Abstract
Differential gene transcription is a fundamental regulatory mechanism of biological systems during development, body homeostasis, and disease. Comparative genomics is believed to be a rapid means for the identification of regulatory sequences in genomes. We tested this approach to identify regulatory sequences that control expression in the midline of the zebrafish embryo. We first isolated a set of genes that are coexpressed in the midline of the zebrafish embryo during somitogenesis stages by gene array analysis and subsequent rescreens by in situ hybridization. We subjected 45 of these genes to Compare and DotPlot analysis to detect conserved sequences in noncoding regions of orthologous loci in the zebrafish and Takifugu genomes. The regions of homology that were scored in nonconserved regions were inserted into expression vectors and tested for their regulatory activity by transient transgenesis in the zebrafish embryo. We identified one conserved region from the connective tissue growth factor gene (ctgf), which was able to drive expression in the midline of the embryo. This region shares sequence similarity with other floor plate/notochord-specific regulatory regions. Our results demonstrate that an unbiased comparative approach is a relevant method for the identification of tissue-specific cis-regulatory sequences in the zebrafish embryo.
Differential gene transcription is essential for many developmental processes and physiological responses. Moreover, evolution of the body plan is thought to have been driven to a large extent by changes in gene expression, and differences in gene expression are believed to determine disease risk (Davidson 2001;Ludwig 2002 and references therein). A detailed knowledge of the cis-regulatory architecture of genomes is thus a central prerequisite for a comprehensive understanding of many fundamental biological processes and has also medical implications.
Despite enormous progress in our understanding of transcriptional regulation in vertebrates (Davidson 2001), we are not able to unequivocally predict the pattern and responsiveness of expression by mere sequence analysis of a gene. This is largely because of the structure and the scattered location of cis-regulatory regions in the huge vertebrate genomes (Davidson 2001). In vertebrates, promoter regions are rarely sufficient to drive faithful gene transcription. Frequently, other regulatory regions contribute to the correct spatial and temporal expression of genes. These regions can be scattered over large distances in noncoding sequences, and they can be located upstream, downstream, and in intronic regions (Davidson 2001). Regulatory regions are usually a composite of multiple transcription-factor-binding sites. The small size and degenerate DNA sequence of these binding sites renders a genome-wide computational analysis very difficult as not only the binding sequence but also the context in which it is placed plays an important role. Delineation of the regulatory architecture of individual loci by reverse genetics and functional assays is tedious and costly. The elucidation of the cis-regulatory wiring of vertebrate genomes by conventional assays will be a formidable task despite the fact that the genome sequences are available.
Comparison of genomes of related species was shown to provide a possible shortcut in the identification of regulatory regions as they form islands of partially conserved sequence in noncoding regions (Hardison 2000;Wasserman et al. 2000;Pennacchio and Rubin 2001). Within the vertebrate lineage, the teleost genomes such as those of pufferfish (Takifugu rubripes), medaka (Oryzias latipes), and zebrafish (Danio rerio) represent promising tools to elucidate the regulatory architecture in mammalian genomes as these genomes have significantly diverged to eliminate functionally irrelevant sequence identities (Aparicio et al. 1995;Muller et al. 2002).
Several studies reported conservation of regulatory sequences in comparisons of mammalian and Takifugu loci (Aparicio et al. 1995;Goode et al. 2003). Takifugu has a very small, compact genome (Aparicio and Brenner 1997). However, Takifugu cannot be maintained in a laboratory environment, preventing characterization of gene expression patterns and functional analysis of putative regulatory regions. In contrast, medaka and zebrafish allow large-scale expression profiling as well as functional assessment of regulatory sequences by transgenesis (Muller et al. 2002). Fast transient transgenic assays in the zebrafish allow scanning large genomic regions for regulatory activity very efficiently (Westerfield et al. 1992;Muller et al. 1999). As in the Takifugu genome, regulatory regions appear to be highly conserved between zebrafish and mammals. For example, three of the five functionally mapped regulatory regions of the zebrafish Sonic hedgehog (shh) locus are structurally highly related in mouse and human shh, and four of the five regulatory regions are conserved in zebrafish and Takifugu shh orthologs (Muller et al. 1999, 2002; Goode et al. 2003; R. Ertzer and U. Strähle, unpubl.). Also, the three functionally characterized regulatory regions of the neurogenin1 (ngn1) gene showed high similarity to the 5′ noncoding region of the mammalian counterpart of ngn1 (Blader et al. 2003). The two enhancers residing in the noncoding sequences between the two distalless genes dlx4 and dlx6 have also highly related sequences in the mouse orthologs (Zerucha et al. 2000). These regions of conservation are in the range of 100-400 bp and have an overall sequence identity between 60% and 80%. Moreover, the orientation and relative position of these elements with respect to one another and the coding sequence were retained, indicating a high selection pressure not only for the regulatory sequences themselves but also for the position of the elements within the locus. Several of these enhancers were tested by transgenesis in zebrafish and mouse demonstrating conserved regulatory function (Muller et al. 1999, 2002;Zerucha et al. 2000). Taken together, the results from the analysis of this albeit limited number of genes indicate that the comparisons of the genome of the zebrafish with that of other vertebrates also have a high potential to elucidate the location of regulatory regions.
Because the number of zebrafish genes whose regulatory structure has been analyzed in depth is rather small, it remains to be seen whether this high conservation of regulatory sequences of teleost genomes is indeed a general phenomenon and can be exploited as a rapid approach to elucidate the regulatory structure of genomes. In fact, there is evidence that regulatory sequences in zebrafish genes were not maintained structurally during evolution. The structure of an enhancer that was mapped to the second intron of the zebrafish netrin1 gene and that mediates floor plate expression in zebrafish has not been conserved (Rastegar et al. 2002;data not shown). Similarly, a regulatory region in the first intron of shh that drives expression in the floor plate of zebrafish embryos is not conserved in the mammalian shh orthologs (Muller et al. 2002).
To assess the wider applicability of comparative approaches to the zebrafish genome sequence, we carried out a systematic search for homologies in nonconserved regions of the zebrafish and Takifugu loci and tested the regulatory activity of these regions by transient transgenesis in the zebrafish. We chose these two teleost genomes to carry out the comparison as we expected a higher yield of conserved sequences within the teleost lineage. We first identified genes expressed in the embryonic midline comprising the axial mesoderm, notochord, and prechordal plate. In addition to providing mechanical support, these midline structures have important regulatory activity that controls cell differentiation in the adjacent neural tube, endoderm, and somites (Pourquie et al. 1993;Placzek 1995;Tanabe and Jessell 1996). We characterized 152 genes that fall into diverse functional groups, ranging from genes involved in signaling, transcriptional regulation, cytoarchitecture, and cell metabolism. Among these, we identified 85 genes that are specifically expressed in the midline. In the compared 45 genes, we found 18 conserved noncoding regions. One homology from the connective tissue growth factor (ctgf) gene drove expression in the midline in injected zebrafish embryos. We demonstrate here that this approach in the zebrafish does allow isolation of regulatory regions. Comparisons of the ctgf region with midline enhancers, which were identified previously, revealed several short sequence stretches, indicating shared protein-binding sites.
