Background: The role of sequence variation among Tm isoforms remains unexplored.
Results: We describe the structures of smooth muscle Tm isoforms.
Conclusion: The formation of overlap complexes may be an intrinsic property of the Tm molecule. Sequence variation may not significantly affect actin binding, but it could determine the gatekeeper function of Tm.
Significance: Work formulates a new understanding of the role of sequence variation in Tm function.
Keywords: Actin, Cytoskeleton, Smooth Muscle, Tropomyosin, X-ray Crystallography, Head-to-tail Overlap Complex, Parallel and Anti-parallel Coiled Coils, Smooth Muscle, Tropomyosin, X-ray Structure
Abstract
A large number of tropomyosin (Tm) isoforms function as gatekeepers of the actin filament, controlling the spatiotemporal access of actin-binding proteins to specialized actin networks. Residues ∼40–80 vary significantly among Tm isoforms, but the impact of sequence variation on Tm structure and interactions with actin is poorly understood, because structural studies have focused on skeletal muscle Tmα. We describe structures of N-terminal fragments of smooth muscle Tmα and Tmβ (sm-Tmα and sm-Tmβ). The 2.0-Å structure of sm-Tmα81 (81-aa) resembles that of skeletal Tmα, displaying a similar super-helical twist matching the contours of the actin filament. The 1.8-Å structure of sm-Tmα98 (98-aa) unexpectedly reveals an antiparallel coiled coil, with the two chains staggered by only 4 amino acids and displaying hydrophobic core interactions similar to those of the parallel dimer. In contrast, the 2.5-Å structure of sm-Tmβ98, containing Gly-Ala-Ser at the N terminus to mimic acetylation, reveals a parallel coiled coil. None of the structures contains coiled-coil stabilizing elements, favoring the formation of head-to-tail overlap complexes in four of five crystallographically independent parallel dimers. These complexes show similarly arranged 4-helix bundles stabilized by hydrophobic interactions, but the extent of the overlap varies between sm-Tmβ98 and sm-Tmα81 from 2 to 3 helical turns. The formation of overlap complexes thus appears to be an intrinsic property of the Tm coiled coil, with the specific nature of hydrophobic contacts determining the extent of the overlap. Overall, the results suggest that sequence variation among Tm isoforms has a limited effect on actin binding but could determine its gatekeeper function.
Introduction
Tropomyosin (Tm)2 is a coiled-coil molecule that associates head-to-tail to form super-helical polymers along the two long pitch helices of the actin filament (1, 2). Like the actin filament, the Tm polymer is polar, with its N- and C-terminal ends directed toward the pointed and barbed ends of the actin filament, respectively. Mammals express more than 40 Tm isoforms (1). It is thought that different Tm isoforms exert specialized “gatekeeper” functions, regulating the spatiotemporal interactions of actin-binding proteins (ABPs) with discrete actin filament networks in cells (3–14). At least two such actin networks can be distinguished in smooth muscle cells, the actomyosin contractile network and the cytoplasmic (or non-muscle) network. Much like in skeletal and cardiac muscles, the actin (or thin) filaments of the actomyosin network in smooth muscle cells act as molecular tracks for the myosin II motor and therefore are directly involved in the generation of contractile force (15–17). The actomyosin network is composed of α- and γ-actin (18), caldesmon, smitin, and telokin, but unlike striated and cardiac muscles, it lacks troponin (17). The cytoplasmic actin network localizes mainly to the cell cortex, and its main function may be to control cell shape and cell-cell communication during contraction and force maintenance (19). This network is thought to be more dynamic and can be viewed to a certain extent as analogous to the actin cytoskeleton of non-muscle cells, composed of β-actin and a myriad of ABPs (20–24). At least five Tm isoforms are expressed in smooth muscle cells; Tmα (also known as Tm6), Tmβ (also known as Tm1), and the non-muscle isoforms Tm2, Tm5NM1, and Tm4 (3). Tmα, the most abundant isoform, is associated with the actomyosin network, and Tmβ appears to be mainly linked to the cytoplasmic network, whereas the location of the non-muscle isoforms remains to be established.
Tm isoforms differ in length and sequence. Long Tm isoforms, including skeletal and smooth muscle α and β isoforms, contain 284 amino acids, organized into seven pseudo-repeats of ∼40 aa, with the rise per pseudo-repeat corresponding to that of one actin subunit of the long pitch helix of the actin filament (1, 6, 7). Short Tm isoforms contain between four and six such repeats (1, 25). Sequence differences among Tm isoforms cluster within pseudo-repeat 2 and at the C terminus. Because structural studies have focused predominantly on skeletal muscle Tmα (26–30), it is unclear how these differences affect the shape and interactions of Tm with the actin filament and other Tm- and actin-binding proteins. For instance, it has been proposed that Tm may be pre-shaped to the contours of the actin filament helix, such as to facilitate binding to the filament (10). Crystal structures of skeletal muscle Tmα support this notion, as they often display a super-helical twist that matches that of the actin filament (26, 31). Whether this is also true of other isoforms remains to be demonstrated. It is also intriguing that the variable C terminus mediates head-to-tail interactions of the Tm polymer. The fact that these interactions are not conserved suggests that there exist multiple possible combinations of head-to-tail complexes, probably characterized by different stabilities and different extents of the overlap. In an attempt to address some of these questions, we present here the crystal structures of three N-terminal fragments of smooth muscle Tm isoforms α and β, revealing the conformation of one of the most variable regions of the Tm sequence. The structures share some features in common with those of skeletal muscle Tmα, particularly the presence of a super-helical twist that appears to match the contours of the actin filament. Contrary to previous studies, none of the structures described here contains coiled-coil stabilizing elements, such as GCN4 leucine zippers or disulfide bonds, leading to some unique observations, including the formation of head-to-tail overlap complexes and one antiparallel coiled-coil dimer.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The cDNA encoding for 98-aa N-terminal fragments of chicken gizzard smooth muscle Tmα and Tmβ were synthesized (GENEWIZ). Chicken gizzard smooth muscle Tm isoforms were selected for this study because they have been more extensively characterized at the biochemical and structural levels. Constructs sm-Tmα81 and sm-Tmα98, containing 81 and 98 N-terminal amino acids, were cloned between the NdeI and EcoRI sites of vector pTYB12 (New England Biolabs), containing a chitin-intein affinity and self-cleavage tag. Construct sm-Tmβ98 was cloned between the BamHI and HindIII sites of vector pRSFDuet-1 (Novagen), incorporating a hexahistidine N-terminal affinity tag. Plasmids were transformed into BL21(DE3) cells (New England Biolabs). Cells were grown in Terrific Broth medium at 37 °C to A600 = 1.0–1.2.
