ASCIZ antibody from parental (−) or GFP-ASCIZ overexpressing (+) U2OS cells, probed with ASCIZ or DYNLL1 antibodies. Input represents ∼5% of lysate relative to IP lanes. F, co-immunoprecipitation of stably over-expressed GFP-ASCIZ with the FLAG antibody in transiently FLAG-Dynll1 transfected (+) or control (−) cells. Input represents ∼10% of lysate relative to IP lanes. G, fluorescence micrographs of GFP-ASCIZ-667 and FLAG-DYNLL1 in untreated or MMS-treated human U2OS cells; a merged image of the MMS-treated cells is shown underneath. H, yeast two-hybrid assays of the indicated ASCIZ fragments fused to the Gal4-activation domain with or without Dynll1 fused to the Gal4-DNA binding domain vector on selective plates for reporter genes (top) and input control plates (bottom). Western blots of the corresponding cell lysates with ASCIZ and DYNLL1 antibodies are shown below. I, two-hybrid assays similar to panel G, using the ASCIZ TAD fragment as a positive control, the TAD-17AQ mutant as a negative control, and indicated TQT motifs individually restored in the context of TAD-17AQ. 15AQ-TQT#6 + 7 denotes the TAD-17AQ fragment restored with the overlapping TQTQT motifs at positions 6 and 7.
Yeast Two-hybrid Assays
PJ69–4A yeast cells were co-transformed with DYNLL1 in the Gal4-DNA binding domain vector pGBT9 and the indicated ASCIZ fragments in the Gal4-activation domain vector pGADGH, or relevant empty vector controls. Co-transformed colonies were selected on plates lacking tryptophan and leucine. Equal aliquots were plated on minus-tryptophan/minus-leucine control plates, as well as high-stringency reporter plates also lacking adenine and histidine with 2 mm 3-amino-1,2,4-triazole as described (22).
RESULTS
ASCIZ Is a Conserved Regulator of DYNLL1 Expression
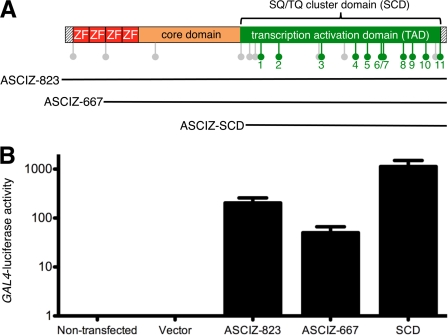
Human ASCIZ contains either 823 amino acid residues with four N-terminal ZnFs or 667 residues with two ZnFs, depending on alternative splicing, and an extended C-terminal SQ/TQ cluster domain with 17 potential ATM/ATR phosphorylation site motifs (Fig. 1A). ASCIZ can activate transcription when tethered to promoters of reporter genes, and in yeast one-hybrid assays, the C-terminal ASCIZ SQ/TQ-cluster-domain was sufficient for reporter activation (14). Similar results were obtained with Gal4DBD-ASCIZ fusion proteins in GAL4-promoter luciferase reporter assays in human cells (Fig. 1B), indicating that the SQ/TQ-cluster functions as the ASCIZ transcription activation domain (TAD).
FIGURE 1.
Role of the ASCIZ SQ/TQ cluster domain (SCD) as a transcription activation domain (TAD) in human cells. A, top, schematic diagram of ASCIZ domain topology. ZF, Zn2+ finger; lollipops indicate SQ/TQ (gray) or TQT motifs (green). Bottom, range of the ASCIZ fragments used for reporter assays; ASCIZ-823 and ASCIZ-667 are the two major human splice variants (NCBI AceView). The following residues were changed to alanine in AQ mutants: 17AQ-421, 441, 449, 459, 493, 577, 585, 622, 649, 672, 702, 704, 740, 765, 785, 804, 811; 18AQ, 17 AQ with additional alanine substitution at 257; 20AQ, 18AQ with additional alanine substitutions at 55 and 147. B, dual luciferase reporter activity (firefly/Renilla ratio) of the indicated fragments cloned into pCDNA3-Gal4DBD in human U2OS cells co-transfected with pFR-Luc and pRL-CMV (100:1 ratio of firefly:Renilla luciferase vectors). Data are means ± S.E. of three independent experiments.