RESULTS
Preparation of a Complex Probe From Midline Cells
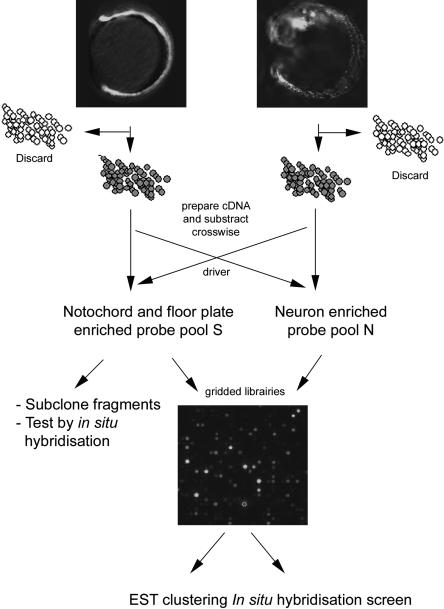
To isolate a representative set of genes expressed in the midline of zebrafish embryos, we used a subtraction strategy that was previously used to identify genes acting downstream from Nodal signals in the gastrula (Dickmeis et al. 2001a). This approach entails several steps of enrichment and selection to minimize the scale of the final screening by in situ hybridization (Fig. 1). We used transgenic embryos that express the green fluorescent protein (gfp) gene under control of the regulatory elements of the shh gene (Albert et al. 2003). At the 6-10-somite stage, these transgenic embryos express the gfp reporter gene in the notochord, the prechordal plate, and in the ventral midline of the neural plate comprising the prospective floor plate and hypothalamus (Fig. 1). Several hundred shh:gfp embryos were dissociated into single cells at the 6-10-somite stage, and GFP-positive cells were enriched with a fluorescence-activated cell sorter (FACS). As a second further enrichment, we carried out a cDNA subtraction using cDNA prepared from the shh:gfp cells as tester cDNA. The driver cDNA was prepared from the GFP-positive cells sorted from a second transgenic line that expressed GFP under control of the ngn1 gene (-8.4ngn1:gfp;Blader et al. 2003). GFP marks primary neurons and is also weakly detectable in the epidermis of these embryos at the 6-10-somite stage (Fig. 1). The forward subtraction (S) of the two cDNA pools normalized the abundance of transcripts and enriched midline-specific genes (Fig. 1). The reverse subtraction (N) was carried out as a control.
Figure 1.
Strategy to enrich for midline genes. Transgenic embryos carrying the -2.2shh:gfpABC (left) or the -8.4ngn1:gfp transgene (right) were dissociated, and GFP-positive cells were sorted with a fluorescence-activated cell sorter. cDNA was prepared from GFP-positive cells and subtracted crosswise before analysis of subclones and microarray hybridization. The complex probe S is enriched for midline specific genes, whereas the reverse subtraction (probe N) serves as a hybridization control.
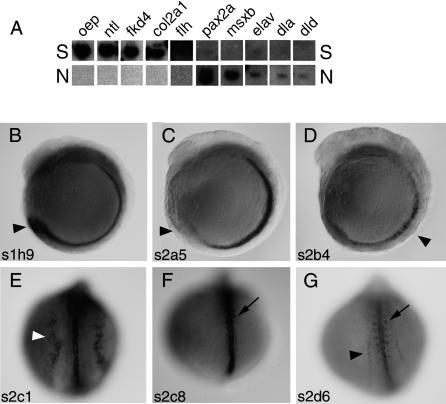
As a first assessment of the distribution of cDNAs in the two resulting probe pools, S and N, we performed Southern blot analysis with a series of genes whose expression patterns are known (Fig. 2A). The pool S that was expected to contain genes expressed in the midline hybridized strongly to cDNAs such as one-eyed pinhead (oep), no tail (ntl), floating head (flh), collagen 2α1, and forkhead4 (fkd4) that are expressed in the midline of the zebrafish embryo (Schulte-Merker et al. 1992;Talbot et al. 1995; Yan et al. 1995;Odenthal and Nusslein-Volhard 1998;Gritsman et al. 1999). Much weaker or no signals were detected with probes that are derived from genes expressed predominantly in the neuroectoderm such as pax2a (Krauss et al. 1991), elav (Kim et al. 1996), msxB (Ekker et al. 1997), deltaA (dla), and deltaD (dld) (Haddon et al. 1998). The reverse pattern of hybridization signals was obtained when the probe pool N was hybridized: The neuroectoderm-specific cDNAs hybridized strongly, whereas the midline genes gave rise to only weak signals (Fig. 2A, bottom row, N). These results indicate that the subtraction resulted in the anticipated enrichment of genes. As a second evaluation, cDNA fragments from the S pool were subcloned and 37 fragments were sequenced (GenBank Accession numbers CD777505-CD777543). These 37 clones corresponded to 33 genes, indicating a low level of redundancy in the complex probe. Next, in situ hybridization was carried out to assess the tissue-specific expression of individual S clones. Of the clones, 40% were expressed in midline tissues including the prechordal plate-derived hatching glands (Fig. 2B-G;data not shown). Taken together, these tests indicate that the subtraction successfully enriched clones that are expressed in the midline.
Figure 2.
Assessment of S and N probes. (A) Southern blot analysis of subtracted probe pools. (S) Forward subtraction (shh:gfp vs. ngn1:gfp line); (N) reverse subtraction (ngn1:gfp vs. shh:gfp line). Blotted cDNAs are oep (one-eyed pinhead), ntl (no tail), fkd4, (forkhead 4), col2a1 (collagen type II α-1), flh (floating head), pax2b, msxb, elav, dla (δA), dld (δD). (B-G) Expression pattern of clones of the S subtraction. (B) Clone s1h9, zebrafish EST fc02c07.y1, shows expression in the notochord and the tail bud (arrowhead). (C) Clone s2a5, zebrafish EST fm63h08.x1, is expressed in the notochord, but excluded from the tail bud (arrowhead). (D) Clone s2b4 is expressed in the notochord (arrowhead). (E) Clone s2c1, procollagen C-terminal enhancer protein 2, is expressed in the notochord and the edge of the lateral plate mesoderm (white arrowhead). (F) Clone s2c8, apolipoprotein H, shows expression in the notochord and the adaxial edge of the somites (arrow). (G) Clone s2d6, zebrafish EST fm91b12.x1, is expressed in the notochord and both the edge of the lateral plate mesoderm (arrowhead) and the adaxial part of the somites (arrow). (B-D) Lateral view, dorsal right; (E-G) dorsal view, anterior up.
Macroarray Analysis
As a next step, the S and N probes were hybridized to a macroarray containing 25,102 cDNAs from 24-h-old whole embryos and adult liver cDNA (MPMGp609; Clark et al. 2001). This library contains a minimally redundant clone set derived from initially 75,000 clones by normalization with oligonucleotide fingerprinting (Clark et al. 1999). Hybridizations with the two complex probes were carried out in triplicate to membranes with two distinct arrangements of spotted clones. The resulting signals were averaged, and the differential intensity of each signal was calculated. A total of 662 cDNAs with a 1.8-fold or higher representation in the hybridizations with the S probe in comparison to the N probe were chosen for further analysis. The cDNAs were sequenced (GenBank accession numbers CD777544-CD778191), and the subsequent BLAST analysis revealed that 487 clones represented 152 genes with homologs in the protein database and that 93 clones were without such homologs. On average, every gene was represented 3.2 times in the set of 487 clones that yielded homology scores in the databases. Large differences in abundance of clones representing the same gene existed, with cathepsin l being the biggest cluster (55 clones) and 78 genes being represented only by one clone.