Expression was induced with the addition of 0.5 mm isopropyl β-d-thiogalactopyranoside for 16 h at 20 °C. Cells expressing the two Tmα constructs were resuspended in chitin column equilibration buffer (20 mm Tris-HCl, pH 8.0, 500 mm NaCl, 1 mm EDTA, 1 mm PMSF) and purified through a chitin affinity column. Self-cleavage of the intein was induced by addition of 50 mm DTT for 16 h. Three amino acids of the pTYB12 vector, Ala-Gly-His, remain at the N terminus of the two Tmα constructs after cleavage. Proteins were then additionally purified on a Superdex-200 gel filtration column (GE Healthcare) in 20 mm Tris-HCl, pH 8.0, 50 mm NaCl, and 1 mm DTT. Cells expressing sm-Tmβ98 were resuspended in 20 mm Tris-HCl, pH 8.0, 300 mm NaCl, 1 mm PMSF, and 20 mm imidazole and purified through a nickel-nitrilotriacetic acid affinity column (Qiagen). Cleavage of the His tag was carried out at 4 °C with addition of tobacco etch virus protease (at a molar ratio of 1:50) in 50 mm Tris-HCl, pH 8.0, 50 mm NaCl, 0.5 mm EDTA, and 1 mm DTT. After cleavage of the His tag, three amino acids remain at the N terminus, Gly-Ala-Ser. These three amino acids have been previously shown to mimic N-acetylation (32). Selenomethionine-substituted sm-Tmα81 and sm-Tmβ98 were obtained by growing cells in SelenoMet medium (Athena Enzyme Systems), supplemented with 70 mg/ml selenomethionine (Acros Organics).
Crystallization, Data Collection, and Structure Determination
The three Tm fragments were concentrated to ∼10 mg/ml using Vivaspin centrifugal devices (Sartorius Stedim Biotech) in 50 mm Tris-HCl, pH 8.0, and 50 mm NaCl, and 1 mm DTT. Crystals were obtained using the hanging drop vapor-diffusion method at 20 °C and under similar conditions (crystallization conditions are listed in Table 1 and supplemental Fig. 1, which also shows representative pictures of the crystals). Crystals were flash-frozen in liquid nitrogen directly from the crystallization drop.
TABLE 1.
Crystallization, crystallographic data, and refinement statistics
Values in parentheses correspond to highest resolution shell. r.m.s.d., root mean square deviation. MPD is 2-methyl-2,4-pentanediol.
| sm-TMα81 | sm-TMα81-Se | sm-TMα98 | sm-TMα98-Pt | sm-TMβ98-Se | |
|---|---|---|---|---|---|
| Crystallization | 0.1 m sodium acetate, pH 4.4, 20 mm CaCl2, 30–33% MPD | 0.1 m sodium acetate, pH 4.0, 40 mm CaCl2, 32–35% MPD | 0.1 m HEPES, pH 7.4, 5 mm CaCl2, 28–31% MPD | ||
| Diffraction data | |||||
| Wavelength | 0.9795 Å | 0.9795 Å | 0.9795 Å | 1.54 Å | 0.9789 Å |
| Space group | P 2 2 2 | P 2 2 2 | P 43 21 2 | P 43 21 2 | P 21 21 2 |
| Unit cell a, b, c | 58.8, 63.2, 101.3 Å | 58.1, 63.0, 101.2 Å | 76.7, 76.7, 79.7 Å | 77.5, 77.5, 79.2 Å | 43.1, 75.7, 137.6 Å |
| Unit cell α, β, γ | 90.0, 90.0, 90.0° | 90.0, 90.0, 90.0° | 90.0, 90.0, 90.0° | 90.0, 90.0, 90.0° | 90.0, 90.0, 90.0° |
| Resolution | 2-40 Å (2.0-2.1 Å) | 2.4-50 Å (2.4-2.5 Å) | 1.8-25 Å (1.8-1.89 Å) | 3.3-35 Å (3.3-3.4 Å) | 2.5-29 Å (2.5-2.6 Å) |
| Completeness | 98.9% (98.2%) | 99.9% (99.8%) | 96.0% (95.5%) | 99.8% (100.0%) | 99.6% (100.0%) |
| Multiplicity | 12.05 (10.40) | 12.8 (13.5) | 18.33 (15.09) | 57.52 (44.18) | 7.09 (7.46) |
| Rmergea | 10.58% (41.98%) | 11.3% (57.9%) | 7.49% (39.60%) | 13.33% (67.26%) | 6.57% (25.49%) |
| I/σ | 14.59 (4.52) | 11.1 (3.3) | 24.90 (5.57) | 26.84 (4.64) | 19.21 (6.11) |
| Refinement | |||||
| Resolution | 2.0-40 Å | 1.8-25 Å | 2.5-29 Å | ||
| No. of reflections | 24,909 | 20,857 | 15,687 | ||
| Completeness | 95% | 92% | 100% | ||
| No. of residues/waters | 330/216 | 202/151 | 400/159 | ||
| Rfactorb | 22.2% | 24.6% | 29.5% | ||
| Rfreec | 26.5% | 28.4% | 33.7% | ||
| r.m.s.d. bonds | 0.009 Å | 0.009 Å | 0.012 Å | ||
| r.m.s.d. angles | 0.94° | 1.14° | 1.42° | ||
| B-factor protein | 43.0 Å2 | 46.8 Å2 | 77.5 Å2 | ||
| B-factor solvent | 42.0 Å2 | 48.4 Å2 | 70.3 Å2 | ||
| Protein Data Bank code | 3U1A | 3U1C | 3U59 | ||
a Rmerge = Σhkl(I − 〈I〉)/SI, where I and 〈I〉 are the observed and mean intensities of all observations of reflection hkl, including its symmetry-related equivalents.
b Rfactor = Σhkl‖Fobs| − |Fcalc‖/Σ|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors of reflection hkl.
c Rfree, Rfactor were calculated for a randomly selected subset of the reflections (5%) that were not used in refinement.