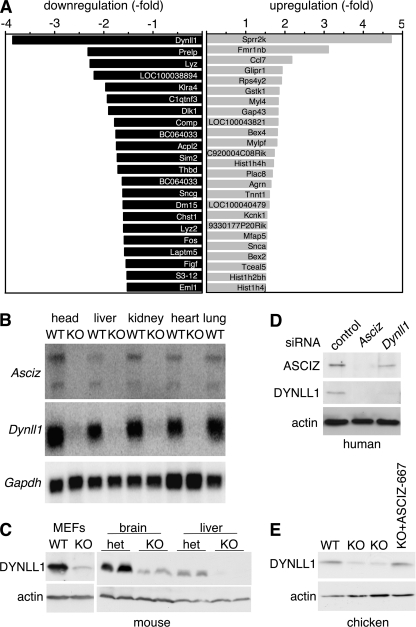
To identify potential transcriptional targets of ASCIZ, we performed an RNA expression array analysis of single passage primary murine embryonic fibroblasts (MEFs) from three independent wild type (WT) and Asciz−/− embryos. Only 22 genes were found to be down-regulated and 25 genes were up-regulated by >1.5-fold (and p < 0.05) in Asciz−/− versus WT MEFs (Fig. 2A), and for selected genes changes were confirmed by Northern blot analysis (supplemental Fig. S1). The most clearly down-regulated gene in ASCIZ-deficient cells was Dynll1 (Fig. 2A), which was confirmed by Northern blots demonstrating 9–15-fold lower Dynll1 mRNA levels in all tested tissues of mutant embryos compared with the WT (Fig. 2B and supplemental Fig. S1). A corresponding dramatic reduction of DYNLL1 protein levels in the absence of ASCIZ was confirmed by Western blot for mouse cells and tissues, as well as human and chicken cells (Fig. 2, C–E). Thus, ASCIZ has a conserved function as a critical regulator of Dynll1 gene expression.
FIGURE 2.
ASCIZ is required for DYNLL1 expression. A, mRNA expression array of single passage MEFs (3 independent embryos per genotype). Only genes whose levels are changed by >1.5-fold with p < 0.05 between WT and ASCIZ-deficient cells are shown. B, Northern blot analysis of the indicated tissues of E14.5 WT and Asciz−/− littermate embryos. C, immunoblot analysis of early-passage primary MEFs of WT and Asciz−/− littermates (left), and whole brain and fetal liver extracts from two E16.5 heterozygous and Asciz−/− littermates each. D, immunoblot analysis of ASCIZ and DYNLL1 levels in Asciz or Dynll1 siRNA-treated human U2OS cells. E, immunoblot analysis of WT and two independent Asciz−/− chicken DT40 clones, and one of the KO clones complemented with the human 667-residue ASCIZ isoform.
ASCIZ Regulates Dynll1 Expression as a ZnF Transcription Factor
To investigate how ASCIZ regulates Dynll1 expression, we first performed luciferase reporter assays. In transiently transfected ASCIZ-deficient primary MEFs, the human Dynll1 promoter was significantly less active than in WT controls (Fig. 3A). In immortalized ASCIZ KO MEFs, Dynll1 promoter activity was increased by more than 30-fold over basal levels upon complementation with the long form of human ASCIZ, but not in similar complementation assays using a ZnF-deleted construct (Fig. 3B). These results indicate that ASCIZ regulates Dynll1 via its promoter and in a ZnF-dependent manner.
To extend this finding, we performed chromatin-immunoprecipitation (ChIP) assays of ASCIZ in untransfected primary MEFs. In these experiments, primary MEFs from ASCIZ-null littermates were used as an antibody specificity control. Only background binding with equal signals for the WT and KO was observed at the Gapdh promoter as a negative control (Fig. 3C). In contrast, two different regions located close to the transcription start site of the endogenous Dynll1 promoter could be efficiently precipitated with the ASCIZ antibody in WT cells at considerately higher levels and in a statistically highly significant manner compared with ASCIZ-deficient control cells (Fig. 3C, regions 1 and 2). The specific ASCIZ ChIP signal was considerably diminished at ∼970 bp distance from the transcription start site (Fig. 3C, region 3) and was reduced to background levels at ∼1900bp away (Fig. 3C, region 4). These data indicate that ASCIZ is bound to the Dynll1 promoter under physiological conditions.
