Background: The SUMO-interacting motif (SIM) mediates SUMO-dependent regulation.
Results: The structure of a SUMO1-specific SIM in complex with SUMO1 is solved, and the SIM sequence requirements are identified by peptide arrays.
Conclusion: SIMs bound in the parallel orientation have more strictly conserved sequence than those in the antiparallel orientation.
Significance: The findings will facilitate the identification of new SIMs and the design of SIM mimetics.
Keywords: NMR, Protein Dynamics, Protein-Protein Interactions, Protein-Peptide Interactions, SUMO, Sumoylation, SUMO-interacting Motif (SIM), High Affinity Binding, Paralogue Specificity
Abstract
The small ubiquitin-like modifiers (SUMOs) regulate many essential cellular functions. Only one type of SUMO-interacting motif (SIM) has been identified that can extend the β-sheet of SUMO as either a parallel or an antiparallel strand. The molecular determinants of the bound orientation and paralogue specificity of a SIM are unclear. To address this question, we have conducted structural studies of SUMO1 in complex with a SUMO1-specific SIM that binds to SUMO1 with high affinity without post-translational modifications using nuclear magnetic resonance methods. In addition, the SIM sequence requirements have been investigated by peptide arrays in comparison with another high affinity SIM that binds in the opposing orientation. We found that antiparallel binding SIMs tolerate more diverse sequences, whereas the parallel binding SIMs prefer the more strict sequences consisting of (I/V)DLT that have a preference in high affinity SUMO2 and -3 binding. Comparison of two high affinity SUMO1-binding SIMs that bind in opposing orientations has revealed common SUMO1-specific interactions needed for high affinity binding. This study has significantly advanced our understanding of the molecular determinants underlining SUMO-SIM recognition.
Introduction
Intricate protein interaction networks are responsible for almost all aspects of cellular functions. These networks are mediated by a small number of common modules, of which the ubiquitin-like proteins are a special type that can be covalently attached to other proteins enzymatically. Conjugation and deconjugation of these modules allows the cell to quickly turn on and off protein-protein interactions (1, 2). The small ubiquitin-like modifier (SUMO)5 family of proteins has recently been established as an important mechanism in regulating disease pathogenesis and many essential cellular functions (3–7). Like ubiquitination and other ubiquitin-like modifications, the attachment of SUMO to cellular proteins is catalyzed by three types of enzymes referred to generally as E1 (activation enzyme), E2 (conjugation enzyme), and E3 (ligase). SUMO modifications can be removed by a family of SUMO-specific proteases. At least three SUMO paralogues, known as SUMO1, -2, and -3, are expressed in human cells (8, 9). SUMO1 has less than 50% sequence identity with the other SUMO isoforms, and SUMO2 and -3 are nearly identical.
The SUMO-interacting (binding) motif (SIM or SBM) sequences are critical to both SUMO conjugation and SUMO-mediated effects. SIMs in E3 ligases control SUMO paralogue-specific modifications (10, 11). In addition, SIMs in substrate proteins promote SUMO modifications (12) or protect the substrates from deconjugation enzymes (13). The SIMs in receptor proteins are responsible for recognizing SUMOylated substrates (14) and thereby determining intracellular trafficking, protein-protein interactions, and localization of SUMOylated substrates. Unlike ubiquitin-mediated protein-protein interactions, which involve many different ubiquitin-binding motifs (15), only one type of SIM is prevalent in SUMO-mediated protein-protein interactions of all SUMO paralogues (14, 16). All SUMO proteins form a ubiquitin-like fold, which contains an α-helix and a β-sheet. The SIM binds to a surface between the α-helix and β-sheet and extends the β-sheet as either a parallel or antiparallel β-strand (17). The known characteristics of SIMs are that they are short (less than 10 amino acid residues) and rich in hydrophobic residues. The molecular mechanism underlining the choice of SIM bound orientation and paralogue specificity is unclear.
Previous studies have shown that a segment spanning the IR1 (internal repeat 1)-M (spacer region) IR2 (internal repeat 2) domains of the nuclear pore protein RanBP2 (Ran-binding protein 2) contains two SIMs. One SIM is located in IR1, and the other is located at the junction of the M and IR2 regions. Further analysis demonstrated that the E3 ligase activity of RanBP2 is dependent on the SIMs (10, 18). The IR1 domain could not pull down SUMO1 or SUMO2 or -3, but it could stimulate modifications involving both SUMO1 and SUMO2, suggesting that this SIM binds the SUMO proteins weakly and does not have paralogue specificity. In contrast, the M-IR2 domain only pulled down SUMO1, and not SUMO2 or -3, and only stimulated SUMO1 modifications (10), indicating its specificity and high affinity for SUMO1. The high affinity and specificity of the M-IR2 SIM to SUMO1 is independent of post-translational modifications, such as phosphorylation, as reported for the Daxx SIM (19).
Structural studies have been reported for only three SIMs, from the IR1 domain of RanBP2, PIASX (protein inhibitor of activated STAT X), and Daxx, in complex with SUMO1 (10, 19–21) and only one SIM from MCAF1 that contains a similar sequence as PIASX in complex with SUMO3 (22). Whereas the MCAF1 SIM is specific for SUMO2/3, the IR1 and PIASX SIMs do not show preferences for a specific SUMO paralogue. The Daxx SIMs were studied by two groups that reported inconsistent findings (19, 21). Thus, structural studies of the SIMs thus far have not included a high affinity SUMO1-specific SIM, for which the affinity and specificity do not depend on post-translational modifications at a site outside of the SIM.
To improve our understanding of the sequence requirements of SIMs for high affinity and high specificity interactions, we carried out NMR analysis of the structure of SUMO1 in complex with the high affinity SUMO1-specific M-IR2 SIM. We found that this SIM binds SUMO1 as an antiparallel β-strand. In addition, pull-down studies with peptide arrays were carried out for the M-IR2 SIM in order to better understand the SIM sequence requirements. Furthermore, the sequence requirements were also investigated for the parallel orientation based on the PIASX SIM, which binds SUMO1 with affinity similar to the M-IR2 SIM. Binding orientation-dependent and -independent features have been identified for high affinity interaction with SUMO1. These studies were supplemented with isothermal titration calorimetry and biochemical pull-down analyses. The molecular insights obtained here provide further understanding of the sequence requirements and mechanism underlying SUMO-SIM recognition.