X-ray datasets (Table 1) were collected from wild type and selenomethionine-substituted crystals using beamline 17-ID of the Industrial Macromolecular Crystallography Association-Collaborative Access Team at the Advance Photon Source (Argonne, IL). The native dataset of sm-Tmα98 was collected at CHESS beamline A1. Data indexing and scaling were performed with the programs XDS/XSCALE (33). Additionally, a dataset was collected from crystals of sm-Tmα98 soaked for 1 h in 0.5 mm K2PtCl4 (added to the crystallization drop) using a Bruker X8 PROSPECTOR x-ray apparatus equipped with an APEX II CCD detector and an IμS microfocus x-ray source (Bruker AXS Inc.). The platinum derivative data were indexed and scaled using the Bruker software PROTEUM2.
All the structures were determined using the single wavelength anomalous dispersion method, because molecular replacement was unsuccessful. The anomalous scattering atom was selenium for the structures of sm-Tm81α and sm-Tm98β and platinum for the structure of sm-Tm98α. The programs SHELXD/SHELXE (34) were used to find the selenium or platinum sites, calculate phases, and perform density modification. Model building and refinement were carried out with the programs Coot (35) and Phenix (Table 1) (36). Rfactor and Rfree of the final models are higher than expected at similar resolutions for globular structures, which is a general feature of Tm coiled-coil structures where most of the chain is solvent-exposed (see for instance Protein Data Bank entries 1IC2, 2B9C, and 2D3E). Nevertheless, the final models are well defined in the electron density maps (supplemental Fig. 1). Illustrations of the structures were prepared with the program PyMOL (Schrödinger).
Molecular Dynamics (MD) Simulations
MD simulations were performed on crystal structures of the sm-Tm fragments determined here and also on chimeras of full-length Tm, where the current structures replaced corresponding segments of an atomic model of smooth muscle Tmα (37). After energy optimization, MD was run for 30 ns at 300 K in implicit solvent mode as described previously (31).
RESULTS
Crystal Structure of sm-Tmα81
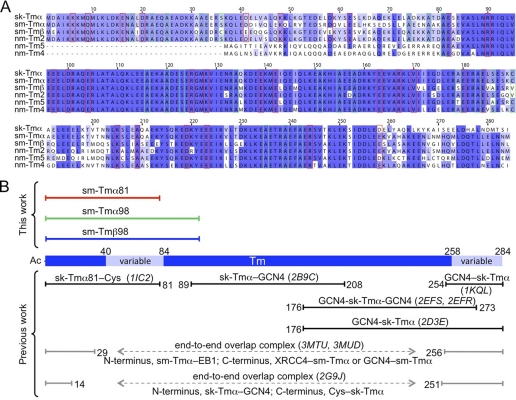
Most sequence variation among Tm isoforms occurs within pseudo-repeat 2 and at the C terminus (Fig. 1A and supplemental Fig. 2). Knowing the structure of these variable regions is important to understand isoform-specific functional differences. Various structures of fragments of skeletal muscle Tmα have been determined that combined reveal nearly the entirety of the molecule (Fig. 1B). These structures all contain coiled-coil stabilizing elements, such as GCN4 leucine zippers or disulfide bonds. One of the structures is that of an 81-aa N-terminal fragment of skeletal muscle Tmα (sk-Tmα81), containing additionally a cysteine disulfide bond at the C terminus to stabilize the coiled coil (26). This structure includes the variable region between residues 40 and 80, corresponding to pseudo-repeat 2 of Tm (Fig. 1B). To assess the effect of sequence variation within this region, we determined the structure of an equivalent 81-aa fragment of smooth muscle Tmα (sm-Tmα81). Despite the similarities with sk-Tmα81, a molecular replacement solution of the structure of sm-Tmα81 could not be obtained, presumably because of slight differences in bending and the elongated nature of the Tm molecule. Thus, the structure of sm-Tmα81 was determined ab initio, using the anomalous signal of a selenomethionine derivative, and refined at 2.0 Å resolution (Table 1).
FIGURE 1.
Sequences and structures of Tm isoforms. A, alignment of the sequences of sk-Tmα, chicken gizzard sm-Tmα, and sm-Tmβ and three non-muscle isoforms expressed in smooth muscle (3). Accession codes are as follows: sk-Tmα, P04268-1; sm-Tmα, P04268-2; sm-Tmβ, P19352-2; nm-Tm2, P09493-3; nm-Tm5, P06753-2; and nm-Tm4, P67936. Residues implicated in interactions with actin (31) and those forming part of alanine clusters (26, 30) are outlined red and green, respectively. B, fragments whose structures were determined here or previously published are shown above and below a diagram of the Tm molecule, where variable regions are highlighted (light blue). For the published structures, Protein Data Bank accession codes are listed in parentheses and the N- and C-terminal coiled-coil stabilizing elements used are indicated (GCN4, Cys bonds or EB1 and XRCC4 fusion proteins).
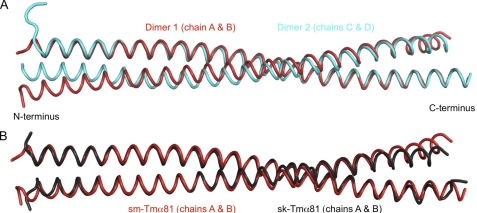
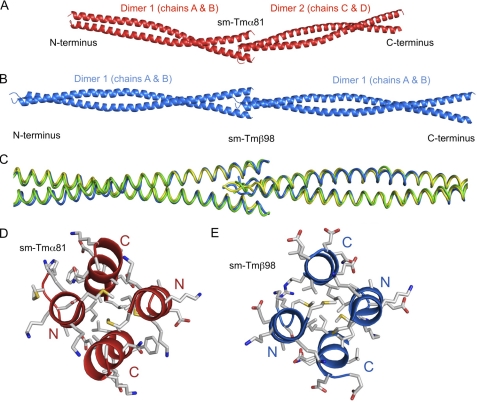
Two dimers are present in the asymmetric unit of the structure of sm-Tmα81 (Fig. 2A). Dimers 1 (chains A and B) and 2 (chains C and D) exhibit canonical coiled-coil conformations between residues 3–70 and 3–74, respectively. Although the secondary structure is still α-helical, both dimers open up slightly toward the N and C termini compared with a canonical coiled coil, possibly because of the lack of N-acetylation and coiled-coil stabilizing elements. The 81-aa fragment includes two of the six alanine clusters of the Tm molecule (26, 30). The alanine clusters are regions where small side chains (mainly Ala but also Ser and Cys) accumulate at heptad positions a and d in the hydrophobic core of the coiled coil (Fig. 1A), allowing for slight axial staggering of the coiled-coil helices relative to one another. Such staggering is responsible for super-helical bending of the Tm coiled coil to match the contours of the actin filament (26). In the current structure, differences between the two dimers in the asymmetric unit become more pronounced around the first alanine cluster, resulting in an overall r.m.s.d. of 1.6 Å for 149 superimposable Cα atoms of the two dimers within a distance cutoff of 2.0 Å (Fig. 2A). This observation is consistent with the notion that the alanine clusters correspond to regions of increased conformational variability (30, 38–40).