We next tested if the reduced Dynll1 expression in the absence of ASCIZ was reversible. Near-normal DYNLL1 protein levels could be restored in primary ASCIZ-deficient MEFs by retroviral complementation with the 823-residue splice variant of human ASCIZ (Fig. 3D), and similar results were observed in ASCIZ-deleted chicken cells complemented with the ZnF-containing human ASCIZ-667 isoform (Fig. 2E). Importantly, similar to the results observed in Dynll1-promoter luciferase reporter assays (Fig. 3B), a truncated ASCIZ version lacking the ZnF-domain was unable to restore endogenous DYNLL1 expression in ASCIZ-deficient MEFs (Fig. 3D). These data, together with our results that endogenous ASCIZ associates with the endogenous Dynll1 promoter (Fig. 3C) and that its absence leads to ∼10-fold reduced Dynll1 mRNA levels in tissues (Fig. 2B), strongly indicate that ASCIZ activates Dynll1 expression as a ZnF transcription factor.
Direct Interaction between ASCIZ and DYNLL1 Proteins
In parallel with the transcription analyses, we performed independent pilot experiments for a microscopy-based high-throughput siRNA screen for modulators of DNA damage-induced ASCIZ focus formation. Strikingly, DYNLL1 was also the strongest hit in the pilot sample comprising the Dharmacon SMARTpool drug targets sublibrary, where its knockdown completely prevented methyl-methane sulfonate (MMS)-induced ASCIZ focus formation (Fig. 4A and supplemental Fig. S2, A and B). The identification of DYNLL1 as the top hit in unrelated screens for targets as well as regulators of ASCIZ prompted us to further investigate the dual relationship between the two proteins. DYNLL1 regulates its numerous targets by preferentially binding to TQT motifs (5, 7). Remarkably, ASCIZ itself contains eleven such TQT motifs within its TAD (Fig. 4, B and C), all of which are conserved in their exact positions between human, mouse and zebrafish ASCIZ (supplemental Fig. S2C), suggesting that DYNLL1 and ASCIZ may directly interact with each other at the protein level.
To test if ASCIZ and DYNLL1 proteins interact with each other, we performed co-immunoprecipitation assays. In human U2OS cells, endogenous ASCIZ was efficiently precipitated with our ASCIZ antibody (Fig. 4D). Interestingly, a small but readily detectable amount of endogenous DYNLL1 could be co-precipitated with the ASCIZ antibody but not with a negative control IgG fraction (Fig. 4D). This interaction was further confirmed in a GFP-ASCIZ-overexpressing cell line where substantially more endogenous DYNLL1 could be co-immunoprecipitated with the ASCIZ antibody compared with the parental cell line (Fig. 4E). In reciprocal experiments, GFP-ASCIZ was readily co-immunoprecipitated with a FLAG antibody following transient transfection with FLAG-Dynll1, but not in the untransfected control (Fig. 4F). Importantly, FLAG-DYNLL1 partially redistributed to DNA damage-induced nuclear foci in a pattern similar to GFP-ASCIZ foci upon MMS treatment (Fig. 4G), consistent with a stable cellular interaction between the two proteins.
The ASCIZ-DYNLL1 interaction was further analyzed in yeast two-hybrid assays. Co-expression of DYNLL1 and the 667-residue ASCIZ isoform in the relevant vectors strongly activated reporter genes under high-stringency conditions, and the TQT-rich ASCIZ TAD was sufficient for this interaction (Fig. 4H). As DYNLL1 can sometimes bind to imperfect TQT sites, including SQ motifs (23–25), we mutated all eleven TQTs (green lollipops in Figs. 1A and 4B; see legend to Fig. 1 for amino acid position numbers) as well as six additional TQ and SQ motifs (gray lollipops in Figs. 1A and 4B) in the TAD to AQ. This 17AQ-mutated TAD fragment completely abrogated the two-hybrid interaction with DYNLL1 despite expression levels comparable to the corresponding WT construct (Fig. 4H). To determine how many ASCIZ TQT motifs are functional DYNLL1-binding sites, each one was individually restored within the 17AQ-TAD fragment in a series of single-TQT add-back constructs. Except for TQT motif #6, which actually forms an overlapping TQTQT motif with TQT#7 (Fig. 4C), each of the single-TQT motifs was sufficient for two-hybrid interaction with DYNLL1 (Fig. 4I). Thus, these data indicate that the ASCIZ TAD may be able to bind ten DYNLL1 molecules simultaneously.