EXPERIMENTAL PROCEDURES
NMR Resonance Assignments and Derivation of Structural Constraints
The 1H resonances of SUMO1-bound M-IR2 (DNEIEVIIVWEKK) SIM peptide (1:1 molar ratio) were assigned using standard homonuclear TOCSY and NOESY spectra using perdeuterated/15N-labeled SUMO1. Such a sample in 100% D2O allowed for unambiguous assignment of the 13C-1H resonances of SUMO1-complexed M-IR2 on natural abundance 13C. The 13Cα shifts were used to deduce the dihedral constraints of the bound SIM for structural calculation.
The structure determination used 13C/15N/2D-enriched SUMO1 in complex with unlabeled synthetic peptide. For the SUMO1·M-IR2 complex, a full suite of sequential assignment experiments, HNCA, HNCOCA, HNCACB, HNCOCACB, and 15N-edited NOESYHSQC, was acquired to obtain the backbone resonance assignments of SUMO1 in the complex. The assigned backbone (15N, 13C, 1H) shifts allowed for calculation of the dihedral constraints of bound SUMO1 using TALOS+ (23). NOE distance constraints used in the structure calculation were obtained from the following data sets at 250 ms mixing time: three-dimensional 15N-edited NOESY-HSQC for intermolecular NOEs and two-dimensional 1H-1H NOESY acquired for complex samples in 90% H2O/10% D2O or 100% D2O for intra-M-IR2 NOEs (Table 1).
TABLE 1.
Structural statistics of the 10 final structures (pH 6.8, 298 K)
| Root mean square deviation from the average structure (residues 20–97 of SUMO1 + residues 2708–2716 of RanBP2) (Å) | |
| Main chain | 0.84 |
| All heavy atoms | 1.10 |
| Total NOE distance constraints | 84 |
| Intra M-IR2 | 51 |
| Intermolecular | 33 |
| Total dihedral constraints | 64 |
| φ SUMO1 (M-IR2) | 21 (11) |
| ψ SUMO1 (M-IR2) | 21 (11) |
| Violations of structural constraints | |
| Distance constraints >0.5 Å | 0 |
| Distance constraints >0.1 Å | 2 |
| Dihedral angle constraints > 5° | 0 |
| Root mean square deviation from idealized covalent geometry | |
| Bonds (Å) | 0.0037 ± 0.00017 |
| Angles (degrees) | 0.65 ± 0.0083 |
| Impropers (degrees) | 0.47 ± 0.0096 |
| Ramachandran analysis | |
| Residues in allowed regions | 89% |
| Residues in marginal regions | 9.8% |
| Residues in disallowed regions | 1.3% |
Structure Calculations
The program HADDOCK (high ambiguity-driven docking) (24) was used for docking calculations for the structure of the complex between SUMO1 and the M-IR2 SIM. No ambiguous constraints were used. For the dihedral constraints, all Φ and Ψ angles predicted by TALOS for M-IR2 (residues 2706–2716) and angles of SUMO1 residues that experienced significant amide chemical shift changes upon binding the M-IR2 SIM (regions 20–27 and 33–56) were used. Similarly, the side chains of these regions were allowed to be flexible in the docking calculation. The HADDOCK calculation started with rigid body dockings of 1000 structures, of which 200 were further refined, and the 10 with the lowest energy were chosen for analysis.
NMR Studies to Extract Binding Affinities
Two-dimensional 15N-1H HSQC NMR spectra of SUMO1 or -2 (100–200 μm) were recorded at 298 K at each incremental titration of the SIM peptide (0.6–2 mm) until no additional chemical shift changes were observed. We extracted the binding dissociation constant (Kd) by integrating line shapes of the one-dimensional 15N slices of the two-dimensional 15N-1H HSQC spectra of SUMO at each titration point and fitted them to the Bloch-McConnell equations for two-state chemical exchange (free versus bound) using the LineShapeKin package (25).
Peptide Array Immunoblotting
Peptide arrays were synthesized using SPOT technology as described previously (26). The peptide array membrane was blocked with 5% skim milk in TBST (0.1 mm Tris-HCl, pH 7.4, 150 mm NaCl, and 0.1% Tween 20) for 1 h. For the PIASX-SIM peptide array, GST-SUMO1 or GST-SUMO3 protein was added directly to the blocking buffer to a final concentration of 1 μg/ml and incubated with the membrane at room temperature for 1 h. The membrane was then washed three times for 5 min each with TBST before a rabbit anti-GST antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) was added. The membrane was allowed to incubate at room temperature for 30 min prior to three 5-min washes with TBST. A goat anti-rabbit GST-horseradish peroxidase conjugate was then added and incubated with the membrane for another 30 min. After three final 5-min washes in TBST, the peptide array membrane was visualized by enhanced chemiluminescence.
For peptide array based on the M-IR2 SIM, competitive SUMO1 and SUMO3 binding was used as described below. Peptide arrays were first treated with blocking buffer (LI-COR) for 2 h and then incubated with SUMO1 and SUMO3 (10 μg/ml) in PBS overnight. The array was then washed three times with 500 mm NaCl in phosphate buffer (pH 7.5) to remove unbound SUMO. Subsequently, mouse anti-SUMO1 (1:1000; Abgent) and rabbit anti-SUMO2/3 (1:2000; Abcam) were incubated with the array for 2 h at room temperature. Unbound antibodies were removed by three washes in PBST. Secondary antibodies (donkey anti-mouse CW800 and donkey anti-rabbit CW680 (1:2000; LI-COR) were incubated with the array for 1 h at room temperature. After three washes with PBST, the array was imaged using the Odyssey imaging system (LI-COR).