FIGURE 2.
Structure of sm-Tmα81. A, superimposition of the two dimers in the asymmetric unit of the structure of sm-Tmα81 (r.m.s.d. of 1.6 Å for 149 superimposable Cα positions within a distance cutoff of 2.0 Å). B, superimposition of the structure of sm-Tmα81 (chains A and B) with that of sk-Tmα81 (chains A and B). The two structures superimpose with r.m.s.d. of 1.0 Å for 143 Cα positions.
Overall, the structure of sm-Tmα81 is similar to sk-Tmα81 (Fig. 2B). Indeed, both structures contain two dimers in the asymmetric unit, and differences between dimers within each structure (r.m.s.d. of 1.6 Å for 150 superimposable Cα atoms of the two dimers of sk-Tmα81 within a distance cutoff of 2.0 Å) are comparable with those between the dimers of the two structures (Cα r.m.s.d. ranging from 1.0 to 2.3 Å for the four possible pairwise comparisons). Overall, the differences between isoforms are more pronounced toward the N and C termini (Fig. 2), which is expected because no coiled-coil stabilizing elements were used in the current structure. Importantly, changes in sequence occurring in pseudo-repeat 2 (amino acids 40–80) have only a limited and local impact on the conformation of sm-Tmα81 compared with sk-Tmα. What is more, residues within this region predicted to become involved in interactions with actin are highly conserved among Tm isoforms (Fig. 1A and supplemental Fig. 2). These residues are exposed, and their conservation suggests that the actin-binding function tends to be preserved among Tm isoforms. MD simulations further show that on average sm-Tmα81 retains a twist during simulation comparable with that of the crystal structure. In MD simulations performed on a model of full-length Tm obtained by splicing the structure of sm-Tmα81 into a previous model of full-length smooth muscle Tmα (37), the average super-helical trajectory of the coiled-coil matches well the shape of the actin filament helix (supplemental Fig. 3).
Crystal Structure of sm-Tmα98
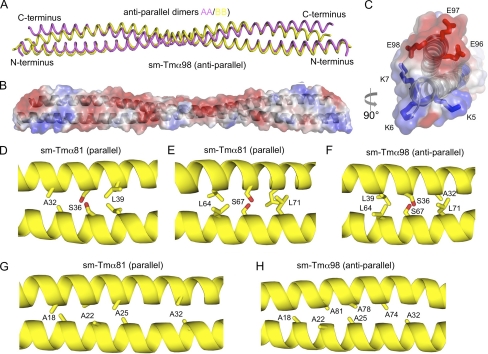
The structure of a 98-aa N-terminal fragment of smooth muscle Tmα (sm-Tmα98) was determined ab initio, using a platinum heavy atom derivative, and refined at 1.8 Å resolution (Table 1). The asymmetric unit of the structure contains two chains, each belonging to a different coiled-coil dimer that is completed by crystal symmetry. Surprisingly, both crystallographically independent dimers form antiparallel coiled coils. The antiparallel dimers differ somewhat (r.m.s.d. of 2.4 Å for 172 equivalent Cα atoms), but overall their conformations and inter-helical contacts are similar (Fig. 3A). The serendipitous occurrence of antiparallel dimers in the structure of sm-Tmα98 appears to be a consequence of our particular choice of construct, because the shorter fragment sm-Tmα81 forms a parallel coiled coil. Indeed, sm-Tmα98 is charged positively at the N terminus and negatively at the C terminus (Fig. 3, B and C). Therefore, it is possible that, in the absence of coiled-coil stabilizing elements, repulsive electrostatic interactions force the two helices apart and favor an antiparallel arrangement. Interestingly, similar inter-helical contacts are observed in the hydrophobic core of the antiparallel and parallel structures of sm-Tmα98 and sm-Tmα81. Thus, the two helices of the antiparallel coiled coil are staggered by only 4 aa, such that hydrophobic residues are masked at the inter-helical interface, somewhat analogous to the parallel dimer. Moreover, residues Ser36 and Ser67 of the two helices form hydrogen-bonding contacts in the hydrophobic core of the parallel coiled-coil dimer, which in the antiparallel coiled coil are replaced by interactions between Ser36 of one helix and Ser67 of the other helix and vice versa (Fig. 3, D–F). A similar inter-helical swap occurs between the two alanine clusters present in this construct (Fig. 3, G and H). Additionally, a number of salt bridges (12 and 8 in the two crystallographically independent dimers) help stabilize the antiparallel dimers. Some of these salt bridges occur toward the N and C termini of the dimer, which might explain why the two helices remain tightly packed toward the ends (Fig. 3, B and C).
FIGURE 3.
Structure of sm-Tmα98. A, overlay of the two antiparallel dimers in the asymmetric unit of the structure of sm-Tmα98 (r.m.s.d. of 2.4 Å for 172 Cα atoms). B and C, two perpendicular views of an electrostatic surface representation (red, negatively charged; blue, positively charged) of the structure of sm-Tmα98. Note the existing charge compensation between positively and negatively charged residues at the N and C termini. D–F, residues Ser36 and Ser67 form hydrogen bonding contacts with themselves in the hydrophobic core of the parallel coiled coil of sm-Tmα81, which are swapped by two identical hydrogen bonding contacts between Ser36 and Ser67 in the antiparallel coiled coil of sm-Tmα98. G and H, two alanine clusters of the parallel coiled coil are swapped in a similar manner in the antiparallel structure.