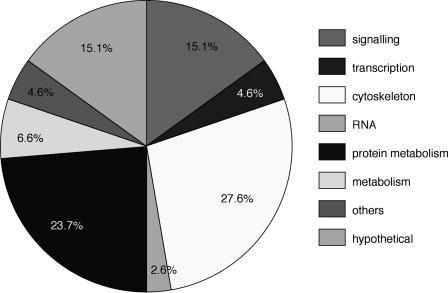
Grouping the genes with homologies according to functional aspects (Fig. 3) revealed that the largest classes are the cytoskeletal/cell adhesion/extracellular matrix (27.6%) and the protein metabolism group (23.7%), followed by proteins in cell signaling (15.1%) and proteins of unknown function (15.1%). Smaller groups contain proteins involved in transcription (4.6%) and RNA metabolism (2.6%), and 4.6% of genes fall into various other functional categories.
Figure 3.
Sequence-deduced, putative function of isolated midline genes. The identified midline genes fall into several groups with different functions ranging from cytoskeleton/cell adhesion/extracellular matrix genes to genes involved in regulatory processes and cell metabolism.
Groups of Coexpressed Genes Identified by In Situ Hybridization
Several of the identified genes were shown previously to be expressed in the axial mesoderm. These include shh (Krauss et al. 1993), no tail (Schulte-Merker et al. 1992), semaphorin Z2 (Halloran et al. 1998), calreticulin (Rubinstein et al. 2000), treb5 (Liang et al. 2001), col2a1 (Yan et al. 1995), calymmin (Cerda et al. 2002), cathepsin l (Mangos et al. 2000), hsp47 (Lele and Krone 1997), and brain subtype creatine kinase (Dickmeis et al. 2001b). To assess the pattern of expression of the remaining clones, an in situ hybridization screen with the entire initial set of cDNA clones was performed. The in situ analysis provided an independent line of proof for the clustering of the clones, assuming that the clones assigned to the same cluster yield the same pattern of expression. Expression was analyzed at 8- and 18-somite stages, and in situ hybridization was carried out using an in situ hybridization robot.
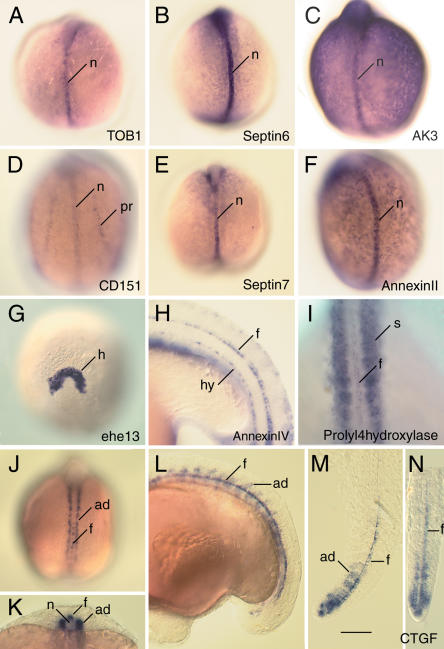
Altogether 68.5% of the genes showed tissue-restricted expression. The most frequently expressing tissue is the notochord, with 47% of the genes at the 6-10- somite stage (Table 1;Fig. 4;Supplemental material) and 33% at the 16-20-somite stage (Table 1;Supplemental material;data not shown). Expression was also frequently detected in the polster, a derivative of the prechordal plate that gives rise to the hatching glands in older stages. At the 6-10-somite and 16-20-somite stages, 30% and 29% of genes, respectively, were expressed in this tissue. Altogether, the genes expressing in axial tissues (notochord, polster) is 55.9% at the 6-10-somite stage and 53.3% at the 16-20-somite stage. At lower frequency, expression was noted in floor plate (11%) and hypochord (7%). Expression in these tissues was only scored in 16-20-somite-stage embryos for technical reasons and because the hypochord has not completely formed at early stages (Eriksson and Lofberg 2000). Several genes whose transcripts were detectable in axial tissues were also expressed in the somites (Fig. 4I;data not shown), in the pronephros (Fig. 4D), or ubiquitously at low levels (Fig. 4C). The connective tissue growth factor (ctgf) gene is expressed in adaxial cells of the somites and the floor plate at the 8-somite stage (Fig. 4J) and 18-h-old embryos (Fig. 4L). These domains of expression become restricted to the tail at 30 hours post-fertilization (hpf;Fig. 4M,N).
Table 1.
In Situ Expression Patterns of Identified Clones
| Pattern groups | 6–10 somite | 16–20 somite |
|---|---|---|
| Hatching glands | 46 (30.3%) | 44 (28.9%) |
| Notochord | 63 (47.4%) | 51 (33.6%) |
| Floor plate | n.d. | 17 (11.2%) |
| Hypochord | n.a. | 11 (7.2%) |
| Somites | 21 (13.8%) | 22 (14.5%) |
| Lateral plate mesoderm/pronephros | 6 (3.9%) | 6 (3.9%) |
| Nervous system/sensory organs | 9 (5.9%) | 13 (8.5%) |
| Sum of axial structuresa | 85 (55.9%) | 81 (53.3%) |
Hatching glands and/or notochord/floor plate/hypochord
Figure 4.
Representative RNA expression patterns of identified midline genes. (A) transducer of erbB-2 1 (Tob1). (B) septin6. (C) adenylate kinase 3 α like 1 (AK3). (D) CD151/tetraspanin (CD151). (E) septin7. (F) annexin II. (G) Homolog of the medaka hatching enzyme ehe 13. (H) annexin IV. (I) prolyl-4- hydroxylase. (J-N) connective tissue growth factor (ctgf). (A-G,I,J) Eight-somite-stage embryos; (H,L,N) 16-18-somite-stage embryos. Orientation of embryos is anterior up, dorsal view(A-G,I,J). Scale bar: 50 μm (A-G,J,L); 25 μm (H,K,M,N). (ad) Adaxial cells; (f) floor plate; (h) hatching gland; (hy) hypochord; (n) notochord; (pr) pronephros; (s) somites.
Comparative Sequence Analysis of Homologous Loci in the Takifugu Genome
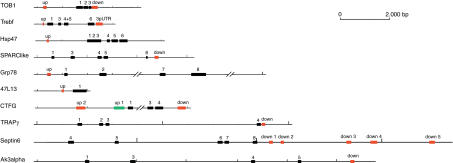
To delineate the putative regulatory sequences that are responsible for expression in the midline of the zebrafish embryo, the genomic sequence of the genes that are expressed in the midline were compared with the orthologous loci of the Takifugu genome. The Takifugu gene with the highest sequence similarity at the protein level was assumed to represent the ortholog, and only those genes (45 genes total) for which an unequivocal score was obtained in the annotated gene set of the Takifugu genome were analyzed further. For each locus, the coding region with the intronic sequences, 10 kb upstream of the ATG and 10 kb downstream from the stop codon, were searched for homologies with the “Compare” and “DotPlot” programs (GCG Wisconsin package). The settings of the search algorithm were kept at the default values, which allowed scoring the regulatory sequences of the zebrafish ngn1 (Blader et al. 2003) and shh genes (Muller et al. 1999, 2000;Albert et al. 2003). Among 45 pairwise comparisons, 18 homologies were detected in the noncoding regions of 10 genes (Fig. 5;Table 2). The size of these regions varied from ∼75 to 680 bp, and the degree of similarity ranged from 63.9% to 88.9% (Table 2). The positions of the regions of homology were found upstream and downstream of the coding regions (Fig. 5).
Figure 5.
Results from DotPlot searches for homologous noncoding sequences by comparison of zebrafish and Takifugu orthologs. The conserved coding regions (black boxes) and the position of homologous noncoding sequence blocks (colored boxes) are illustrated. The genes are oriented with the 5′-end to the left. The region up1 of the ctgf genethat drives expression in the midline when inserted in reporter constructs is highlighted in green.
Table 2.