Immunocytochemistry
MCF-7 cells were maintained in DMEM (Invitrogen) supplemented with 10% (v/v) fetal bovine serum (Irvine Scientific), 1 mm sodium pyruvate, nonessential amino acids, and 100 μg/ml penicillin/streptomycin (Irvine Scientific) and Normocin (InvivoGen). Cells were transfected using Lipofectamine 2000 (according to the manufacturer's instructions) with pcDNA-FLAG-SIM1-GFP and pcDNA-FLAG-SIM2-GFP in an 8-well chamber slide. After 48 h of transfection, cells were fixed and permeabilized. SIM1 and SIM2 fusion proteins were recognized by anti-FLAG M2 (monoclonal, 1:1000; Stratagene) and FITC-conjugated anti-mouse antibodies. Rabbit anti-SUMO1 (1:200; Boston Biochem), rabbit anti-SUMO2/3 (1:1000; Abcam), and Texas Red-conjugated donkey anti-rabbit (1:200; Jackson ImmunoResearch) antibodies were used to determine the localization of SUMO1 and SUMO2/3 in cells. Cells were imaged using the LSM510 confocal microscope.
RESULTS
Structure of Highly Specific SUMO1-binding SIM in Complex with SUMO1
As discussed in the Introduction, the M-IR2 domain of RanBP2 harbors a high affinity SIM that is specific to SUMO1 in the absence of a post-translational modification. We conducted solution structural studies of this SIM in complex with SUMO1. The boundary of the SIM spans the segment DNEKECIIVWEKK (residues 2705–2717), identified from chemical shift perturbation analysis of 13C/15N-labeled M-IR2 domain in complex with SUMO-1 and confirmed by a synthetic peptide (supplemental Table S1). In addition, substituting Cys in this segment with a Val to avoid potential complications due to oxidation of the Cys side chain -SH group did not change the SUMO1 binding affinity and specificity (supplemental Fig. S1). We also substituted the first N-terminal Lys of this peptide to an Ile (K2708I) in order to resolve resonance overlap. Consistent with the biochemical pull-down data (supplemental Fig. S1), the K2708I peptide did not alter binding specificity to SUMO1, as indicated by the nearly identical SUMO NMR spectra between SUMO bound to K2708I and to the wild-type SIM (supplemental Figs. S2 and S3). Measurement of the binding affinity by isothermal titration calorimetry was not successful due to a small enthalpy change, suggesting an entropy-driven mechanism for the recognition. Thus, using NMR chemical shift perturbation and line shape analysis, the Kd was estimated to be ∼1.8 μm for SUMO1 and 22 μm for SUMO2, consistent with the high affinity and specificity of M-IR2 for SUMO1 (Fig. 4C and supplemental Table S1). Structure determination was carried out using the K2708I peptide (DNEIEVIIVWEKK).
FIGURE 4.
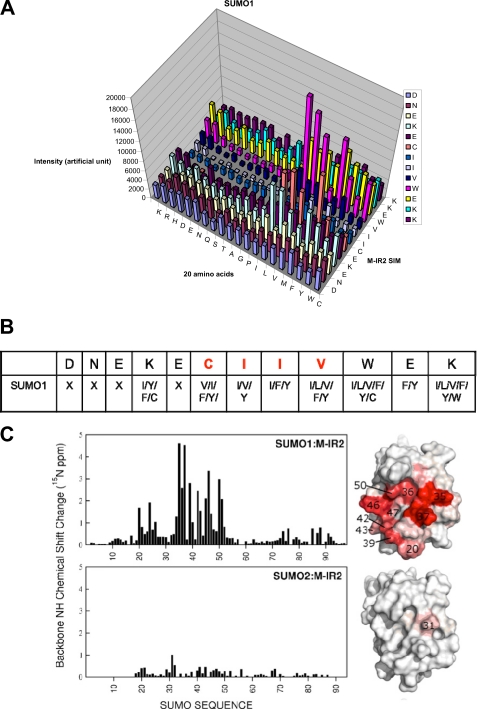
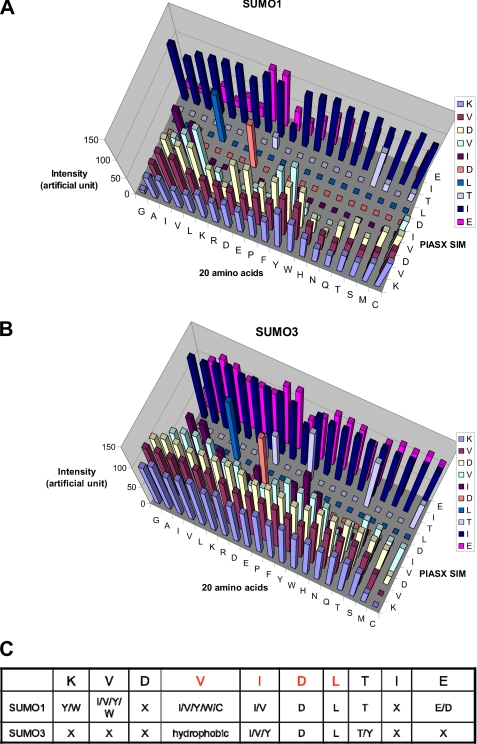
Sequence requirements of the M-IR2 SIM. A, quantification of SUMO1 interactions with peptides from a peptide array based on the M-IR2 SIM (which binds in the antiparallel orientation). The SIM sequence is listed vertically with the N terminus at the bottom, and the 20 amino acid substitutions are listed horizontally. The intensity of each spot indicates the binding affinity toward SUMO, with taller bars indicating stronger interaction. B, summary of the amino acid preferences for binding SUMO1. X, no particular residue type is preferred. Amino acid residues are indicated by their one-letter codes. Red type indicates conserved residues. C, chemical shift perturbation analysis for the SUMO1-specific M-IR2 SIM with SUMO1. Changes in the backbone amide (15N-1H) chemical shift of SUMO are mapped on the surface representations of SUMO on the right and indicated by a white to red gradient to indicate zero to significant changes.