Crystal Structure of sm-Tmβ98
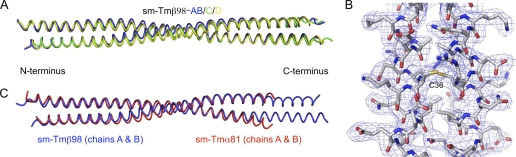
The region between residues 35 and 85 of sm-Tmβ differs quite significantly from both smooth and skeletal muscle Tmα (Fig. 1A). We determined the structure of sm-Tmβ98 using the anomalous signal of a selenomethionine derivative and refined the structure at 2.5 Å resolution (Table 1). In this case, we added three amino acids at the N terminus, Gly-Ala-Ser, which were previously shown to mimic N-acetylation (32) and were thus expected to help preserve a native coiled-coil conformation at the N terminus. The asymmetric unit of the crystal structure contains four chains, two of which form a complete dimer and the other two form half-dimers that are completed by crystal symmetry for a total of three crystallographically independent coiled-coil dimers. In contrast to the structure of sm-Tmα98, all three dimers in the structure of sm-Tmβ98 are parallel. There are differences in bending between the three dimers resulting in r.m.s.d. between dimers ranging from 1.0 to 1.2 Å for 190 Cα positions (Fig. 4A). Because the electron density is better defined for the complete dimer (i.e. chains A and B, Fig. 4B), our analysis is based on this coiled coil.
FIGURE 4.
Structure of sm-Tmβ98. A, superimposition of the three dimers in the asymmetric unit of the structure of sm-Tmβ98 (r.m.s.d. between dimers range from 1.0 to 1.2 Å for 190 superimposable Cα positions within a distance cutoff of 2.0 Å). B, 2Fo − Fc electron density map contoured at 1.0σ in the region around residue Cys36. C, superimposition of the structures of sm-Tmα81 and sm-Tmβ98 (r.m.s.d. of 1.4 Å for 155 Cα atoms).
The structure of sm-Tmβ98 overlays well with that of sm-Tmα81 (r.m.s.d. of 1.4 Å for 155 Cα atoms used in the superimposition based on a distance cutoff of 2.0 Å), except at the C terminus where the structure of sm-Tmα81 is more open (Fig. 4C). One significant difference, however, between sm-Tmβ and other Tm isoforms is the presence of a Cys residue at position 36 (Ser in both smooth and skeletal muscle Tmα, Fig. 1A), which we anticipated would form a disulfide bond in the hydrophobic core. Yet to our surprise, residue Cys36 does not form a disulfide bond despite the fact that the side chains of this residue from both helices face each other and the Sγ atoms are in close proximity (3.86 versus 2.03 Å for an ideal disulfide bond; Fig. 4B (41)). This result was confirmed by mass spectrometry analysis of dissolved crystals, showing that most of the protein in the crystals is not covalently cross-linked (supplemental Fig. 4). This finding is in agreement with a recent study that found that Tm is in a reduced state in rabbit psoas and rat cardiac muscles (42, 43). A reduced state of the Tm molecule is also consistent with slight inter-helical staggering near regions of alanine clusters (26). Indeed, such staggering could not occur if cysteine residues in the hydrophobic core were covalently bound. Coincidentally, Cys36 forms part of the first alanine cluster (Fig. 1A).
Overlap Complexes
Head-to-tail overlap complexes are generated by crystal symmetry in the two parallel coiled-coil structures reported here, sm-Tmα81 and sm-Tmβ98 (Fig. 5). Combined, these structures contain five crystallographically independent dimers, which form four unique N to C termini overlap complexes; one in the structure of sm-Tmα81 and three in that of sm-Tmβ98. In the structure of sm-Tmα81, dimer 1 forms an overlap complex with dimer 2 (Fig. 5A), whereas in the structure of sm-Tmβ98, each crystallographically independent dimer forms an overlap complex with its own symmetry mate (Fig. 5, B and C). The head-to-tail complexes observed here are similar in nature to previously reported structures (32, 44) in that they all form four-helix bundles stabilized by hydrophobic interactions. However, there are also differences in bending and in the extent of the overlap between the complexes of sm-Tmα81 and sm-Tmβ98 (Fig. 5) and with previously published structures.
FIGURE 5.
Head-to-tail overlap complexes. A, head-to-tail overlap complex formed between dimers 1 and 2 in the structure of sm-Tmα81. B, head-to-tail overlap complex formed between dimes 1 and its symmetry mate in the structure of sm-Tmβ98. C, superimposition of the three overlap complexes formed by the dimers of the structure sm-Tmβ98 and their respective symmetry mates. D and E, four-helix bundles formed by the overlap complexes of sm-Tmα81 and sm-Tmβ98 viewed along the longitudinal axis of the coiled coil. Note that the distances between the N- and C-terminal helices are similar. N-terminal residues Met1 and Met8 make part of the hydrophobic interface in all the overlap complexes characterized thus far.
In previous studies, it was concluded that the C-terminal coiled coil opens to allow insertion of the N-terminal coiled coil (32, 44). In the complexes observed here, however, the degree of opening is somewhat similar at both ends, and because the extent of the overlap is relatively short, the interaction actually occurs with very little opening (Fig. 5, D and E). In the two previously reported structures of the overlap complex of chicken smooth muscle Tmα, the extent of the overlap varied from 12 to 15 residues, and both structures were bent, albeit to different degrees (32). In the case of rat striated muscle Tmα, 11 residues formed the overlap complex, which had mostly a straight conformation (44). By comparison, in the current structure of sm-Tmβ98 the extent of the overlap is ∼8 residues (i.e. slightly more than two helical turns) for all three crystallographically independent dimers, and the conformation of the complex is mostly straight, more closely resembling that of rat striated muscle Tmα (44). In the structure of sm-Tmα81, the overlap complex involves 11 residues (i.e. three helical turns), similar to the structure of rat striated muscle Tmα (44), but the conformation is slightly bent, which is more similar to the complexes of chicken smooth muscle Tmα (32). In all the structures, hydrophobic interactions of core residues help stabilize the overlap complex, and the contact interface displays pseudo-2-fold symmetry (Fig. 5).