Summary of Transient Analysis of Conserved Noncoding Sequences
| Gene | Homology region | % identity (length in base pairs) | Tested | Vector | Expression |
|---|---|---|---|---|---|
| Hsp47 | Upstream | 72.1% (87) | + | Net/TK | – |
| Trebf | Upstream | 69.4% (89) | + | Net/TK | – |
| 3′-UTR | 69.3% (405) | + | Net/TK | – | |
| 47L13 | Upstream | 74.7% (82) | + | Net/TK | – |
| TOB1 | Upstream | 78.4% (125) | + | Net/TK | – |
| Downstream | 88.9% (228) | – | – | – | |
| CTGF | Upstream 1 | 63.9% (680) | + | Net/TK | Notochord/floor plate |
| Upstream 2 | 66.5% (252) | + | Net/TK | – | |
| Downstream | 76.9% (313) | – | – | – | |
| Septin 6 | Downstream 1 | 71.9% (95) | + | Net | – |
| Downstream 2 | 74.0% (102) | – | – | – | |
| Downstream 3 | 79.6% (102) | – | – | – | |
| Downstream 4 | 82.9% (82) | – | – | – | |
| Downstream 5 | 67.7% (291) | + | Net | – | |
| SPARC | Downstream | 65.1% (171) | – | – | – |
| Trapγ | Downstream | 88.0% (75) | + | Net | – |
| Adenylate Kinase 3 | Downstream | 65.2% (121) | – | – | – |
| GRP78 | Upstream | 73.0% (165) | – | – | – |
Functional Analysis of Identified Homology Regions
To test whether these regions of homology mediate gene expression in the zebrafish embryo, we inserted PCR fragments spanning the regions of homology downstream from a reporter gene comprising the Herpes simplex virus thymidine kinase TATA-box directing expression of a green fluorescent reporter (gfp) gene (Rastegar et al. 2002). This expression vector works well in transient expression experiments and has a very low basal activity without inserted additional regulatory sequences (Rastegar et al. 2002). We inserted conserved fragments also downstream from an netrin1:gfp reporter gene. The promoter of the netrin1 gene is weakly active in the neural tube of the zebrafish and was used previously to map the regulatory sequences of the netrin1 locus (Rastegar et al. 2002).
The activity of the homology regions was tested by a transient expression protocol (Westerfield et al. 1992; Muller et al. 1999;Rastegar et al. 2002). This protocol provides a fast means to identify sequences with a regulatory potential in the midline of the zebrafish embryo, even though the reporter expression in the G0 embryos is highly mosaic. We analyzed 10 homologies downstream from the netrin1 and/or tk promoters in 24-h-old embryos (Table 2). The homology region up1 from the ctgf gene drove expression of the tk:gfp reporter in notochord and floor plate cells (18%, n = 85;Fig. 6B). It was less efficient downstream from the netrin1 expression system (data not shown). The floor plate activity matches the expression pattern of the endogenous ctgf gene (Fig. 4J-N), indicating that this region is a bona fide cis-regulator of the endogenous ctgf gene. However, we did not detect expression of the endogenous ctgf gene in the notochord. This implies that a repressor element, which prevents expression in the notochord, is missing in our constructs.
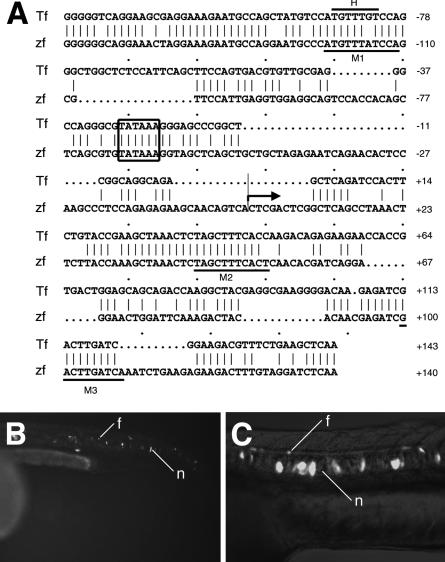
Figure 6.
The up1 homology of the ctgf gene mediates midline expression in the zebrafish. (A) Comparison of the up1 homology from the Takifugu and the zebrafish ctgf gene. A putative TATA box and the end of the zebrafish ctgf cDNA are indicated. Underlined sequences represent three regions (M1 to M3) partially conserved in the floor plate/notochord enhancers ar-A, ar-B, and ar-C of shh and the FPE of netrin1. H indicates a putative FoxA2/Axial/HNF3β binding site. (B) Transient transgenic embryo expressing GFP under control of the up1 region from the tk:gfp cassette. (C) Transient transgenic embryo expressing GFP from the ctgf:gfp transgene. (f) Floor plate; (n) notochord.
The up1 homology of the ctgf gene is located immediately upstream of and overlaps with the first exon of the gene. This indicates that this region comprises the promoter of the gene. In agreement, a well-conserved TATA-box is located 69 bp upstream of the reported 5′-end of the mRNA (Fig. 6A). We thus inserted this region upstream of the gfp reporter gene in place of the netrin1 or tk promoters. We detected strong expression in the notochord (41%, n = 100) and floor plate (31%, n = 100) of 24-h-old embryos injected with the ctgf:gfp construct (Fig. 6C). These results indicate that the up1 region includes the promoter of the ctgf gene. This region seems to function, however, also as an enhancer because it can also act in a downstream position.
The other homology regions tested did not yield significant expression in injected embryos (Table 2;data not shown). Either these regions do not have transcription regulatory activity in the 24-h-old embryo or they are not active in the specific context of our reporter constructs as they are, for example, repressor elements. It is also possible that they have functions other than transcription regulation (Glazko et al. 2003;Mistry et al. 2003).
The Regulatory Region of the ctgf Gene Shares Homology With Other Floor Plate/Notochord Enhancers
We searched the ctgf up1 region for potential binding sites of known regulators of midline development (Rastegar et al. 2002). We found an FoxA2 (Axial, HNF3β) binding site (Transfac database) at position -121, which is, like the TATA-box, highly conserved between the Takifugu and zebrafish ctgf sequences.
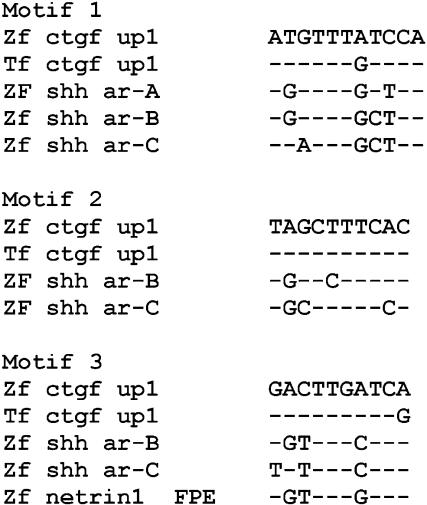
It can be assumed that genes with similar expression profiles use similar sets of regulatory factors. Several regulatory regions with activity in the floor plate or notochord were previously identified in the zebrafish shh and netrin1 genes (Muller et al. 1999;Rastegar et al. 2002;Albert et al. 2003). We compared the sequence of the ctgf up1 region with the floor plate/notochord enhancers ar-A, ar-B, and ar-C of the shh, and the FPE of the netrin1 gene (Figs. 6A and 7). A conserved motif (M1) is located at the position of the putative HNF3β-binding site. Related sequences were found in all three shh enhancers (Fig. 7). In particular, the homology to M1 resides in a region (subregion H1) of ar-C that is necessary for function (Muller et al. 2002; F. Müller and U. Strähle, unpubl.). Two other sequence stretches (M2 and M3) in ctgf up1 were found in the shh and netrin enhancers. The latter has similarity with the M1 motif.