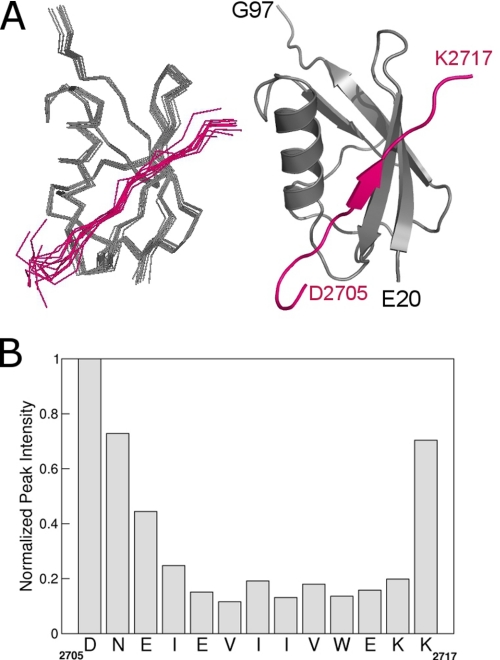
The bound SIM showed a well defined conformation in the middle of the peptide, with flexible termini, as shown by superimposition of the 10 best structures (Fig. 1A and Table 1). This is consistent with changes of line widths and chemical shifts of the 1Hα and 13Cα resonances between the free and bound peptide (Fig. 1B). Residues Asp-2705, Asn-2706, and Lys-2717 had sharp resonances, suggesting that they undergo fast dynamics and do not form tight interactions with SUMO1 (Fig. 1B). However, the negatively charged residues Asp-2705 to Glu-2707 at the N terminus are adjacent to a positively charged patch on the SUMO1 surface, and thus, these residues probably contribute to the electrostatic interactions.
FIGURE 1.
Solution structure of SUMO1 bound to a SUMO1-specific SIM. A, backbone Cα trace (left) of the structural ensemble and ribbon representation (right) of SUMO1 (gray) bound to the SIM peptide (magenta) from the M-IR2 region of RanBP2. B, peak intensity of the backbone 13Cα-1Hα NMR resonances of the bound SIM showing differential line-broadening effects due to formation of the SIM-SUMO1 complex. SIM residues that undergo fast dynamics have sharper resonances and higher peak intensities than those that are tightly bound to SUMO1.
Molecular Details of SUMO1-SIM Interactions
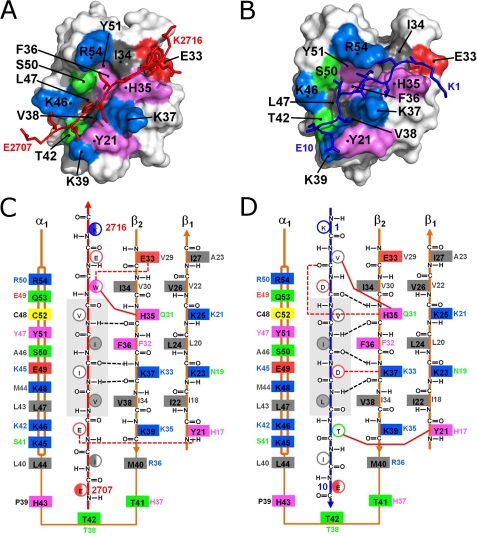
In order to identify features that contribute to the high affinity SUMO1 binding, the structure was compared with that of another high affinity SUMO1-binding SIM that bound in the opposing orientation, the SIM from PIASX, as we previously characterized (16). Both the M-IR2 and PIASX SIMs have Kd values in the low micromolar range for binding SUMO1, and their affinities for SUMO1 are among the strongest of the characterized SIMs. Both SIM peptides bind between the α-helix and a β-strand of SUMO1, but they extend the β-sheet in opposing orientations (Fig. 2). The antiparallel binding M-IR2 SIM backbone displayed an extended conformation, whereas the parallel binding PIASX SIM showed a curved backbone that appeared to accommodate optimal side chain contacts. This comparison indicates that there are key differences in the binding mechanisms for these two high affinity SIMs.
FIGURE 2.
Structural insights into SIM-SUMO1 interactions of two high-affinity SIMs bound in opposing orientations. A and B, surface representations of SUMO1 in complex with the M-IR2 (A) and PIASX (B) SIMs. The interacting residues in SUMO1 are colored according to amino acid type: positive (blue), negative (red), aromatic (violet), hydrophobic (gray), and polar (green). The red and dark blue stick models represent the bound M-IR2 and PIASX SIMs, respectively. C and D, schematic of the interactions between SUMO1 and the M-IR2 (C) and PIASX (D) SIMs, illustrating paralogue- and/or orientation-specific interactions. The two SIMs extend the β-sheet of SUMO1 in opposing orientations, antiparallel for M-IR2 and parallel for PIASX. The side chains of the SIMs are shown as circles, with open circles representing side chains pointing to the surface and filled circles representing side chains pointing toward the interior of the SIM binding groove on SUMO. The multiple conformations of Trp and the flexible terminal residues in the M-IR2 SIM are represented as half-filled circles. Listed beside each side chain of the SUMO1 residues (solid boxes) is the corresponding residue in SUMO2. All residues are colored as described in A and B. Black dashed lines indicate backbone hydrogen bonds between the SUMO1 β-strand and SIMs; red dashed lines represent hydrogen bonds or salt bridges involving side chains. Solid red lines represent key side chain interactions discussed in the “Results” section for Molecular Details of SUMO1-SIM Interactions. The central core region of the SIMs is shaded in gray.
