Background: The physiological importance of mitophagy in yeast has been largely unexplored.
Results: Mitochondrial DNA deletion frequently occurs in mitophagy-deficient cells during nitrogen starvation because of overproduction of the reactive oxygen species from unregulated mitochondria.
Conclusion: Mitophagy prevents excess reactive oxygen species production and mitochondrial DNA mutation.
Significance: Our findings provide insight into mitophagy-related disorders such as Parkinson disease.
Keywords: Autophagy, Mitochondria, Protein Degradation, Reactive Oxygen Species (ROS), Yeast, Atg32, Mitophagy
Abstract
In mammalian cells, the autophagy-dependent degradation of mitochondria (mitophagy) is thought to maintain mitochondrial quality by eliminating damaged mitochondria. However, the physiological importance of mitophagy has not been clarified in yeast. Here, we investigated the physiological role of mitophagy in yeast using mitophagy-deficient atg32- or atg11-knock-out cells. When wild-type yeast cells in respiratory growth encounter nitrogen starvation, mitophagy is initiated, excess mitochondria are degraded, and reactive oxygen species (ROS) production from mitochondria is suppressed; as a result, the mitochondria escape oxidative damage. On the other hand, in nitrogen-starved mitophagy-deficient yeast, excess mitochondria are not degraded and the undegraded mitochondria spontaneously age and produce surplus ROS. The surplus ROS damage the mitochondria themselves and the damaged mitochondria produce more ROS in a vicious circle, ultimately leading to mitochondrial DNA deletion and the so-called “petite-mutant” phenotype. Cells strictly regulate mitochondrial quantity and quality because mitochondria produce both necessary energy and harmful ROS. Mitophagy contributes to this process by eliminating the mitochondria to a basal level to fulfill cellular energy requirements and preventing excess ROS production.
Introduction
Autophagy is a protein degradation process that involves the formation of a double-membrane compartment, termed the autophagosome, which engulfs cytoplasmic components nonselectively and fuses with the lysosome/vacuole for degradation (1). This is important for various physiological processes such as cell survival under starvation, intracellular clearance, development, the immune response, and aging (2).
Recent studies have revealed that autophagy can degrade mitochondria selectively. This autophagic mitochondrial degradation is called mitophagy (3, 4). Recently, the mitochondrial protein Atg32, which is specifically required for mitophagy, was identified in yeast, and studies of Atg32 have revealed the molecular processes of mitochondrial selection by the autophagic machineries (5–9). In brief, when mitophagy is initiated, the mitochondrial outer-membrane protein Atg32 is recognized and bound by the cytosolic adaptor protein Atg11. Atg11 recruits the mitochondria to the pre-autophagosomal structure/phagophore assembly site where the phagophores, the initial sequestering membrane structures, are generated to uptake and degrade mitochondria (10, 11). Although this molecular mechanism is known, the physiological importance of mitophagy in yeast has been largely unexplored.
It is believed that mitophagy in mammalian cells functions in mitochondrial quality control. Loss-of-function mutations in the PARK2 and PARK6 genes, which encode Parkin and PINK1, respectively, cause Parkinson disease. Recent studies have linked Parkin and PINK1 to mitophagy (12–14). Cytosolically synthesized PINK1 can target and stably localize on damaged mitochondria, and mitochondrial PINK1 translocates the cytosolic E3 ubiquitin ligase Parkin to the mitochondria (15–19). Finally, the damaged mitochondria are degraded by Parkin-mediated mitophagy, although the details of this process have not been revealed. These findings imply that PINK1/Parkin-mediated mitophagy plays a very important role in the quality control of mitochondria, eliminating damaged mitochondria selectively, and that dysfunction of this mitophagic process causes Parkinson disease (12–14).
The importance of mitophagy in mammalian cells suggests that mitophagy also plays an important physiological role in yeast. Accordingly, we examined the phenotypes associated with mitophagy-deficient atg32Δ and atg11Δ cells and found that mitochondrial DNA (mtDNA)2 deletion frequently occurs in mitophagy-deficient cells during nitrogen starvation, especially if the cells are in respiratory growth before starvation. Further analysis revealed that when wild-type cells encounter nitrogen starvation, they initiate mitophagy and quickly eliminate mitochondria that have proliferated during respiratory growth. As a result, cellular reactive oxygen species (ROS) production, which occurs mainly in mitochondria, is suppressed. On the other hand, in mitophagy-deficient cells, undegraded mitochondria produce excess ROS during nitrogen starvation. ROS damage mitochondria and damaged mitochondria produce more ROS, and finally, overproduced ROS cause mtDNA deletion. Ultimately, cells with mtDNA deletion generate small colonies even on fermentable medium, and this phenotype is called “petite” (20). From these findings, we conclude that mitophagy prevents mtDNA deletion by eliminating ROS-producing mitochondria during nitrogen starvation.