Figure 7.
Conserved sequence motifs in the up1 region of ctgf. The sequence of up1 of zebrafish (zf) and Takifugu (Tf) ctgf; ar-A, ar-B, ar-C of zebrafish shh and the FPE of zebrafish netrin1 were searched for related sequence stretches using the MEME algorithm. Three conserved motifs (M1 to M3) were detected.
DISCUSSION
Here we have tested the applicability of the zebrafish genome in comparative searches for cis-regulatory elements. We identified 85 genes that are expressed in midline tissues comprising notochord, prechordal plate, and floor plate. In a second step, we used the previously observed conservation of regulatory regions between orthologous loci in zebrafish and Takifugu. This approach was appealing as it has the advantage of yielding an enrichment of regulatory sequences over predicted nonfunctional sequences and was expected to facilitate detection of shared transcription-factor-binding sites. In a search of 45 genes expressed in the midline, we found 10 genes with significant homologies. One of these conserved regions directed gene expression in the midline of the zebrafish embryo. Our results demonstrate that the unbiased approach of comparing between orthologous loci of the zebrafish and Takifugu genomes can yield regulatory elements.
The Embryonic Midline Expresses Collagens and Collagen-Processing Enzymes
The zebrafish midline depends on the activity of Nodal signals (Feldman et al. 1998). We have previously carried out a screen for Nodal-dependent genes at the early gastrula stage (Dickmeis et al. 2001a). At this early stage, the embryonic fate of blastomeres is induced but not irreversibly determined as heterotopic transplantation can reprogram cell fate according to the new location (Ho and Kimmel 1993). In contrast, many cells including the cells of the body axis have become committed to a cell fate irreversibly by the 6-10-somite stage. When one compares the distribution of the gene functions in the two screens, several important changes can be noted that reflect this commitment to a differentiated state. Whereas the relative number of genes involved in transcription regulation decreased by 5.7-fold, the abundance of genes with a role in the cytoskeleton, cell adhesion, and extracellular matrix increased 2.5-fold in the somitogenesis gene set. Similarly, genes involved in protein metabolism are twice as abundant in the midline of the 6-10-somite stage embryos in comparison with early gastrula embryos expressing the constitutively active form of the Nodal receptor TARAM-A (Fig. 3;Dickmeis et al. 2001a). Genes with a putative function in cell communication were equally abundant in both screens.
When examined for specific biochemical pathways, several collagens (col1a1,col2a1, col5a1, col9a1, col9a2, col9a3, and col11a1) and collagen-processing enzymes were detected in the somitogenesis but not in the gastrula stage screen. The processing genes include procollagen-lysine; 2-oxoglutarate 5-dioxygenase 1; prolyl-4-hydroxylase;the peptidyl-prolyl isomerase; fk506 binding protein;and the heat-shock proteins hsp47 and glucose regulated protein 78 (grp78). The abundance of these enzymes in the midline is likely to reflect the mechanical support function of the notochord for the early embryo.
The Promoter Region of the ctgf Gene Controls Floor Plate and Notochord Expression
The ctgf gene encodes a secreted protein, which is a member of the CCN family (Brigstock et al. 2003). Knockout mice exhibit skeletal dysmorphisms and have reduced cell proliferation, extracellular matrix remodeling, and impaired angiogenesis during chondrogenesis (Ivkovic et al. 2003). CTGF was implicated as a modulator of TGFβ signaling (Abreu et al. 2002). It inhibits the activity of BMP signals and can enhance signalling by low levels of TGF-β1. In this respect, expression of zebrafish ctgf in the floor plate is intriguing as the differentiation of the floor plate in zebrafish depends on the TGFβ related Nodal factor Cyclops (Rebagliati et al. 1998;Sampath et al. 1998;Muller et al. 2000). Moreover, BMPs are known inhibitors of floor plate differentiation (Liem et al. 1995). ctgf may thus play also a crucial role in the differentiation of this structure.
The conserved up1 region confers notochord and floor plate expression to the tk:gfp reporter gene. The region overlaps with the first exon of the ctgf gene and is thus likely to represent the promoter of this gene. In agreement, it harbors a conserved TATA-box 69 bp upstream of the 5′-end of the isolated ctgf cDNA and is able to direct expression of gfp from ctgf:gfp constructs. The related region of human ctgf directs expression in tissue culture cells (Eguchi et al. 2001), providing further support for the notion that up1 has promoter activity. However, our results indicate that it can also act at a distance, as it directed midline expression of the Tk promoter when inserted downstream of the reporter transcription unit.
The endogenous ctgf gene is not expressed at detectable levels in the notochord. This expression in the notochord in the transient expression assays used here appears thus to be an ectopic activity of the up1 region. This has previously been noted also for the promoter region of the tiggywinkle hedgehog (twhh) gene (Du et al. 1997). Like ctgf, twhh mRNA is present in the floor plate of zebrafish embryos. However, when the twhh promoter was inserted upstream of a reporter gene, also notochord expression was observed. This indicates that repressor elements are a common feature of floor plate genes and function to prevent notochord expression. These are predicted to be absent from the up1 fragment.
Several features in the up1 fragment are shared with previously identified notochord/floor plate enhancers. Motif1 overlaps with an FoxA2/HNF3β/Axial DNA-binding site (Strähle et al. 1993; Chang et al. 1997). Knockdown of FoxA2 by antisense morpholinos abolishes floor plate differentiation, indicating a crucial role of FoxA2 in floor plate development in the zebrafish (Rastegar et al. 2002). The fact that FoxA2 sites are present in floor plate enhancers of shh (Chang et al. 1997;Müller et al. 1999), netrin1 (Rastegar et al. 2002), and ctgf indicates that FoxA2 is involved in the control of medial floor plate genes in general. It is, however, unlikely to be sufficient for the control of these genes (Strähle et al. 1996). The other conserved sequence stretches may bind additional factors that cooperate with FoxA2.
Zebrafish/Takifugu Locus Comparisons Permit Detection of Regulatory Elements
We reasoned that comparisons within the teleost lineage would be a most efficient way to detect conserved regulatory elements. Zebrafish and Takifugu are separated by only 100 million years of independent evolution, whereas 400-450 million years elapsed since the last common ancestor of fish and mammals. Ten genes from the tested 45 genes showed at least one homology, and one homology (of 10 tested homologies from eight genes) was functional in our assay system. Our transient test system may have failed to detect regulatory activity in particular in the case of repressors or regulatory regions that function only in the context of other sequences in the locus of origin. Conserved regions can also represent sequences that are involved in other processes such as, for example, replication or matrix attachment.
The frequency with which we detected homologous sequences is lower than expected from the previous analysis of a small sample of zebrafish genes (Zerucha et al. 2000;Hans and Campos-Ortega 2002;Muller et al. 2002;Blader et al. 2003). The regulatory sequences in at least 35/45 genes have evolved in the Takifugu and zebrafish genomes to such an extent that they cannot be detected by straight sequence comparisons. Most of the regulatory regions of the shh and ngn1 genes are strongly conserved in the Takifugu, mouse, and human genomes and can easily be scored using the same parameters and algorithms. We thus regard it as unlikely that we failed to detect homologous noncoding sequences for technical reasons. We rather believe that the inability to detect significant homologies in a large proportion of the genes analyzed is a reflection of the low structural conservation of the regulatory regions. Similar observations were made in comparisons of the mammalian and Takifugu interleukin locus (Loots et al. 2000). Obviously regulatory regions evolve at different speeds in different loci and species. In these cases, the DNA-binding sites have presumably been strongly rearranged, or sequences between the sites have changed substantially so that they escape detection by algorithms using pairwise alignment of long sequences.