The 2710VIIV2713 segment of the M-IR2 SIM and 4VIDL7 segment of the PIASX SIM form interactions with the same residues on SUMO1, which include residues 35–39 on the β-strand and residues 46, 47, 50, and 54 on the α-helix (Fig. 2). The hydrophobic SUMO residues Phe-36 and Leu-47 form the bottom of the binding groove with residues Lys-37, Lys-46, and Ser-50 forming the sides. For both SIMs, three notable backbone hydrogen bonds are formed between the core residues of the SIMs and residues His-35 and Lys-37 of the SUMO1 β-strand. Ile-5 and Leu-7 of the parallel-binding SIM (PIASX SIM) form contacts with the hydrophobic groove of SUMO1 that consists of the side chains of Phe-36 and Leu-47, whereas Val-2710 and Ile-2712 of the antiparallel binding SIM (M-IR2 SIM) form equivalent contacts with SUMO1. Asp-6 of the PIASX SIM and Ile-2711 of M-IR2 point toward the surface, but Asp-6 forms a salt bridge with Lys-37, whereas the hydrophobic Ile-2711 does not. Both Val-4 (PIASX) and Val-2713 (M-IR2) interact with His-35. As shown at the top of Fig. 2, C and D, Trp-2714 (M-IR2) and Val-2 (PIASX) both interact with His-35 of SUMO1. His-35 also interacts with a hydrophobic residue in the SIM-N of Daxx (21), although it does not appear to participate in binding the SIM-C of Daxx (19), which was reported to undergo dynamic exchange between parallel and antiparallel bound orientations (21).
The β-strand adjacent to the β-strand that forms the SIM-binding groove contributes to binding of both SIMs. Glu-2709 (M-IR2) forms a hydrogen bond with the aromatic -OH group of SUMO1 Tyr-21, but the equivalent H17 side chain of SUMO2/3, which lacks the -OH group, may not form this hydrogen bond with the Glu-2709 side chain (Fig. 2C). In the PIASX SIM, which binds all SUMO paralogues with similar affinities, the T8 methyl group forms hydrophobic interactions with the SUMO1 Tyr-21 aromatic ring (Fig. 2D) (16). His-17 of SUMO2/3 is expected to form similar interactions with Thr-8. Similar contacts involving Tyr-21 were not observed in the Daxx SIM-C complex with SUMO1, which is the only other SUMO1-SIM structure available to date (19, 21).
Flexibilities at SUMO-SIM Interface
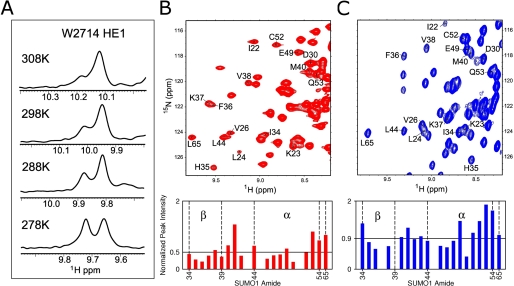
NMR resonances contain information regarding conformational flexibilities. The interface of the parallel M-IR2 SIM in complex with SUMO1 is more flexible than that of the antiparallel PIASX SIM in complex with SUMO1. Upon forming a complex with SUMO1, a key interacting residue, Trp-2714 of the M-IR2 SIM peptide, displayed two resonances for the aromatic side chain epsilon HN, indicating that this side chain is in at least two distinct conformational states that are in exchange on a time scale slower than the NMR chemical shift (Fig. 3A). This is not due to exchange between the parallel and antiparallel orientations, as observed for the Daxx SIM (21), because other SIM residues did not produce two sets of resonances, and no evidence of another bound orientation was observed in intermolecular NOEs. The intensity of each peak was proportional to the population of each conformational state, and their relative ratios changed with temperature, consistent with the temperature-based modulations of exchange kinetics and population between the different conformational states due to changes of the energy barrier (Fig. 3A). Additionally, more severe line-broadening effects of SUMO1 resonances were observed when SUMO1 was in complex with the M-IR2 SIM than with the PIASX SIM (Fig. 3, B and C) (16). The line-broadening effect on SUMO1 upon binding the M-IR2 SIM was most pronounced on the β-strand residues His-35, Phe-36, and Lys-37. Because both SIMs of M-IR2 and PIASX have Kd values in the low micromolar range for binding SUMO1, the enhanced broadening by the M-IR2 SIM indicates additional dynamic processes on the microsecond to millisecond time scale that were absent in the PIASX SIM. Retaining conformational flexibility in the complex can contribute to the high affinity interaction of the M-IR2 SIM with SUMO1 by reducing the entropic cost of complex formation.
FIGURE 3.
Conformational dynamics in the SUMO1 interaction with SIMs bound in parallel and antiparallel orientations. A, temperature dependence of the 1H NMR peaks of the two conformers of the side chain tryptophan amine group of SUMO1-bound M-IR2 SIM. B and C, top panels show sections of the two-dimensional 15N-1H HSQC spectra at 298 K of SUMO1 saturated by SIMs of M-IR2 (red) and PIASX (blue). The labeled residues represent part of the SIM recognition region on SUMO1, except for Leu-65 (L65), which is not at the binding interface and was used as an internal control for relative peak intensity analysis. The selective broadening of resonances encompassing the recognition α-helix and β-sheet of SUMO1 was normalized against Leu-65 and is shown in the bar charts. Differential peak intensities reflect differences in line-broadening effects, due to chemical exchange dynamics on the microsecond to millisecond time scale. Missing bars indicate overlapped resonances. The average peak intensity is depicted by horizontal lines.
Examination of SIM Sequence Requirements by Peptide Array Analysis
To identify key interactions and examine the SIM sequence requirement, we conducted peptide arrays based on the M-IR2 SIM (2705DNEKECIIVWEKK2717), for which each SIM residue was replaced with each of the 20 naturally occurring amino acid residues. To investigate the SUMO paralogue specificity, SUMO1 and SUMO3 were used in competition for binding the peptide array, followed by simultaneous identification of bound SUMO1 and SUMO3 by their respective antibodies (Fig. 4A and supplemental Fig. S5). However, the peptides in the array did not bind SUMO2/3 well, suggesting that single amino acid substitution did not lead to enhanced SUMO2/3 binding. The peptide array results were validated by introducing mutations into the entire M-IR2 domain, followed by pull-down experiments (supplemental Fig. S1). In addition, a series of peptides were synthesized, and their interactions with either SUMO1 or SUMO2 were investigated by NMR studies (supplemental Table S1 and Fig. S4). This array did not show a strict sequence requirement of the M-IR2 SIM. The essential requirement is that the central segment of five residues, WVIIV, be made of bulky hydrophobic or aromatic residues with propensities to form β-strands (27) (Fig. 4B). These residues contact the key residues in SUMO1, including Phe-36, Lys-37, Lys-46, Leu-47, and Ser-50 (Fig. 2C). The specificity of the M-IR2 SIMs for SUMO1 is also demonstrated by the much larger chemical shift perturbation generated upon binding SUMO1 than binding SUMO3 (Fig. 4C).