EXPERIMENTAL PROCEDURES
Strains, Culture Media, and Antibodies
The yeast strains used in this study are listed in Table 1. Yeast cells were grown in rich medium (YPD; 1% yeast extract, 2% peptone, 2% glucose), lactate medium (YPL; 1% yeast extract, 2% peptone, 2% lactate), or ethanol glycerol medium (YPEG; 1% yeast extract, 2% peptone, 3% ethanol, 3% glycerol). Nitrogen starvation experiments were performed in synthetic minimal medium lacking nitrogen (SD-N; 0.17% yeast nitrogen base without amino acids and ammonium sulfate, 2% glucose). Anti-Cox2 antibodies (Invitrogen), anti-Por1 antibodies (Invitrogen), and anti-Pgk1 antibodies (Nordic Immunological Laboratories, Eindhoven, The Netherlands) were used for immunoblotting.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| SEY6210 | MATα his3-Δ200 leu2–3,112 lys2–801 | 29 |
| trp1-Δ901 ura3–52 suc2-Δ9 GAL | ||
| TKYM139 | SEY6210 atg32Δ::LEU2 | This study |
| WHY1 | SEY6210 atg1Δ::HIS5 s.p. | 30 |
| YTS147 | SEY6210 atg11Δ::LEU2 | 8 |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Open Biosystems, Huntsville, AL |
| BYATG1 | BY4742 atg1Δ::KAN | Open Biosystems |
| BYATG11 | BY4742 atg11Δ::KAN | Open Biosystems |
| BYATG32 | BY4742 atg32Δ::KAN | Open Biosystems |
Assessment of Viability by Colony Formation Assay
The cellular viability assay was performed as described previously with some modifications (21). Briefly, cells grown in YPD or YPL to mid-log phase were washed twice with water and cultured in SD-N. On the indicated days, the cells contained in 0.02 μl of SD-N were inoculated onto YPD plates. After 2 days, the number and size of the resulting colonies were measured.
Measurement of ROS
Cells were incubated in staining buffer (10 μm dihydroethidium (DHE), 100 mm HEPES, pH 7.5, 5% glucose) for 10 min at room temperature and then loaded onto a flow cytometer (BD Biosciences) to observe the oxidized DHE fluorescence. To detect protein oxidization, we used an OxyBlotTM Protein Oxidation Detection kit (Millipore, Billerica, MA) according to the manufacturer's instructions.
Southern Blotting
Cells were cultured in YPD medium to mid-log phase, and whole cell DNA was extracted by standard methods. Five milligrams of whole cell DNA was digested with BamHI (Takara, Kyoto, Japan) or ScaI (Takara), separated on a 0.8% agarose gel, and transferred onto Hybond-N+ nylon membrane (GE Healthcare). The COX1 and COB coding regions were amplified by PCR using primers 5′-GGTATGGCAGGAACAGCAAT-3′ and 5′-ACAGCCCTCCAAATGTCAAC-3′ for COX1, and 5′-ATGATTGGCCGGTTTAATTG-3′ and 5′-TATGGGAGTTCCCACAAAGC-3′ for COB. The PCR products were labeled with alkaline phosphatase using AlkPhos Direct Labeling reagents (GE Healthcare) and used as probes on the blots. The signals were detected with CDP-star (GE Healthcare), according to the manufacturer's instructions.