DISCUSSION
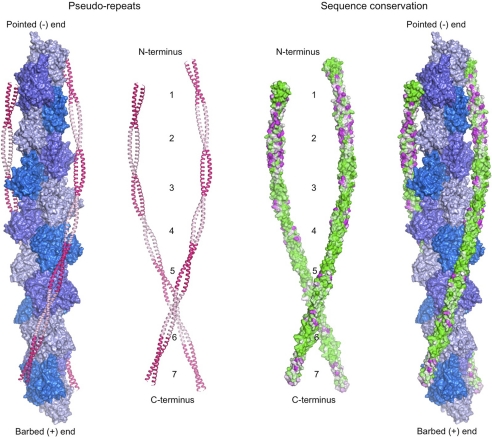
Previous structural studies have focused on skeletal muscle Tmα, such that the structural impact of sequence variation among Tm isoforms remained unexplored. An analysis of 66 Tm sequences from different species (supplemental Fig. 2) identifies pseudo-repeat 2 and the C terminus as the most variable regions among Tm isoforms. Here, we determined crystal structures of three N-terminal fragments of smooth muscle Tm isoforms α and β, including the variable region in pseudo-repeat 2. The structures show that sequence variation within this region does not generally affect the overall curvature of the Tm molecule. When sequence conservation is plotted, using the program ConSurf (45), on the surface of a full-length model of the Tm molecule derived from the crystal structures and MD simulations, sequence variation in pseudo-repeat 2 appears to be uniformly distributed on the convex (exposed) and concave (actin-bound) surfaces of the Tm molecule (Fig. 6). Closer inspection, however, reveals that residues previously identified to be directly involved in actin binding (31) are highly conserved (supplemental Fig. 2). Combined, the conservation of these residues and the preservation of an overall curvature pre-shaped to the contours of the actin filament (10) suggest that the actin-binding function is highly conserved among Tm isoforms.
FIGURE 6.
Sequence conservation and Tm pseudo-repeats. Model of full-length Tm derived from the crystal structure of sm-Tmα81 and a published model of sm-Tmα (37) using MD simulations. Tm coiled coils, bound symmetrically along the two long pitch helices of the actin filament, are colored according to pseudo-repeat spacing (left) and amino acid conservation (right). Conservation decreases from green to magenta. Actin subunits are colored along the short pitch helix using three alternating shades of blue. Conservation scores were calculated with the program ConSurf (45) and displayed with the program PyMOL (DeLano Scientific LLC). The sequence alignment used to calculate the conservation scores is shown in supplemental Fig. 2.
What then is the role of sequence variation in pseudo-repeat 2 and at the C terminus of Tm isoforms? An idea that is gaining significant traction is that the large number of Tm isoforms expressed in mammals (1) may contribute to the segregation of actin filaments into discrete networks with specialized cellular functions (3–14). At the molecular level, this idea would imply that either different Tm isoforms interact with different surfaces of the actin filament, exposing actin-binding sites in an isoform-dependent manner, or that ABPs interact specifically with certain Tm isoforms. As pointed out above, our work suggests that sequence variation among Tm isoforms may not have a significant effect on their actin-binding function, and most isoforms are likely to bind along a similar surface on actin. We thus favor a model according to which ABPs recognize sequence variation on the Tm or acto-Tm surface, by direct interaction with the two variable regions in pseudo-repeat 2 and at the C terminus (or head-to-tail complex). In other words, the two variable regions are directly responsible for the gatekeeper role of Tm and may also determine the spacing for the interaction of certain ABPs, including myosin (46), with the actin filament. Very little is still known about the preferences of ABPs for specific Tm isoforms, but one well known example is striated muscle troponin T, which has two main sk-Tmα-binding sites, including one along the variable C terminus and overlap complex (47).
The head-to-tail overlap complex emerges as one of the most variable regions of the Tm molecule, implying that no single structure is fully representative of this important linkage. Here, we observed head-to-tail complexes in the structures of the parallel dimers of sm-Tmα and sm-Tmβ. Their occurrence here was likely made possible by the lack of N- or C-terminal coiled-coil stabilizing elements. The absence of such stabilizing elements was also likely responsible for the observation here of an antiparallel Tm coiled coil in the structure of sm-Tmα98. It thus appears that the Tm coiled coil has inherent low helix-helix affinity, permitting the opening of the helices to form overlap complexes, which distinguishes it from stable coiled coils such as GCN4. Although strictly speaking the overlap complexes observed here are not native, because the C termini are truncated in these constructs, they offer important new information, particularly considering the apparent intrinsic variability and functional plasticity (53) of this linkage. Consistent with previous observations (32, 44), the overlap complexes of sm-Tmα and sm-Tmβ consist of four-helix bundles stabilized by hydrophobic interactions. Of particular interest are residues Met1 and Met8 (Fig. 5, D and E), which are highly conserved (supplemental Fig. 2) and make part of the head-to-tail interface in all the structures determined thus far. Methionine residues are recognized for their finger-like flexibility at hydrophobic interfaces (48) and could therefore help adapt the conserved N terminus to sequence variation at the C terminus. Despite general similarities, there are also significant differences in bending and in the extent of the overlap among the existing structures of head-to-tail overlap complexes. In particular, in the published structures (32, 44) the extent of the overlap varies from 12 to 15 residues, whereas the length of the overlap complexes observed here is 8 to 11 residues. Coincidentally, an overlap length of 8–11 residues was similarly suggested based on the low resolution (7 Å) structure of full-length Tm (49), and it was also predicted based on the need for consecutive Tm molecules to remain in-phase along the long pitch helix of the actin filament (50). It is finally important to note that despite the intrinsic plasticity of the overlap complex, its conservation within a particular Tm isoform appears to be critical, as evidenced by the observation that some of the mutations causing nemaline myopathy localize to this region (1, 51, 52).
Supplementary Material
Acknowledgments
Use of IMCA-CAT beamline 17-ID was supported by the Industrial Macromolecular Crystallography Association through a contract with the Hauptman-Woodward Medical Research Institute. The Advanced Photon Source was supported by the Department of Energy Contract W-31-109-Eng-38. Crystal data collection at CHESS was supported by National Science Foundation Grant DMR-0936384 and National Institutes of Health Grant RR-01646.