A similar peptide array experiment was conducted for the PIASX SIM, which binds SUMO1 in the opposing orientation. Because the PIASX SIM (1KVDVIDLTIE10) binds to all SUMO paralogues with similar affinities (14, 17), the peptide array was synthesized in two identical copies on cellulose membranes (28); one array was used for binding SUMO1, and the other was used for binding SUMO3. Each position of the PIASX SIM (vertical sequences, Fig. 5, A and B, and supplemental Fig. S6) was substituted by each of the 20 amino acid residues (horizontal sequences). Because SUMO2 and -3 have nearly identical amino acid sequences in the SIM-binding surface, SUMO3 was used as a representative of the two. Binding of GST-tagged SUMO1 or SUMO3 was detected by an anti-GST Western blot, and the relative binding affinities are indicated by the signal strength (Fig. 5, A and B, and supplemental Fig. S6). The peptide array results were validated by measuring the binding affinities of synthetic peptides to SUMO1 and SUMO2 by isothermal titration calorimetry (supplemental Table S2 and supplemental Fig. S7). In striking contrast to the M-IR2 SIM, residues in the middle of the PIASX SIM were very restricted, with the Asp-6 and Leu-7 residues being strictly required for high affinity binding to both SUMO1 and SUMO2/3 (Fig. 5C). This array has identified the sequence requirements of SUMO2/3-specific SIM. Substitution of Val-2 of the PIASX SIM by a Gly or an Ala significantly reduced SUMO1 but not SUMO2 binding affinity (Fig. 5 and supplemental Fig. S6). The sequence requirements for binding SUMO-2/3, (I/V)DLT, identified by peptide array, have been validated by recent findings of SUMO2/3-specific SIM sequences (11, 12, 22).
FIGURE 5.
Sequence Requirements of the PIASX SIM. Quantification of SUMO1 (A) or SUMO3 (B) interactions with peptides from a peptide array based on the PIASX SIM (which binds in the parallel orientation). The SIM sequence is listed vertically with the N terminus at the bottom, and the 20 amino acid substitutions are listed horizontally. The intensity of each spot defines the binding affinity of the mutant to SUMO, with taller bars indicating stronger interaction. C, summary of the amino acid preferences for binding SUMO1 and SUMO3 by the PIASX SIM. X, no particular residue type is preferred. Amino acid residues are indicated by their one-letter codes. Red type indicates conserved residues.
SUMO Paralogue-specific SIMs Confer Paralogue Specificity in Cells
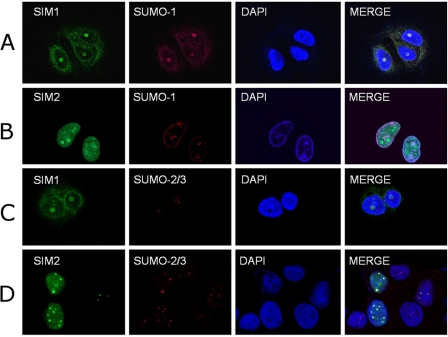
To examine the cellular SUMO paralogue specificity of the SIMs identified from the studies described above, colocalization of the SIMs with SUMO1 and SUMO2/3 was investigated in cells. MCF-7 cells were transfected with pcDNA-FLAG-SIM1-GFP and pcDNA-FLAG-SIM2-GFP, for which SIM1 corresponds to the SUMO1-specific SIM (M-IR2) used for structure determination discussed above, and SIM2 corresponds to the Val-2 to Gly mutation of the PIASX SIM that demonstrated specificity for SUMO2/3. The SIM1 and SIM2 fusion proteins were identified with anti-FLAG M2 and FITC-conjugated anti-mouse antibodies, whereas rabbit anti-SUMO1, rabbit anti-SUMO2/3, and Texas Red-conjugated donkey anti-rabbit antibodies were used to identify SUMO-1 and SUMO2/3. The subcellular localizations of the SUMO paralogues are distinct (29); SUMO1 mainly associates with the nuclear membrane and nucleoli, whereas SUMO2 and -3 localize to PML nuclear bodies and the nucleoplasm. The paralogue-specific SIMs specifically colocalized with SUMO1 and SUMO2/3 in these locations in MCF-7 cells. SIM1 was mainly recruited to the nuclear membrane and nucleoli, whereas SIM2 colocalized strongly with nuclear foci that contained SUMO2, some of which also contained SUMO1 (Fig. 6). In addition, in 293T cells, a cell line with faster proliferation, ∼50% of SIM1 was also present in the nucleoplasm. The difference in SIM1 localization in rapidly proliferating cells was probably due to dynamic SUMO1 localization at different points in the cell cycle (30, 31). In contrast to SIM1, the subcellular localization of SIM2 in 293T and MCF-7 cells was similar. Taken together, these results suggest that the paralogue-specific SIMs identified by the studies described above confer the same paralogue specificity in cells and can be used to probe SUMO paralogue-specific protein-protein interactions, as shown previously (32).
FIGURE 6.
In vivo SUMO paralogue specificity of the SIMs. A and B, colocalization of FLAG-tagged and GFP-fused SUMO1-specific SIM (SIM1; green) (A) and SUMO2/3-specific SIM (SIM2; green) (B) with SUMO1 (red) in MCF-7 cells. The SIM1 used is identical to that used for the structure determination (Fig. 1). SIM2 is the V2G mutant of the PIASX SIM. C and D, colocalization of SIM1 (green) (C) or SIM2 (green) (D) with SUMO2/3 (red) in MCF-7 cells. A–D, nuclear DNA was stained with DAPI (blue). Scale bars, 5 μm.