DYNLL1 Binding Inhibits ASCIZ Transcriptional Activity
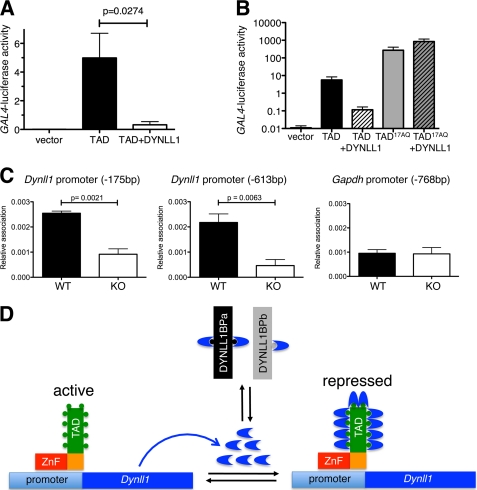
Why would ASCIZ need so many conserved DYNLL1-binding sites? Given that all TQT motifs are located within the ASCIZ TAD, we hypothesized that DYNLL1 binding may be part of an auto-regulatory feedback loop. Supporting this hypothesis, the transcriptional activity of the ASCIZ TAD in GAL4-promoter luciferase reporter assays was reduced by >10-fold when Dynll1 was simultaneously overexpressed (Fig. 5A). Interestingly, the 17AQ-TAD that does not bind DYNLL1 efficiently (Fig. 4H, I) had >10-fold higher activity in these GAL4-reporter assays compared with the WT sequence, and 17AQ-TAD activity was also completely resistant to feedback inhibition by co-overexpressed DYNLL1 (Fig. 5B). Similarly, complementation of Asciz−/− MEFs with the 20AQ-mutated long-form of ASCIZ restored Dynll1-promoter reporter activity (Fig. 3B) as well as endogenous DYNLL1 protein to considerably higher levels than the WT cDNA (Fig. 3D, compare lanes 4 and 6). Finally, we also tested if endogenous DYNLL1 protein was able to associate with its own promoter in an ASCIZ-dependent manner. Similar to ASCIZ ChIP assays (Fig. 3C), only low background levels of the Gapdh promoter as a negative control could be immunoprecipitated with the DYNLL1 antibody (Fig. 5C). Interestingly, two separate regions of the endogenous Dynll1 promoter could be precipitated with the DYNLL1 antibody at significantly higher levels in WT compared with ASCIZ-deficient primary MEFs (Fig. 5C), indicating that DYNLL1 may bind to the ASCIZ TAD while bound at its own promoter (Fig. 5D).
FIGURE 5.
Autoregulation of DYNLL1 expression through feedback inhibition of the ASCIZ TAD. A, relative transcriptional activity of the isolated ASCIZ TAD tethered to the GAL4-luciferase reporter via the yeast Gal4-DNA binding domain in the absence or presence of overexpressed DYNLL1 in human U2OS cells; empty vector control is pCDNA3-Gal4DBD. Values are means ± S.E. of five independent experiments. B, similar experiments to panel A, also including the 17AQ-mutated ASCIZ TAD; n = 3 independent experiments. C, ChIP using the DYNLL1 antibody and quantitative real-time PCR analysis of the indicated regions of the Dynll1 or Gapdh promoters. Values are means ± S.E. of three to four different sets of WT and Asciz−/− littermate early-passage MEFs. D, proposed model of ASCIZ-dependent Dynll1 autoregulation. ASCIZ binds to the Dynll1 promoter via its ZnF domain and activates transcription via its TAD, resulting in expression of DYNLL1 protein (blue arcs). The equilibrium with other DYNLL1-binding proteins (DYNLL1BPa/b) determines how much DYNLL1 can bind to ASCIZ. At high levels of free DYNLL1, saturated binding to ten TQT motifs (circles) in the ASCIZ TAD represses Dynll1 transcription. Transcription is de-repressed when excess DYNLL1 is degraded or sequestered by increasing levels of DYNLL1-binding proteins.