This work was supported, in whole or in part, by National Institutes of Health Grant P01 HL086655 (to R. D. and W. L.).

This article contains supplemental Figs. S1–S4.
The atomic coordinates and structure factors (codes 3U1A, 3U1C, and 3U59) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- Tm
- tropomyosin
- sm
- smooth muscle
- MD
- molecular dynamics
- aa
- amino acid
- ABP
- actin-binding protein
- r.m.s.d.
- root mean square deviation
- sk
- skeletal muscle.
REFERENCES
- 1. Gunning P., O'Neill G., Hardeman E. (2008) Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol. Rev. 88, 1–35 [DOI] [PubMed] [Google Scholar]
- 2. Tobacman L. S. (2008) Cooperative binding of tropomyosin to actin. Adv. Exp. Med. Biol. 644, 85–94 [DOI] [PubMed] [Google Scholar]
- 3. Gallant C., Appel S., Graceffa P., Leavis P., Lin J. J., Gunning P. W., Schevzov G., Chaponnier C., DeGnore J., Lehman W., Morgan K. G. (2011) Tropomyosin variants describe distinct functional subcellular domains in differentiated vascular smooth muscle cells. Am. J. Physiol. Cell Physiol. 300, C1356–C1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Skau C. T., Courson D. S., Bestul A. J., Winkelman J. D., Rock R. S., Sirotkin V., Kovar D. R. (2011) Actin filament bundling by fimbrin is important for endocytosis, cytokinesis, and polarization in fission yeast. J. Biol. Chem. 286, 26964–26977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tojkander S., Gateva G., Schevzov G., Hotulainen P., Naumanen P., Martin C., Gunning P. W., Lappalainen P. (2011) A molecular pathway for myosin II recruitment to stress fibers. Curr. Biol. 21, 539–550 [DOI] [PubMed] [Google Scholar]
- 6. Dominguez R. (2011) Tropomyosin. The gatekeeper's view of the actin filament revealed. Biophys. J. 100, 797–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang C. L., Coluccio L. M. (2010) New insights into the regulation of the actin cytoskeleton by tropomyosin. Int. Rev. Cell Mol. Biol. 281, 91–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lord M. (2011) Cytoskeletal regulation. Sorting out stress fibers with tropomyosin. Curr. Biol. 21, R255–R257 [DOI] [PubMed] [Google Scholar]
- 9. Martin C., Gunning P. (2008) Isoform sorting of tropomyosins. Adv. Exp. Med. Biol. 644, 187–200 [DOI] [PubMed] [Google Scholar]
- 10. Holmes K. C., Lehman W. (2008) Gestalt-binding of tropomyosin to actin filaments. J. Muscle Res. Cell Motil. 29, 213–219 [DOI] [PubMed] [Google Scholar]
- 11. Blanchoin L., Pollard T. D., Hitchcock-DeGregori S. E. (2001) Inhibition of the Arp2/3 complex-nucleated actin polymerization and branch formation by tropomyosin. Curr. Biol. 11, 1300–1304 [DOI] [PubMed] [Google Scholar]
- 12. Ishikawa R., Yamashiro S., Kohama K., Matsumura F. (1998) Regulation of actin binding and actin bundling activities of fascin by caldesmon coupled with tropomyosin. J. Biol. Chem. 273, 26991–26997 [DOI] [PubMed] [Google Scholar]
- 13. Hitchcock-DeGregori S. E. (2008) Tropomyosin. Function follows structure. Adv. Exp. Med. Biol. 644, 60–72 [DOI] [PubMed] [Google Scholar]
- 14. Gunning P., Hardeman E., Jeffrey P., Weinberger R. (1998) Creating intracellular structural domains. Spatial segregation of actin and tropomyosin isoforms in neurons. BioEssays 20, 892–900 [DOI] [PubMed] [Google Scholar]
- 15. Ali F., Paré P. D., Seow C. Y. (2005) Models of contractile units and their assembly in smooth muscle. Can. J. Physiol. Pharmacol. 83, 825–831 [DOI] [PubMed] [Google Scholar]
- 16. Seow C. Y., Fredberg J. J. (2011) Emergence of airway smooth muscle functions related to structural malleability. J. Appl. Physiol. 110, 1130–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Craig R., Woodhead J. L. (2006) Structure and function of myosin filaments. Curr. Opin Struct. Biol. 16, 204–212 [DOI] [PubMed] [Google Scholar]
- 18. Perrin B. J., Ervasti J. M. (2010) The actin gene family. Function follows isoform. Cytoskeleton 67, 630–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim H. R., Appel S., Vetterkind S., Gangopadhyay S. S., Morgan K. G. (2008) Smooth muscle signaling pathways in health and disease. J. Cell. Mol. Med. 12, 2165–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gunst S. J., Zhang W. (2008) Actin cytoskeletal dynamics in smooth muscle. A new paradigm for the regulation of smooth muscle contraction. Am. J. Physiol. Cell Physiol. 295, C576–C587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gerthoffer W. T. (2005) Actin cytoskeletal dynamics in smooth muscle contraction. Can. J. Physiol. Pharmacol. 83, 851–856 [DOI] [PubMed] [Google Scholar]
- 22. Conley C. A. (2001) Leiomodin and tropomodulin in smooth muscle. Am. J. Physiol. Cell Physiol. 280, C1645–C1656 [DOI] [PubMed] [Google Scholar]
- 23. Saito S. Y., Hori M., Ozaki H., Karaki H. (1996) Cytochalasin D inhibits smooth muscle contraction by directly inhibiting contractile apparatus. J. Smooth Muscle Res. 32, 51–60 [DOI] [PubMed] [Google Scholar]
- 24. Mehta D., Gunst S. J. (1999) Actin polymerization stimulated by contractile activation regulates force development in canine tracheal smooth muscle. J. Physiol. 519, 829–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hitchcock-DeGregori S. E., An Y. (1996) Integral repeats and a continuous coiled coil are required for binding of striated muscle tropomyosin to the regulated actin filament. J. Biol. Chem. 271, 3600–3603 [DOI] [PubMed] [Google Scholar]