Thus other strategies have to be used to elucidate the cis-regulatory architecture of the zebrafish in a comprehensive fashion. It is possible to search the genome for clusters of known binding sites of transcription factors as used in the study of transcriptional networks in Drosophila (Markstein et al. 2002). However, in many instances the transcription factors involved in cell-specific gene expression are not known. Prior identification of short conserved sequence stretches using the approach described here and subsequent search for clusters of these sequences in coexpressed genes may provide a possible avenue into identification of further, less well-conserved regulatory regions. Our results suggest that Takifugu is not a good model for identifying regulatory regions at a large scale in the zebrafish genome. The genomic sequence of a more closely related teleost may be more helpful in this respect.
In summary, however, our work shows that a comparative search for regulatory sequences using the Takifugu and zebrafish genomic sequences permits identification of regulatory sequences. The central question is why the regulatory sequences of certain genes are more conserved than others. It is intriguing that the zebrafish genes whose regulatory sequences were shown to be conserved so far are developmental key regulators (Zerucha et al. 2000;Hans and Campos-Ortega 2002;Muller et al. 2002;Blader et al. 2003;this report). Also, comparison of the Takifugu regulatory genes such as, for example, hoxb-4, wnt1, sox9, scl, and shh genes, with those of mammals revealed structurally and functionally conserved regulatory elements (Aparicio et al. 1995; Gellner and Brenner 1999;Bagheri-Fam et al. 2001;Barton et al. 2001;Goode et al. 2003). Maybe requirements for precise spatial localization of binding sites are more important in such genes than they are in genes acting further downstream in differentiation and maintenance pathways.
METHODS
Fish Stocks
The wild-type line is an intercross between the AB line (University of Oregon, Eugene) and the wtOX line, which was purchased from the Goldfish Bowl (Oxford, UK) and has been bred for several years in our laboratory. Fish were bred and transgenic embryos were collected as described (Westerfield 1993). The transgenic lines -2.2shh:gfpABC (shh:gfp) and -8.4ngn1:gfp (ngn1:gfp) were reported previously (Albert et al. 2003;Blader et al. 2003).
Cell sorting, Complex Probes, and Array Hybridization
Cells were dissociated as described in Dickmeis et al. (2001a). For the dissociation of the shh:gfp line, 2-3 mL of collagenase P (Boehringer Mannheim;2 mg/mL in Hank's Balanced Salt Solution: 8 g/L NaCl, 0.4 g/L KCl, 0.14 g/L CaCl2, 0.1 g/L MgSO4 × 7 H2O, 0.1 g/L MgCl2 × 6 H2O, 0.06 g/L KH2PO4, 1 g/L glucose, 0.02 g/L phenol red, 0.35 g/L NaHCO3) was added after the initial dissociation with trypsin to facilitate separation of notochord cells. Fluorescence-activated cell sorting was carried out with an Elite cell sorter (Coulter). Generation of complex probes from the sorted cells, nylon macroarray hybridization, and statistical analysis of the image data were carried out as described in Dickmeis et al. (2001a).
Southern blotting and sequencing were carried out following standard procedures (Sambrook and Russel 2001).
In Situ Hybridization
Templates for in vitro transcription were generated as described in Dickmeis et al. (2001a). Digoxigenin (DIG)-labeled antisense RNA probes were prepared with a DIG labeling mix (Roche) and T3 RNA polymerase (Stratagene) following the manufacturer's instructions with minor modifications: DIG-probes were synthesized in 96-well PCR plates (Costar) and immediately diluted 1:5 in hybridization buffer (Oxtoby and Jowett 1993) without prior precipitation. Embryos were treated with proteinase K (Oxtoby and Jowett 1993). In situ hybridization and washing steps were carried out with a robot (InsituPro, Intavis) according to the manufacturer's instructions. Staining was carried out manually in 24-well culture dishes as described (Oxtoby and Jowett 1993).
Sequence Comparison
We used the “gscope” platform to find the most homologous mRNAs and proteins in the databases for each of our clones. Gscope is an integration and analysis platform for genome investigation and database searches that automatically runs various analysis and correlations (R. Ripp, L. Bianchetti, Y. Brelivet, A. Carles, F. Chalmel, A. Landenois, O. Lecompte, T. Mochel, L. Moulinier, A. Muller, et al., unpubl.).
Homology searches were carried out with the Compare, DotPlot, and BestFit programs of the GCG Wisconsin package using the default parameters of the program. Comparison of the ctgf up1, the 250-bp ar-C, the 500-bp ar-B, ar-A of shh (Muller et al. 1999), and the 896-bp FPE of netrin1 (Rastegar et al. 2002) were carried out with the program MEME of the GCG package.
Expression Analysis
Regions of homology were amplified from the zebrafish genome by PCR and resulting fragments were either inserted downstream of the tk:gfp or the netrin1:gfp cassettes. tk:gfp contains the TATA-box of the Herpes simplex virus thymidine kinase gene in front of a gfp cassette, and netrin1:gfp drives gfp expression with the zebrafish netrin1 promoter (Rastegar et al. 2002). Standard cloning techniques were used, and details are available upon request.
Acknowledgments
We thank D. Biellmann and C. Vialle for fish care and artwork. We are indebted to Nick Foulkes for a critical reading of the manuscript. We acknowledge the help of the project students Matthias Corrotte and Simon Jochum. This work was supported by Boehringer Ingelheim and AFM fellowships to T.D. We are also grateful to the Institut National de la Santé et de la Recherche Medicale, the Centre National de la Recherche Scientifique, the Hôpital Universitaire de Strasbourg, AFM, ARC, ACI, AICR, and the Max Planck Society.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1819204. Article published online before print in January 2004.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession nos.AY428026-AY428035, CD777505-CD777543, and CD777544-CD778191.]