DISCUSSION
Here, we have identified ASCIZ as a highly specialized and phylogenetically conserved “designated” transcription factor for Dynll1 expression. Dynll1 is clearly the most dramatically down-regulated gene in the absence of ASCIZ, and ASCIZ is able to directly sense free DYNLL1 levels via a remarkably large number of TQT motifs within its TAD. Saturated DYNLL1 binding may simply obscure the ASCIZ TAD and thereby prevent access of co-activators and the transcription machinery (Fig. 5D). Alternatively, binding of individual DYNLL1 dimers to two different binding sites within a single ASCIZ molecule may induce intramolecular rearrangements of the TAD into a repressed state. This ability to directly sense free DYNLL1 levels provides a dynamic mechanism to readily adjust Dynll1 expression to the needs of individual cells depending on tissue-, stage-, or cell cycle-specific complements of other DYNLL1-binding proteins. Consistent with this model, total DYNLL1 levels in the developing brain are higher than in fetal livers, yet, in each tissue ∼10-fold differences between ASCIZ-expressing and ASCIZ-deficient cells are maintained (Fig. 2C). Negative auto-regulation of gene expression is a widespread biological phenomenon, where transcription factors can directly repress their own promoters (e.g. lambda repressor (26) or myc (27)), or can be an indirect process where the induced gene products inhibit their transcriptional activators in the cytoplasm (e.g. in case of axin2 and β-catenin (28)). However, to our knowledge, the ASCIZ-DYNLL1 interaction reported here represents the first example of negative auto-regulation of gene expression where a gene product directly inhibits the main transcriptional activator while bound at its own promoter (Fig. 5D).
While DYNLL1 acts as a negative feedback regulator of ASCIZ transcriptional activity (Fig. 5), DYNLL1 appears to be a positive regulator of DNA damage-related ASCIZ functions (Fig. 4, A and G), indicating that the functional interactions between the two proteins may be context-dependent. As DNA damage-induced foci are large macromolecular structures, it would be conceivable that DYNLL1 functions in this context as molecular glue to generate ASCIZ oligomers, which would be aided by the large number of ASCIZ DYNLL1-binding sites. From a DNA damage perspective, it is interesting to note that TQT motifs in intrinsically disordered regions are not only preferred DYNLL1-binding sites, but also match the phosphorylation site preference for ATM/ATR-like checkpoint kinases (29, 30). For example, a bona fide DYNLL1 binding site 1171TQT of the DNA damage scaffold 53BP1 (31) is also an established ionizing radiation-induced ATM phosphorylation site in vivo (29). Thus, DYNLL1 binding to TQT motifs may in some cases be modulated by DNA damage-induced phosphorylation, or conversely, phosphorylation may depend on whether or not these sites are bound by DYNLL1. It should be noted that during the revision of this manuscript, Rapali et al. independently reported the identification of ASCIZ as a high-affinity, multi-site DYNLL1-binding protein and the co-localization of the two proteins in DNA damage-induced foci upon MMS treatment (32).
Here, we have shown that ASCIZ associates with the endogenous Dynll1 promoter and regulates its expression in a ZnF-dependent manner. In the future, it will be interesting to determine the precise sequence element ASCIZ binds to, and to determine if its ZnFs bind this element directly or indirectly. Finally, as Dynll1 is the main transcriptional target of ASCIZ, lower DYNLL1 levels are likely to play a critical role in the various phenotypes of ASCIZ-deficient mice. However, given the complexity and enormous number of probable DYNLL1 targets (7), it may be challenging to decipher which particular DYNLL1 downstream effectors are involved in specific phenotypes caused by the absence of ASCIZ.
Supplementary Material
Acknowledgments
We thank the Australian Genome Research Facility for microarray analysis, Kaylene Simpson and the Victorian Centre for Functional Genomics for siRNA screening support, Angela Tam for help with mutagenesis, Hamsa Puthalakath for reagents and discussions, and Tony Mason and Ian Smyth for discussions and comments on the manuscript.
This work was supported by grants and a Senior Research Fellowship from the National Health and Medical Research Council (NHMRC) of Australia (to J. H.), a PhD scholarship from the Cooperative Research Centre Cancer Therapeutics (to S. J.), and in part by the Victorian Government's Operational Infrastructure Support Program.

This article contains supplemental Figs. S1 and S2.
- DYNLL
- dynein light chain
- ASCIZ
- ATM substrate Chk2-interacting Zn2+ finger
- ChIP
- chromatin immunoprecipitation
- DBD
- DNA binding domain
- MEF
- murine embryonic fibroblast
- MMS
- methylmethane sulfonate
- TAD
- transcription activation domain
- ZnF
- Zn2+ finger.