RESULTS
Mitophagy-deficient Cells Generate Small Colonies
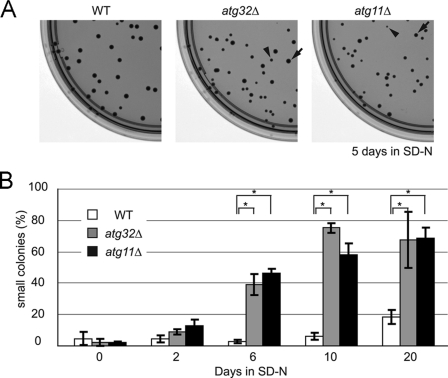
We have previously examined cell viability during nitrogen starvation in atg32Δ cells, and found that both wild-type and atg32Δ cells grew to a similar level (7). In our previous experiment, we pre-cultured cells in a fermentable carbon source-containing medium (YPD: yeast extract, peptone, and dextrose) before nitrogen starvation. We speculated that more drastic conditions might be required to elucidate a phenotype in mitophagy-deficient cells. Here, we pre-cultured cells in a nonfermentable carbon source-containing medium (YPL: yeast extract, peptone, and lactate) in which mitochondria proliferate for respiratory growth, and then shifted the cells to nitrogen starvation medium (SD-N) and observed the cell viability according to the duration of nitrogen starvation. Contrary to our expectations, both the wild-type (SEY6210) and mitophagy-deficient atg32Δ and atg11Δ cells survived at similar levels during nitrogen starvation, whereas macroautophagy-deficient atg1Δ cells became almost nonviable after 12 days of nitrogen starvation (supplemental Fig. S1A). During this experiment, we found that mitophagy-deficient cells tended to form smaller colonies than wild-type cells when inoculated onto a YPD plate after nitrogen starvation. For example, after 5 days of nitrogen starvation, almost all the wild-type cells formed uniformly normal-sized colonies on a YPD plate, whereas atg32Δ and atg11Δ cells also formed relatively small colonies (hereafter we call the normal-sized colonies “large colonies” to distinguish them from the small colonies) (Fig. 1A). As a control, we cultured wild-type, atg32Δ, and atg11Δ cells without starvation for 5 days and then inoculated the cells onto YPD plates. Both wild-type and mitophagy-deficient cells barely formed small colonies (wild-type, 1.88 ± 0.62%; atg32Δ, 2.12 ± 1.19%; atg11Δ, 1.63 ± 0.50%; supplemental Fig. S1B). We pre-cultured wild-type, atg32Δ, and atg11Δ cells in YPL medium and then shifted them to SD-N for up to 20 days, then inoculated the cells onto YPD plates and observed the proportion of small colonies, defined as those with a diameter less than 50% of that of a typical large colony. As shown in Fig. 1B, the proportion of small colonies was ∼40% for both atg32Δ and atg11Δ cells after 6 days of nitrogen starvation, and the proportion was further increased with a longer duration of nitrogen starvation. In contrast, in wild-type cells, the proportion of small colonies was less than 5% even after 10 days of nitrogen starvation, and it increased up to ∼20% after 20 days of nitrogen starvation.
FIGURE 1.
Mitophagy-deficient cells generate small colonies after nitrogen starvation. A, wild-type (WT), atg32Δ, and atg11Δ cells were pre-cultured in YPL to mid-log phase and then shifted to SD-N for 5 days. Cells were inoculated onto YPD plates and the size of colonies formed was observed (typical small and large colonies are indicated by arrowheads and arrows, respectively). B, WT, atg32Δ, and atg11Δ cells were precultured in YPL to mid-log phase and then shifted to SD-N. After the indicated number of days of nitrogen starvation, cells were inoculated onto YPD plates and the proportion of small colonies formed was observed (more than 80 colonies were measured for each experiment). The values represent the mean ± S.D. of three experiments. *, p < 0.001 by paired t test.
To confirm the phenotype observed above, we repeated the same experiment using a different background strain (BY4742). Surprisingly, even wild-type cells almost lost their viability after 14 days of nitrogen starvation (supplemental Fig. S2A). Mitophagy-deficient atg32Δ and atg11Δ cells lost their viability earlier than wild-type cells did, but the difference was not significant (supplemental Fig. S2A). As for SEY6210 cells, atg32Δ and atg11Δ cells of BY4742 tended to form small colonies on YPD plates after nitrogen starvation, with the frequency depending on the duration of starvation (supplemental Fig. S2, B and C). Because cells that formed small colonies generated small colonies again when they were inoculated onto new YPD plates, we speculated that inheritable damage, such as genomic DNA or mtDNA mutation, might have occurred in these cells.