References
- Abreu, J.G., Ketpura, N.I., Reversade, B., and De Robertis, E.M. 2002. Connective-tissue growth factor (CTGF) modulates cell signalling by BMP and TGF-β. Nat. Cell Biol. 4: 599-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert, S., Muller, F., Fischer, N., Biellmann, D., Neumann, C., Blader, P., and Strahle, U. 2003. Cyclops-independent floor plate differentiation in zebrafish embryos. Dev. Dyn. 226: 59-66. [DOI] [PubMed] [Google Scholar]
- Aparicio, S. and Brenner, S. 1997. How good a model is the Fugu genome? Nature 387: 140. [DOI] [PubMed] [Google Scholar]
- Aparicio, S., Morrison, A., Gould, A., Gilthorpe, J., Chaudhuri, C., Rigby, P., Krumlauf, R., and Brenner, S. 1995. Detecting conserved regulatory elements with the model genome of the Japanese puffer fish, Fugu rubripes. Proc. Natl. Acad. Sci. 92: 1684-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagheri-Fam, S., Ferraz, C., Demaille, J., Scherer, G., and Pfeifer, D. 2001. Comparative genomics of the SOX9 region in human and Fugu rubripes: Conservation of short regulatory sequence elements within large intergenic regions. Genomics 78: 73-82. [DOI] [PubMed] [Google Scholar]
- Barton, L.M., Gottgens, B., Gering, M., Gilbert, J.G., Grafham, D., Rogers, J., Bentley, D., Patient, R., and Green, A.R. 2001. Regulation of the stem cell leukemia (SCL) gene: A tale of two fishes. Proc. Natl. Acad. Sci. 98: 6747-6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blader, P., Plessy, C., and Strahle, U. 2003. Multiple regulatory elements with spatially and temporally distinct activities control neurogenin1 expression in primary neurons of the zebrafish embryo. Mech. Dev. 120: 211-218. [DOI] [PubMed] [Google Scholar]
- Brigstock, D.R., Goldschmeding, R., Katsube, K.I., Lam, S.C., Lau, L.F., Lyons, K., Naus, C., Perbal, B., Riser, B., Takigawa, M., et al. 2003. Proposal for a unified CCN nomenclature. Mol. Pathol. 56: 127-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerda, J., Grund, C., Franke, W.W., and Brand, M. 2002. Molecular characterization of Calymmin, a novel notochord sheath-associated extracellular matrix protein in the zebrafish embryo. Dev. Dyn. 224: 200-209. [DOI] [PubMed] [Google Scholar]
- Chang, B.-E., Fischer, N., Blader, P., Ingham, P., and Strähle, U. 1997. Axial (HNF3β) and retinoic acid receptors are regulators of the zebrafish sonic hedgehog promoter. EMBO J. 16: 3955-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, M.D., Panopoulou, G.D., Cahill, D.J., Bussow, K., and Lehrach, H. 1999. Construction and analysis of arrayed cDNA libraries. Methods Enzymol. 303: 205-233. [DOI] [PubMed] [Google Scholar]
- Clark, M.D., Hennig, S., Herwig, R., Clifton, S.W., Marra, M.A., Lehrach, H., Johnson, S.L., and the WU-GSC EST Group. 2001. An oligonucleotide fingerprint normalized and expressed sequence tag characterized zebrafish cDNA library. Genome Res. 11: 1594-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson, E. 2001. Genomic regulatory systems:Development and evolution. Academic Press, San Diego, CA.
- Dickmeis, T., Aanstad, P., Clark, M., Fischer, N., Herwig, R., Mourrain, P., Blader, P., Rosa, F., Lehrach, H., and Strahle, U. 2001a. Identification of nodal signaling targets by array analysis of induced complex probes. Dev. Dyn. 222: 571-580. [DOI] [PubMed] [Google Scholar]
- Dickmeis, T., Rastegar, S., Aanstad, P., Clark, M., Fischer, N., Plessy, C., Rosa, F., Korzh, V., and Strahle, U. 2001b. Expression of brain subtype creatine kinase in the zebrafish embryo. Mech. Dev. 109: 409-412. [DOI] [PubMed] [Google Scholar]
- Du, S.J., Devoto, S.H., Westerfield, M., and Moon, R.T. 1997. Positive and negative regulation of muscle cell identity by members of the hedgehog and TGF-β gene families. J. Cell Biol. 139: 145-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi, T., Kubota, S., Kondo, S., Shimo, T., Hattori, T., Nakanishi, T., Kuboki, T., Yatani, H., and Takigawa, M. 2001. Regulatory mechanism of human connective tissue growth factor (CTGF/Hcs24) gene expression in a human chondrocytic cell line, HCS-2/8. J. Biochem. (Tokyo) 130: 79-87. [DOI] [PubMed] [Google Scholar]
- Ekker, M., Akimenko, M.A., Allende, M.L., Smith, R., Drouin, G., Langille, R.M., Weinberg, E.S., and Westerfield, M. 1997. Relationships among msx gene structure and function in zebrafish and other vertebrates. Mol. Biol. Evol. 14: 1008-1022. [DOI] [PubMed] [Google Scholar]
- Eriksson, J. and Lofberg, J. 2000. Development of the hypochord and dorsal aorta in the zebrafish embryo (Danio rerio). J. Morphol. 244: 167-176. [DOI] [PubMed] [Google Scholar]
- Feldman, B., Gates, M.A., Egan, E.S., Dougan, S.T., Rennebeck, G., Sirotkin, H.I., Schier, A.F., and Talbot, W.S. 1998. Zebrafish organizer development and germ-layer formation require nodal-related signals. Nature 395: 181-185. [DOI] [PubMed] [Google Scholar]
- Gellner, K. and Brenner, S. 1999. Analysis of 148 kb of genomic DNA around the wnt1 locus of Fugu rubripes. Genome Res. 9: 251-258. [PMC free article] [PubMed] [Google Scholar]
- Glazko, G.V., Koonin, E.V., Rogozin, I.B., and Shabalina, S.A. 2003. A significant fraction of conserved noncoding DNA in human and mouse consists of predicted matrix attachment regions. Trends Genet. 19: 119-124. [DOI] [PubMed] [Google Scholar]
- Goode, D.K., Snell, P.K., and Elgar, G.K. 2003. Comparative analysis of vertebrate Shh genes identifies novel conserved non-coding sequence. Mamm. Genome 14: 192-201. [DOI] [PubMed] [Google Scholar]
- Gritsman, K., Zhang, J., Cheng, S., Heckscher, E., Talbot, W.S., and Schier, A.F. 1999. The EGF-CFC protein one-eyed pinhead is essential for nodal signaling. Cell 97: 121-132. [DOI] [PubMed] [Google Scholar]
- Haddon, C., Smithers, L., Schneider-Maunoury, S., Coche, T., Henrique, D., and Lewis, J. 1998. Multiple δ genes and lateral inhibition in zebrafish primary neurogenesis. Development 125: 359-370. [DOI] [PubMed] [Google Scholar]
- Halloran, M.C., Severance, S.M., Yee, C.S., Gemza, D.L., and Kuwada, J.Y. 1998. Molecular cloning and expression of two novel zebrafish semaphorins. Mech. Dev. 76: 165-168. [DOI] [PubMed] [Google Scholar]
- Hans, S. and Campos-Ortega, J.A. 2002. On the organisation of the regulatory region of the zebrafish δD gene. Development 129: 4773-4784. [DOI] [PubMed] [Google Scholar]
- Hardison, R.C. 2000. Conserved noncoding sequences are reliable guides to regulatory elements. Trends Genet. 16: 369-372. [DOI] [PubMed] [Google Scholar]
- Ho, R.K. and Kimmel, C.B. 1993. Commitment of cell fate in the early zebrafish embryo. Science 261: 109-111. [DOI] [PubMed] [Google Scholar]
- Ivkovic, S., Yoon, B.S., Popoff, S.N., Safadi, F.F., Libuda, D.E., Stephenson, R.C., Daluiski, A., and Lyons, K.M. 2003. Connective tissue growth factor coordinates chondrogenesis and angiogenesis during skeletal development. Development 130: 2779-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, C.H., Ueshima, E., Muraoko, O., Tanaka, H., Yeo, S.-Y., Huh, T.-L., and Miki, N. 1996. Zebrafish elav/HuC homologue as a very early neuronal marker. Neurosci. Lett. 216: 109-112. [DOI] [PubMed] [Google Scholar]