DISCUSSION
Insights into Orientation-dependent SIM Sequence Requirements
The combination of structural analysis and peptide arrays described here has revealed striking differences in the sequence requirements of the SIMs bound in the opposing orientations. This is probably due to the fact that antiparallel β-strand orientation usually has a more favorable hydrogen bond geometry than the parallel β-strand. That the antiparallel M-IR2 SIM tolerates more diverse amino acid sequences than the parallel PIASX SIM indicates that backbone hydrogen bonds, as well as the β-strand propensity of the SIM residues (27), make more significant contributions than specific side chain interactions to formation of the complex. In contrast, the side chain interactions between the PIASX SIM and SUMO1 appear to make more significant contributions to binding than the M-IR2 SIM, as suggested by the more strict sequence requirements. The importance of side chain interactions in the SUMO1 complex with the PIASX SIM relative to that with the M-IR2 SIM is also evident from the structures, in which each of the PIASX SIM side chains fits well into a specific pocket, whereas SIM backbone “twists” to accommodate the side chain contacts (Fig. 2B). In contrast, the backbone of the M-IR2 SIM appears to be more linear and “relaxed” (Fig. 2A). The less restricted SIM residues in the M-IR2 SIM as compared with the PIASX SIM are also consistent with the greater conformational flexibility observed at the interfaces of SUMO1 with M-IR2 than with PIASX.
Common Features for High Affinity SUMO1 Binding
Comparison of the structures of SUMO1 in complexes with the M-IR2 and PIASX SIMs, which both bind to SUMO1 with similarly high affinities, has also suggested key interactions for high affinity SUMO1 recognition. Although the M-IR2 and PIASX SIMs bind in opposing orientations, the interaction of Trp-2714 of M-IR2 or Val-2 of PIASX with His-35 of SUMO1 is required for SUMO1 binding affinity. As shown by peptide arrays, substitution of Val-2 by bulky and hydrophobic residues (Ile, Tyr, and Trp) is required for maintaining the binding affinity for SUMO1 but not for SUMO2 (Fig. 5 and supplemental Fig. S6). Similarly, substitution of Trp-2714 by residues that are not bulky and hydrophobic resulted in severe reduction of the binding affinity to SUMO1 (Fig. 4). Because His-35 is not homologous among the SUMO paralogues, the findings suggest that it is a key residue for specificity and affinity for SUMO1. All SUMO2/3-specific SIM sequences identified to date contain the highly conserved (V/I)DLT sequence and not the hydrophobic residue required for interactions with His-35 (11, 12, 22).
Paralogue Specificity of SUMO-SIM Interactions
Although the M-IR2 SIM has preference for SUMO1, it also binds SUMO2 or -3 but with 10-fold lower affinity (supplemental Figs. S2–S4). The central residues of the M-IR2 SIM bind the same SUMO1 core residues, including Phe-36, Lys-37, Lys-46, Leu-47, and Ser-50, that also participate in binding other SIMs, regardless of the bound orientation. These residues are identical across the SUMO paralogues. Thus, it is expected that the SIMs with preference for one SUMO paralogue can also bind another paralogue although with a lower affinity. The 10-fold difference of the M-IR2 SIM for binding SUMO1 compared with SUMO2 or -3 corresponds to ∼1.4 kcal/mol in free energy. Such a difference can result from factors not detected in the structures. For example, formation of a SUMO-SIM complex probably involves the removal of water molecules bound to the hydrophobic surface of a SUMO protein. If the number of water molecules removed from SUMO1 and SUMO2 or -3 differs by one, the difference in the free energy of the interaction can be as much as 2 kcal/mol (33), more than enough to account for the observed difference in specificity. Further studies are required to understand the differences of the different SUMO paralogues.
In summary, this study has significantly improved our understanding of the molecular mechanism underlying SIM sequence requirements and preferences for SUMO1. The M-IR2 SIM confers SUMO1 specificity in cells, and the V2G substitution of the PIASX SIM confers SUMO2/3 specificity in cells. Thus, the molecular insights obtained can form the basis for the design of SUMO paralogue-specific reagents that will be useful for cellular studies and future medical applications.
Supplementary Material
Acknowledgment
We thank Dr. Weidong Hu for assistance with NMR data acquisition.
This work was supported, in whole or in part, by National Institutes of Health Grants R01GM074748 and R01GM086171 (to Y. C.) and F32CA134180 (to Y. L.).

This article contains supplemental Tables S1 and S2 and Figs. S1–S6.
The atomic coordinates and structure factors (code 2LAS) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
All 1H, 13C, and 15N chemical shifts for the complex of SUMO1 and M-IR2 peptide are deposited in the Biological Magnetic Resonance Bank (http://www.bmrb.wisc.edu) under BMRB accession number 17536.
- SUMO
- small ubiquitin-like modifier
- SIM
- SUMO-interacting motif
- SIM1
- SUMO1-specific SIM from M-IR2
- SIM2
- SUMO2/3-specific V2G mutant SIM from PIASX
- NOE
- nuclear Overhauser effect.