mtDNA Deletion in Mitophagy-deficient Yeast Cells
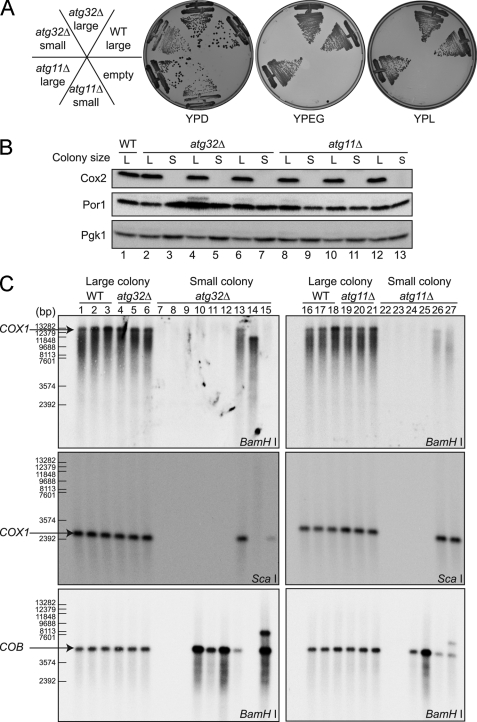
We presumed that a mitochondrial defect was the most likely cause of small colony generation. To test this, we reinoculated both large and small colonies derived from wild-type, atg32Δ, and atg11Δ strains onto YPL and YPEG (yeast extract, peptone, ethanol, and glycerol) plates, on which mitochondria-dependent respiration is essential for cellular growth. Although all cells from large colonies could grow on YPL and YPEG plates, cells from small colonies could not grow on YPL or YPEG plates (Fig. 2A), suggesting that the latter cells have a mitochondrial defect. To examine the mitochondrial defect further, we investigated the expression levels of mitochondrial proteins in large and small colony-forming cells. Surprisingly, cells forming small colonies did not express any of the mtDNA-encoded mitochondrial inner membrane protein Cox2, whereas cells forming either large or small colonies expressed the nuclear genome-encoded mitochondrial outer membrane protein Por1 to a similar extent (Fig. 2B). This finding suggested that there was a defect in intramitochondrial transcription or translation, or the mtDNA itself in the cells that formed small colonies. We then examined the mtDNA by Southern blotting. Whole cell DNA prepared from cells forming either small or large colonies was digested with BamHI or ScaI, Southern blotted, and probed with mtDNA-derived probes: the COX1 or COB coding regions. For all large colony-forming cells, we detected bands of the expected size for both the COX1 and COB probes (Fig. 2C, lanes 1–6 and 16–21). On the other hand, for small colony-forming cells, atg32Δ or atg11Δ, we identified several patterns of results, suggesting that mtDNA deletion had occurred in these cells. In some cells, we could not detect any bands for the COX1 or the COB probe (Fig. 2C, lanes 7–9, 22, and 23), suggesting that both COX1- and COB-encoding mtDNA regions were deleted, or the whole mtDNA genome was lost in these cells. In other cells, we could not detect any bands using the COX1 probe, whereas the COB probe generated a band of the expected size (Fig. 2C, lanes 10–12, 24, and 25), suggesting that at least the COX1 region of the mtDNA genome was deleted in these cells. In addition, there were several atypical cases. In the cells shown in Fig. 2C, lane 14, we observed a smaller than expected band for the COX1 probe on BamHI-digested DNA and no bands on ScaI-digested DNA; similarly we observed no bands for the COB probe on BamHI-digested DNA. In the cells shown in Fig. 2C, lanes 15 and 27, we observed a larger than expected band for the COB probe in addition to the expected size band. Finally, in the cells shown in Fig. 2C, lanes 13 and 26, we observed the expected size bands for both the COX1 and COB probes when DNA was digested with BamHI, but the signals were very weak compared with those of the other samples, even though we used the same amount of DNA for each (supplemental Fig. S3A). These findings suggest that mtDNA deletion frequently occurs in mitophagy-deficient cells during nitrogen starvation. Next, we observed mtDNA in wild-type cells after 20 days starvation. All large colony-forming cells expressed Cox2 protein and had intact mtDNA, whereas small colony-forming cells, which comprised fewer than 20% of all colonies, did not express the Cox2 protein and had deleted mtDNA (supplemental Fig. S3, B and C). From these findings, we concluded that mtDNA deletion occurs to some degree during starvation even in wild-type cells; however, it is augmented in mitophagy-deficient cells.
FIGURE 2.