- Krauss, S., Johansen, T., Korzh, V., and Fjose, A. 1991. Expression of the zebrafish paired box gene pax[zf-b] during early neurogenesis. Development 113: 1193-1206. [DOI] [PubMed] [Google Scholar]
- Krauss, S., Concordet, J.-P., and Ingham, P.W. 1993. A functionally conserved homolog of the Drosophila segment polarity gene hedgehog is expressed in tissues with polarizing activity in zebrafish embryos. Cell 75: 1431-1444. [DOI] [PubMed] [Google Scholar]
- Lele, Z. and Krone, P.H. 1997. Expression of genes encoding the collagen-binding heat shock protein (Hsp47) and type II collagen in developing zebrafish embryos. Mech. Dev. 61: 89-98. [DOI] [PubMed] [Google Scholar]
- Liang, L., Li, M., Wang, Y., Zhao, C., Zhao, Z., and Meng, A. 2001. The zygotic expression of zebrafish trebf during embryogenesis is restricted to the embryonic shield and its derivatives. Dev. Genes Evol. 211: 445-448. [DOI] [PubMed] [Google Scholar]
- Liem, K.F., Tremml, G., Roelink, H., and Jessell, T.M. 1995. Dorsal differentiation of neural plate cells induced by BMP-mediated signals from epidermal ectoderm. Cell 82: 969-979. [DOI] [PubMed] [Google Scholar]
- Loots, G.G., Locksley, R.M., Blankespoor, C.M., Wang, Z.E., Miller, W., Rubin, E.M., and Frazer, K.A. 2000. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science 288: 136-140. [DOI] [PubMed] [Google Scholar]
- Ludwig, M.Z. 2002. Functional evolution of noncoding DNA. Curr. Opin. Genet. Dev. 12: 634-639. [DOI] [PubMed] [Google Scholar]
- Mangos, S., Krawetz, R., and Kelly, G.M. 2000. The Translocon-associated protein β (TRAPβ) in zebrafish embryogenesis. I. Enhanced expression of transcripts in notochord and hatching gland precursors. Mol. Cell. Biochem. 215: 93-101. [DOI] [PubMed] [Google Scholar]
- Markstein, M., Markstein, P., Markstein, V., and Levine, M.S. 2002. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc. Natl. Acad. Sci. 99: 763-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry, N., Harrington, W., Lasda, E., Wagner, E.J., and Garcia-Blanco, M.A. 2003. Of urchins and men: Evolution of an alternative splicing unit in fibroblast growth factor receptor genes. RNA 9: 209-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, F., Chang, B., Albert, S., Fischer, N., Tora, L., and Strahle, U. 1999. Intronic enhancers control expression of zebrafish sonic hedgehog in floor plate and notochord. Development 126: 2103-2116. [DOI] [PubMed] [Google Scholar]
- Muller, F., Albert, S., Blader, P., Fischer, N., Hallonet, M., and Strahle, U. 2000. Direct action of the nodal-related signal cyclops in induction of Sonic hedgehog in the ventral midline of the CNS. Development 127: 3889-3897. [DOI] [PubMed] [Google Scholar]
- Muller, F., Blader, P., and Strahle, U. 2002. Search for enhancers: Teleost models in comparative genomic and transgenic analysis of cis regulatory elements. Bioessays 24: 564-572. [DOI] [PubMed] [Google Scholar]
- Odenthal, J. and Nusslein-Volhard, C. 1998. fork head domain genes in zebrafish. Dev. Genes Evol. 208: 245-258. [DOI] [PubMed] [Google Scholar]
- Oxtoby, E. and Jowett, T. 1993. Cloning of the zebrafish krox-20 gene (krx-20) and its expression during hindbrain development. Nucl. Acids Res. 21: 1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio, L.A. and Rubin, E.M. 2001. Genomic strategies to identify mammalian regulatory sequences. Nat. Rev. Genet. 2: 100-109. [DOI] [PubMed] [Google Scholar]
- Placzek, M. 1995. The role of the notochord and floor plate in inductive interactions. Curr. Opin. Genet. Dev. 5: 499-506. [DOI] [PubMed] [Google Scholar]
- Pourquie, O., Coltey, M., Teillet, M.A., Ordahl, C., and Le Douarin, N. 1993. Control of dorsoventral patterning of somitic derivatives by notochord and floor plate. Proc. Natl. Acad. Sci. 90: 5242-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastegar, S., Albert, S., Le Roux, I., Fischer, N., Blader, P., Muller, F., and Strahle, U. 2002. A floor plate enhancer of the zebrafish netrin1 gene requires Cyclops (Nodal) signalling and the winged helix transcription factor FoxA2. Dev. Biol. 252: 1-14. [DOI] [PubMed] [Google Scholar]
- Rebagliati, M.R., Toyama, R., Fricke, C., Haffter, P., and Dawid, I.B. 1998. Zebrafish nodal related genes are implicated in axial patterning and establishing left-right asymmetry. Dev. Biol. 199: 261-272. [DOI] [PubMed] [Google Scholar]
- Rubinstein, A.L., Lee, D., Luo, R., Henion, P.D., and Halpern, M.E. 2000. Genes dependent on zebrafish cyclops function identified by AFLP differential gene expression screen. Genesis 26: 86-97. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. and Russel, D.W. 2001. Molecular cloning:A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sampath, K., Rubinstein, A.L., Cheng, A.M., Liang, J.O., Fekany, K., Solnica-Krezel, L., Korzh, V., Halpern, M.E., and Wright, C.V. 1998. Induction of the zebrafish ventral brain and floorplate requires cyclops/nodal signalling. Nature 395: 185-189. [DOI] [PubMed] [Google Scholar]
- Schulte-Merker, S., Ho, R.K., Herrmann, B.G., and Nüsslein-Volhard, C. 1992. The protein product of the zebrafish homologue of the mouse T gene is expressed in nuclei of the germ ring and the notochord of the early embryo. Development 116: 1021-1032. [DOI] [PubMed] [Google Scholar]
- Strähle, U., Blader, P., Henrique, D., and Ingham, P. 1993. Axial, a zebrafish gene expressed along the developing body axis, shows altered expression in cyclops mutant embryos. Genes & Dev. 7: 1436-1446. [DOI] [PubMed] [Google Scholar]
- Strahle, U., Blader, P., and Ingham, P.W. 1996. Expression of axial and Sonic hedgehog in wildtype and midline defective zebrafish embryos. Int. J. Dev. Biol. 40: 929-940. [PubMed] [Google Scholar]
- Talbot, W.S., Trevarrow, B., Halpern, M.E., Melby, M.E., Farr, A.E., Postlethwait, J.H., Jowett, T., Kimmel, C.B., and Kimelman, D. 1995. Requirement for the homeobox gene floating head in zebrafish development. Nature 378: 150-157. [DOI] [PubMed] [Google Scholar]
- Tanabe, Y. and Jessell, T.M. 1996. Diversity and pattern in the developing spinal cord. Science 274: 1115-1123. [DOI] [PubMed] [Google Scholar]
- Wasserman, W.W., Palumbo, M., Thompson, W., Fickett, J.W., and Lawrence, C.E. 2000. Human-mouse genome comparisons to locate regulatory sites. Nat. Genet. 26: 225-228. [DOI] [PubMed] [Google Scholar]
- Westerfield, M. 1993. The zebrafish book. University of Oregon Press, Eugene.
- Westerfield, M., Wegener, J., Jegalian, B.G., DeRobertis, E.M., and Püschel, A.W. 1992. Specific activation of mammalian Hox promoters in mosaic transgenic zebrafish. Genes & Dev. 6: 591-598. [DOI] [PubMed] [Google Scholar]
- Yan, Y.L., Hatta, K., Riggleman, B., and Postlethwait, J.H. 1995. Expression of a type II collagen gene in the zebrafish embryonic axis. Dev. Dyn. 203: 363-376. [DOI] [PubMed] [Google Scholar]
- Zerucha, T., Stuhmer, T., Hatch, G., Park, B.K., Long, Q., Yu, G., Gambarotta, A., Schultz, J.R., Rubenstein, J.L., and Ekker, M. 2000. A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J. Neurosci. 20: 709-721. [DOI] [PMC free article] [PubMed] [Google Scholar]