REFERENCES
- 1. King S. M., Patel-King R. S. (1995) The M(r) = 8,000 and 11,000 outer arm dynein light chains from Chlamydomonas flagella have cytoplasmic homologues. J. Biol. Chem. 270, 11445–11452 [DOI] [PubMed] [Google Scholar]
- 2. Williams J. C., Roulhac P. L., Roy A. G., Vallee R. B., Fitzgerald M. C., Hendrickson W. A. (2007) Structural and thermodynamic characterization of a cytoplasmic dynein light chain-intermediate chain complex. Proc. Natl. Acad. Sci. U.S.A. 104, 10028–10033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nyarko A., Barbar E. (2011) Light chain-dependent self-association of dynein intermediate chain. J. Biol. Chem. 286, 1556–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Varma D., Dawn A., Ghosh-Roy A., Weil S. J., Ori-McKenney K. M., Zhao Y., Keen J., Vallee R. B., Williams J. C. (2010) Development and application of in vivo molecular traps reveals that dynein light chain occupancy differentially affects dynein-mediated processes. Proc. Natl. Acad. Sci. U.S.A. 107, 3493–3498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barbar E. (2008) Dynein light chain LC8 is a dimerization hub essential in diverse protein networks. Biochemistry 47, 503–508 [DOI] [PubMed] [Google Scholar]
- 6. King S. M. (2008) Dynein-independent functions of DYNLL1/LC8: redox state sensing and transcriptional control. Sci. Signal 1, pe51. [DOI] [PubMed] [Google Scholar]
- 7. Rapali P., Radnai L., Süveges D., Harmat V., Tölgyesi F., Wahlgren W. Y., Katona G., Nyitray L., Pál G. (2011) Directed evolution reveals the binding motif preference of the LC8/DYNLL hub protein and predicts large numbers of novel binders in the human proteome. PLoS One 6, e18818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jaffrey S. R., Snyder S. H. (1996) PIN: an associated protein inhibitor of neuronal nitric-oxide synthase. Science 274, 774–777 [DOI] [PubMed] [Google Scholar]
- 9. Puthalakath H., Huang D. C., O'Reilly L. A., King S. M., Strasser A. (1999) The proapoptotic activity of the Bcl-2 family member Bim is regulated by interaction with the dynein motor complex. Mol. Cell 3, 287–296 [DOI] [PubMed] [Google Scholar]
- 10. Dick T., Ray K., Salz H. K., Chia W. (1996) Cytoplasmic dynein (ddlc1) mutations cause morphogenetic defects and apoptotic cell death in Drosophila melanogaster. Mol. Cell. Biol. 16, 1966–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Batlevi Y., Martin D. N., Pandey U. B., Simon C. R., Powers C. M., Taylor J. P., Baehrecke E. H. (2010) Dynein light chain 1 is required for autophagy, protein clearance, and cell death in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 107, 742–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McNees C. J., Conlan L. A., Tenis N., Heierhorst J. (2005) ASCIZ regulates lesion-specific Rad51 focus formation and apoptosis after methylating DNA damage. EMBO J. 24, 2447–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oka H., Sakai W., Sonoda E., Nakamura J., Asagoshi K., Wilson S. H., Kobayashi M., Yamamoto K., Heierhorst J., Takeda S., Taniguchi Y. (2008) DNA damage response protein ASCIZ links base excision repair with immunoglobulin gene conversion. Biochem. Biophys. Res. Commun. 371, 225–229 [DOI] [PubMed] [Google Scholar]
- 14. Jurado S., Smyth I., van Denderen B., Tenis N., Hammet A., Hewitt K., Ng J. L., McNees C. J., Kozlov S. V., Oka H., Kobayashi M., Conlan L. A., Cole T. J., Yamamoto K., Taniguchi Y., Takeda S., Lavin M. F., Heierhorst J. (2010) Dual functions of ASCIZ in the DNA base damage response and pulmonary organogenesis. PLoS Genet 6, e1001170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kanu N., Penicud K., Hristova M., Wong B., Irvine E., Plattner F., Raivich G., Behrens A. (2010) The ATM cofactor ATMIN protects against oxidative stress and accumulation of DNA damage in the aging brain. J. Biol. Chem. 285, 38534–38542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Loizou J. I., Sancho R., Kanu N., Bolland D. J., Yang F., Rada C., Corcoran A. E., Behrens A. (2011) ATMIN is required for maintenance of genomic stability and suppression of B cell lymphoma. Cancer Cell 19, 587–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu X., Zha S. (2011) ATMIN: a new tumor suppressor in developing B cells. Cancer Cell 19, 569–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Heierhorst J., Smyth I., Jurado S. (2011) A breathtaking phenotype: unexpected roles of the DNA base damage response protein ASCIZ as a key regulator of early lung development. Cell Cycle 10, 1222–1224 [DOI] [PubMed] [Google Scholar]