Mitochondrial DNA deletion occurs in mitophagy-deficient strains during nitrogen starvation. A, large and small colonies formed after 10 days of starvation were inoculated onto YPD, YPL, and YPEG plates and grown for 2–3 days at 30 °C. atg32Δ and atg11Δ small colony-derived cells only were unable to grow on YPL or YPEG. B, large and small colonies formed after 6 or 10 days starvation were cultured in YPD to mid-log phase (lanes 1–5 and 8–11, 6 days in SD-N; lanes 6, 7, 12, and 13, 10 days in SD-N). Cell lysates were immunoblotted with anti-Cox2, anti-Por1, and anti-Pgk1 (loading control) antibodies. atg32Δ and atg11Δ small colony-derived cells only did not produce the mitochondrially encoded protein Cox2. C, large and small colonies formed after 5 or 6 days starvation were cultured in YPD to mid-log phase. Whole cell DNA was digested by BamHI or ScaI and analyzed by Southern blotting with COX1 and COB mtDNA probes. The atg32Δ and atg11Δ small colonies showed absent or aberrant COX1 and/or COB bands. WT, wild-type.
Reactive Oxygen Species Cause mtDNA Deletion
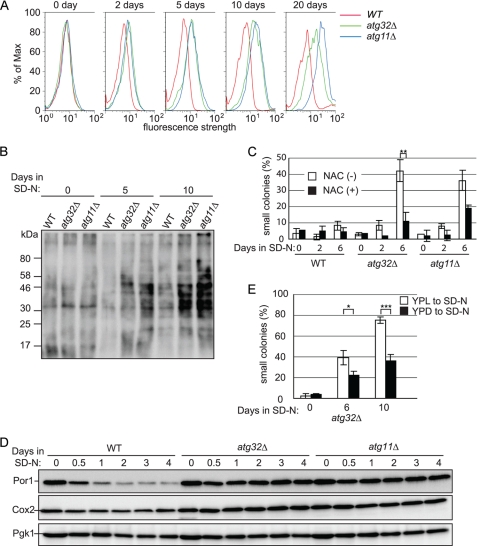
We hypothesized that, in mitophagy-deficient cells during nitrogen starvation, ROS would be overproduced, and that excess ROS would cause mtDNA deletion. To investigate this, we observed cellular ROS during nitrogen starvation by staining cells with the superoxide indicator DHE, detecting oxidized DHE fluorescence by flow cytometry. Cellular ROS production was the same in wild-type, atg32Δ, and atg11Δ cells before nitrogen starvation (Fig. 3A, 0 days). As we expected, cellular ROS production gradually increased in atg32Δ and atg11Δ cells compared with wild-type cells, depending on the duration of nitrogen starvation (Fig. 3A and supplemental Fig. S4, A and B). We further examined cellular ROS production by observing protein oxidation in mitophagy-deficient cells during nitrogen starvation using an OxyBlot protein oxidation detection assay. Consistent with flow cytometry results, the levels of oxidized protein increased dramatically in atg32Δ and atg11Δ cells after nitrogen starvation, compared with wild-type cells (Fig. 3B).
FIGURE 3.
ROS in mitophagy-deficient cells cause mtDNA deletion during nitrogen starvation. A, wild-type (WT), atg32Δ, and atg11Δ cells were precultured in YPL to mid-log phase and then shifted to SD-N. After the indicated number of days of nitrogen starvation, cells were incubated with the superoxide indicator DHE, and oxidized DHE fluorescence was observed using flow cytometry. B, wild-type, atg32Δ, and atg11Δ cells were precultured in YPL to mid-log phase and then shifted to SD-N. After the indicated number of days of nitrogen starvation, oxidized proteins were observed by OxyBlot assay. C, WT, atg32Δ, and atg11Δ cells were precultured in YPL to mid-log phase and then shifted to SD-N containing 1 mm of the ROS scavenger N-acetylcysteine (NAC) or dimethyl sulfoxide (control). After the indicated number of days of nitrogen starvation, cells were inoculated onto YPD plates and the proportion of small colonies formed on YPD plates was calculated (more than 80 colonies were measured for each experiment). The values represent the mean ± S.D. of three experiments. D, WT, atg32Δ, and atg11Δ cells were precultured in YPL to mid-log phase and then shifted to SD-N. Cells were collected after the indicated number of days of nitrogen starvation and cell lysates were immunoblotted with anti-Cox2, anti-Por1, and anti-Pgk1 (loading control) antibodies. E, WT and atg32Δ cells were precultured in YPD or YPL to mid-log phase and then shifted to SD-N. After the indicated number of days of nitrogen starvation, cells were inoculated onto YPD plates and the proportion of small colonies formed was calculated (more than 80 colonies were measured for each experiment). The values represent the mean ± S.D. of three experiments. ***, p < 0.001; **, p < 0.005; *, p < 0.01 by paired t test.