- 26. Brown J. H., Kim K. H., Jun G., Greenfield N. J., Dominguez R., Volkmann N., Hitchcock-DeGregori S. E., Cohen C. (2001) Deciphering the design of the tropomyosin molecule. Proc. Natl. Acad. Sci. U.S.A. 98, 8496–8501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brown J. H., Cohen C. (2005) Regulation of muscle contraction by tropomyosin and troponin. How structure illuminates function. Adv. Protein Chem. 71, 121–159 [DOI] [PubMed] [Google Scholar]
- 28. Li Y., Mui S., Brown J. H., Strand J., Reshetnikova L., Tobacman L. S., Cohen C. (2002) The crystal structure of the C-terminal fragment of striated-muscle α-tropomyosin reveals a key troponin T recognition site. Proc. Natl. Acad. Sci. U.S.A. 99, 7378–7383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Greenfield N. J., Swapna G. V., Huang Y., Palm T., Graboski S., Montelione G. T., Hitchcock-DeGregori S. E. (2003) The structure of the carboxyl terminus of striated α-tropomyosin in solution reveals an unusual parallel arrangement of interacting α-helices. Biochemistry 42, 614–619 [DOI] [PubMed] [Google Scholar]
- 30. Minakata S., Maeda K., Oda N., Wakabayashi K., Nitanai Y., Maéda Y. (2008) Two-crystal structures of tropomyosin C-terminal fragment 176–273. Exposure of the hydrophobic core to the solvent destabilizes the tropomyosin molecule. Biophys. J. 95, 710–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li X. E., Tobacman L. S., Mun J. Y., Craig R., Fischer S., Lehman W. (2011) Tropomyosin position on F-actin revealed by EM reconstruction and computational chemistry. Biophys. J. 100, 1005–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Frye J., Klenchin V. A., Rayment I. (2010) Structure of the tropomyosin overlap complex from chicken smooth muscle. Insight into the diversity of N-terminal recognition. Biochemistry 49, 4908–4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sheldrick G. M. (2008) A short history of SHELX. Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 35. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sousa D., Cammarato A., Jang K., Graceffa P., Tobacman L. S., Li X. E., Lehman W. (2010) Electron microscopy and persistence length analysis of semi-rigid smooth muscle tropomyosin strands. Biophys. J. 99, 862–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brown J. H., Zhou Z., Reshetnikova L., Robinson H., Yammani R. D., Tobacman L. S., Cohen C. (2005) Structure of the mid-region of tropomyosin. Bending and binding sites for actin. Proc. Natl. Acad. Sci. U.S.A. 102, 18878–18883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Singh A., Hitchcock-DeGregori S. E. (2003) Local destabilization of the tropomyosin coiled coil gives the molecular flexibility required for actin binding. Biochemistry 42, 14114–14121 [DOI] [PubMed] [Google Scholar]
- 40. Tripet B., Wagschal K., Lavigne P., Mant C. T., Hodges R. S. (2000) Effects of side-chain characteristics on stability and oligomerization state of a de novo-designed model coiled coil. 20 amino acid substitutions in position “d”. J. Mol. Biol. 300, 377–402 [DOI] [PubMed] [Google Scholar]
- 41. Engh R. A., Huber R. (1991) Accurate bond and angle parameters for x-ray protein structure refinement. Acta Crystallogr. A 47, 392–400 [Google Scholar]
- 42. Lehrer S. S., Ly S., Fuchs F. (2011) Tropomyosin is in a reduced state in rabbit psoas muscle. J. Muscle Res. Cell Motil. 32, 19–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lehrer S. S., Ly S., Fuchs F. (2011) Tropomyosin is in a reduced state in rat cardiac muscle. J. Muscle Res. Cell Motil. 32, 63–64 [DOI] [PubMed] [Google Scholar]
- 44. Greenfield N. J., Huang Y. J., Swapna G. V., Bhattacharya A., Rapp B., Singh A., Montelione G. T., Hitchcock-DeGregori S. E. (2006) Solution NMR structure of the junction between tropomyosin molecules. Implications for actin binding and regulation. J. Mol. Biol. 364, 80–96 [DOI] [PubMed] [Google Scholar]
- 45. Ashkenazy H., Erez E., Martz E., Pupko T., Ben-Tal N. (2010) ConSurf 2010. Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38, W529–W533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lehrer S. S., Golitsina N. L., Geeves M. A. (1997) Actin-tropomyosin activation of myosin subfragment 1 ATPase and thin filament cooperativity. The role of tropomyosin flexibility and end-to-end interactions. Biochemistry 36, 13449–13454 [DOI] [PubMed] [Google Scholar]
- 47. Mak A. S., Smillie L. B. (1981) Structural interpretation of the two-site binding of troponin on the muscle thin filament. J. Mol. Biol. 149, 541–550 [DOI] [PubMed] [Google Scholar]
- 48. O'Neil K. T., DeGrado W. F. (1990) How calmodulin binds its targets. Sequence independent recognition of amphiphilic α-helices. Trends Biochem. Sci. 15, 59–64 [DOI] [PubMed] [Google Scholar]
- 49. Whitby F. G., Phillips G. N., Jr. (2000) Crystal structure of tropomyosin at 7 Angstroms resolution. Proteins 38, 49–59 [PubMed] [Google Scholar]
- 50. McLachlan A. D., Stewart M. (1975) Tropomyosin coiled-coil interactions. Evidence for an unstaggered structure. J. Mol. Biol. 98, 293–304 [DOI] [PubMed] [Google Scholar]
- 51. Laing N. G., Wilton S. D., Akkari P. A., Dorosz S., Boundy K., Kneebone C., Blumbergs P., White S., Watkins H., Love D. R. (1995) A mutation in the α tropomyosin gene TPM3 associated with autosomal dominant nemaline myopathy. Nat. Genet. 9, 75–79 [DOI] [PubMed] [Google Scholar]
- 52. Clarkson E., Costa C. F., Machesky L. M. (2004) Congenital myopathies. Diseases of the actin cytoskeleton. J. Pathol. 204, 407–417 [DOI] [PubMed] [Google Scholar]
- 53. Paulucci A. A., Katsuyama A. M., Sousa A. D., Farah C. S. (2004) A specific C-terminal deletion in tropomyosin results in a stronger head-to-tail interaction and increased polymerization. Eur. J. Biochem. 271, 589–600 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.