- 19. Pear W. S., Miller J. P., Xu L., Pui J. C., Soffer B., Quackenbush R. C., Pendergast A. M., Bronson R., Aster J. C., Scott M. L., Baltimore D. (1998) Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood 92, 3780–3792 [PubMed] [Google Scholar]
- 20. Perdomo J., Crossley M. (2002) The Ikaros family protein Eos associates with C-terminal-binding protein corepressors. Eur. J. Biochem. 269, 5885–5892 [DOI] [PubMed] [Google Scholar]
- 21. Rayala S. K., den Hollander P., Balasenthil S., Yang Z., Broaddus R. R., Kumar R. (2005) Functional regulation of oestrogen receptor pathway by the dynein light chain 1. EMBO Rep. 6, 538–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pike B. L., Yongkiettrakul S., Tsai M. D., Heierhorst J. (2004) Mdt1, a novel Rad53 FHA1 domain-interacting protein, modulates DNA damage tolerance and G(2)/M cell cycle progression in Saccharomyces cerevisiae. Mol. Cell. Biol. 24, 2779–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Navarro C., Puthalakath H., Adams J. M., Strasser A., Lehmann R. (2004) Egalitarian binds dynein light chain to establish oocyte polarity and maintain oocyte fate. Nat. Cell Biol. 6, 427–435 [DOI] [PubMed] [Google Scholar]
- 24. Dorsett M., Schedl T. (2009) A role for dynein in the inhibition of germ cell proliferative fate. Mol. Cell. Biol. 29, 6128–6139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fejtova A., Davydova D., Bischof F., Lazarevic V., Altrock W. D., Romorini S., Schöne C., Zuschratter W., Kreutz M. R., Garner C. C., Ziv N. E., Gundelfinger E. D. (2009) Dynein light chain regulates axonal trafficking and synaptic levels of Bassoon. J. Cell Biol. 185, 341–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Crews S. T., Pearson J. C. (2009) Transcriptional autoregulation in development. Curr. Biol. 19, R241–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Penn L. J., Brooks M. W., Laufer E. M., Land H. (1990) Negative autoregulation of c-myc transcription. EMBO J. 9, 1113–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jho E. H., Zhang T., Domon C., Joo C. K., Freund J. N., Costantini F. (2002) Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell. Biol. 22, 1172–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Matsuoka S., Ballif B. A., Smogorzewska A., McDonald E. R., 3rd, Hurov K. E., Luo J., Bakalarski C. E., Zhao Z., Solimini N., Lerenthal Y., Shiloh Y., Gygi S. P., Elledge S. J. (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316, 1160–1166 [DOI] [PubMed] [Google Scholar]
- 30. Traven A., Heierhorst J. (2005) SQ/TQ cluster domains: concentrated ATM/ATR kinase phosphorylation site regions in DNA-damage-response proteins. Bioessays 27, 397–407 [DOI] [PubMed] [Google Scholar]
- 31. Lo K. W., Kan H. M., Chan L. N., Xu W. G., Wang K. P., Wu Z., Sheng M., Zhang M. (2005) The 8-kDa dynein light chain binds to p53-binding protein 1 and mediates DNA damage-induced p53 nuclear accumulation. J. Biol. Chem. 280, 8172–8179 [DOI] [PubMed] [Google Scholar]
- 32. Rapali P., García-Mayoral M. F., Martínez-Moreno M., Tárnok K., Schlett K., Albar J. P., Bruix M., Nyitray L., Rodriguez-Crespo I. (2011) LC8 dynein light chain (DYNLL1) binds to the C-terminal domain of ATM-interacting protein (ATMIN/ASCIZ) and regulates its subcellular localization. Biochem. Biophys. Res. Commun. 414, 493–498 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.