REFERENCES
- 1. Varshavsky A. (1997) The ubiquitin system. Trends Biochem. Sci. 22, 383–387 [DOI] [PubMed] [Google Scholar]
- 2. Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 3. Kerscher O., Felberbaum R., Hochstrasser M. (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22, 159–180 [DOI] [PubMed] [Google Scholar]
- 4. Martin S., Wilkinson K. A., Nishimune A., Henley J. M. (2007) Emerging extranuclear roles of protein SUMOylation in neuronal function and dysfunction. Nat. Rev. Neurosci. 8, 948–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ulrich H. D. (2009) Preface. Ubiquitin, SUMO, and the maintenance of genome stability. DNA Repair 8, 429. [DOI] [PubMed] [Google Scholar]
- 6. Mo Y. Y., Yu Y., Theodosiou E., Ee P. L., Beck W. T. (2005) A role for Ubc9 in tumorigenesis. Oncogene 24, 2677–2683 [DOI] [PubMed] [Google Scholar]
- 7. Meulmeester E., Melchior F. (2008) Cell biology. SUMO. Nature 452, 709–711 [DOI] [PubMed] [Google Scholar]
- 8. Hay R. T. (2001) Protein modification by SUMO. Trends Biochem. Sci. 26, 332–333 [DOI] [PubMed] [Google Scholar]
- 9. Müller S., Hoege C., Pyrowolakis G., Jentsch S. (2001) SUMO, ubiquitin's mysterious cousin. Nat. Rev. Mol. Cell Biol. 2, 202–210 [DOI] [PubMed] [Google Scholar]
- 10. Tatham M. H., Kim S., Jaffray E., Song J., Chen Y., Hay R. T. (2005) Unique binding interactions among Ubc9, SUMO, and RanBP2 reveal a mechanism for SUMO paralog selection. Nat. Struct. Mol. Biol. 12, 67–74 [DOI] [PubMed] [Google Scholar]
- 11. Chang P. C., Izumiya Y., Wu C. Y., Fitzgerald L. D., Campbell M., Ellison T. J., Lam K. S., Luciw P. A., Kung H. J. (2010) Kaposi's sarcoma-associated herpesvirus (KSHV) encodes a SUMO E3 ligase that is SIM-dependent and SUMO-2/3-specific. J. Biol. Chem. 285, 5266–5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Meulmeester E., Kunze M., Hsiao H. H., Urlaub H., Melchior F. (2008) Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol. Cell 30, 610–619 [DOI] [PubMed] [Google Scholar]
- 13. Zhu S., Goeres J., Sixt K. M., Békés M., Zhang X. D., Salvesen G. S., Matunis M. J. (2009) Protection from isopeptidase-mediated deconjugation regulates paralog-selective sumoylation of RanGAP1. Mol. Cell 33, 570–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Song J., Durrin L. K., Wilkinson T. A., Krontiris T. G., Chen Y. (2004) Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 14373–14378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hicke L., Schubert H. L., Hill C. P. (2005) Ubiquitin-binding domains. Nat. Rev. Mol. Cell Biol. 6, 610–621 [DOI] [PubMed] [Google Scholar]
- 16. Song J., Zhang Z., Hu W., Chen Y. (2005) Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif. A reversal of the bound orientation. J. Biol. Chem. 280, 40122–40129 [DOI] [PubMed] [Google Scholar]
- 17. Song J., Park J. K., Lee J. J., Choi Y. S., Ryu K. S., Kim J. H., Kim E., Lee K. J., Jeon Y. H., Kim E. E. (2009) Structure and interaction of ubiquitin-associated domain of human Fas-associated factor 1. Protein Sci. 18, 2265–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Truong K., Su Y., Song J., Chen Y. (2011) Entropy-driven mechanism of an E3 ligase. Biochemistry 50, 5757–5766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chang C. C., Naik M. T., Huang Y. S., Jeng J. C., Liao P. H., Kuo H. Y., Ho C. C., Hsieh Y. L., Lin C. H., Huang N. J., Naik N. M., Kung C. C., Lin S. Y., Chen R. H., Chang K. S., Huang T. H., Shih H. M. (2011) Structural and functional roles of Daxx SIM phosphorylation in SUMO paralog-selective binding and apoptosis modulation. Mol. Cell 42, 62–74 [DOI] [PubMed] [Google Scholar]
- 20. Reverter D., Lima C. D. (2005) Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature 435, 687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Escobar-Cabrera E., Okon M., Lau D. K., Dart C. F., Bonvin A. M., McIntosh L. P. (2011) Characterizing the N- and C-terminal small ubiquitin-like modifier (SUMO)-interacting motifs of the scaffold protein DAXX. J. Biol. Chem. 286, 19816–19829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sekiyama N., Ikegami T., Yamane T., Ikeguchi M., Uchimura Y., Baba D., Ariyoshi M., Tochio H., Saitoh H., Shirakawa M. (2008) Structure of the small ubiquitin-like modifier (SUMO)-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3. J. Biol. Chem. 283, 35966–35975 [DOI] [PubMed] [Google Scholar]
- 23. Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) TALOS+. A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dominguez C., Boelens R., Bonvin A. M. (2003) HADDOCK. A protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 125, 1731–1737 [DOI] [PubMed] [Google Scholar]
- 25. Kovrigin E. L., Loria J. P. (2006) Enzyme dynamics along the reaction coordinate. Critical role of a conserved residue. Biochemistry 45, 2636–2647 [DOI] [PubMed] [Google Scholar]
- 26. Li S. S., Wu C. (2009) Using peptide array to identify binding motifs and interaction networks for modular domains. Methods Mol. Biol. 570, 67–76 [DOI] [PubMed] [Google Scholar]
- 27. Chou P. Y., Fasman G. D. (1978) Prediction of the secondary structure of proteins from their amino acid sequence. Adv. Enzymol. Relat. Areas Mol. Biol. 47, 45–148 [DOI] [PubMed] [Google Scholar]
- 28. Frank R. (2002) The SPOT-synthesis technique. Synthetic peptide arrays on membrane supports. Principles and applications. J. Immunol. Methods 267, 13–26 [DOI] [PubMed] [Google Scholar]
- 29. Ayaydin F., Dasso M. (2004) Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol. Biol. Cell 15, 5208–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dasso M. (2008) Emerging roles of the SUMO pathway in mitosis. Cell Div. 3, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Joseph J., Liu S. T., Jablonski S. A., Yen T. J., Dasso M. (2004) The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr. Biol. 14, 611–617 [DOI] [PubMed] [Google Scholar]
- 32. Li Y. J., Stark J. M., Chen D. J., Ann D. K., Chen Y. (2010) Role of SUMO:SIM-mediated protein-protein interaction in non-homologous end joining. Oncogene 29, 3509–3518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dunitz J. D. (1994) The entropic cost of bound water in crystals and biomolecules. Science 264, 670. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.