If the overproduction of ROS in mitophagy-deficient cells was the direct cause of mtDNA damage and small colony formation, elimination of the cellular ROS may rescue the phenotype. To examine this possibility, we cultured cells with the ROS scavenger N-acetylcysteine and observed the size of colonies generated after nitrogen starvation. As shown in Fig. 3C and supplemental Fig. S5, addition of N-acetylcysteine dramatically decreased the proportion of small colonies after nitrogen starvation in mitophagy-deficient atg32Δ and atg11Δ cells, whereas it showed no effect in wild-type cells. From these findings, we confirmed that ROS, which are overproduced in mitophagy-deficient cells, damaged the mtDNA and caused mtDNA deletion during nitrogen starvation, causing the cells to generate small colonies on YPD plates.
Mitochondria are the major source of cellular ROS. It is highly possible that mitochondria that are not degraded by mitophagy are the source of the excess ROS in mitophagy-deficient cells. Thus, we investigated the cellular number of mitochondria using Por1 and Cox2 as mitochondrial markers. In wild-type cells, the Por1 signal quickly decreased and the Cox2 signal gradually decreased as a result of mitophagy upon nitrogen starvation, whereas in mitophagy-deficient atg32Δ and atg11Δ cells, both Por1 and Cox2 expression was barely affected by nitrogen starvation (Fig. 3D). This finding is consistent with our hypothesis regarding cellular ROS production: in wild-type cells, ROS production slightly decreased following the degradation of mitochondria during nitrogen starvation (supplemental Fig. S4B), whereas in atg32Δ cells, ROS production gradually increased during nitrogen starvation (supplemental Fig. S4B), presumably because the undegraded mitochondria became old and damaged and produced more ROS. We concluded that wild-type cells quickly degrade excess mitochondria by mitophagy during nitrogen starvation, preventing overproduction of ROS and subsequent mtDNA mutation, whereas in mitophagy-deficient cells that cannot degrade excess mitochondria, ROS produced from undegraded mitochondria cause mtDNA mutation during nitrogen starvation.
In the studies described above, we pre-cultured cells in YPL to proliferate the mitochondria before shifting to the nitrogen-starvation state. If the cells were pre-cultured in YPD, the small colony phenotype might be inhibited in mitophagy-deficient cells during nitrogen starvation, because the number of ROS producing mitochondria is smaller when cells are cultured in YPD compared with in YPL. To test this, we pre-cultured atg32Δ cells in either YPD or YPL, and observed the proportion of small colonies formed on YPD plates after nitrogen starvation. As expected, the proportion of small colonies was dramatically decreased in atg32Δ cells after nitrogen starvation when the cells were precultured in YPD compared with those precultured in YPL (22 and 39% at 6 days, 35 and 75% at 10 days of starvation, when precultured in YPD and YPL, respectively; Fig. 3E). This supports our notion that the ROS produced by undegraded mitochondria during nitrogen starvation cause mtDNA mutation in mitophagy-deficient cells.
DISCUSSION
Although the presence of mitochondria in lysosomes or vacuoles was first reported in 1957 in mammalian cells (22) and in 1992 in yeast (23), the physiological relevance of mitochondrial degradation by autophagy has been unclear. Very recently, in mammalian cells, several studies have suggested that PINK1/parkin-dependent mitophagy selectively degrades depolarized mitochondria (12, 18, 19), implying that mitophagy contributes to mitochondrial quality control. On the other hand, very little is known about the physiological importance of mitophagy in yeast. When we identified the mitophagy-specific gene ATG32 in yeast, we tried to identify the physiological role of mitophagy using atg32Δ cells; we could not, however, identify any phenotypes associated with these cells (7). In this study, we have now identified the physiological relevance of mitophagy in yeast. When yeast cells in respiratory growth encounter nitrogen starvation, the cells initiate mitophagy, degrade excess mitochondria, and suppress ROS production from mitochondria, and as a result, the cells escape the severe oxidative damage that causes mtDNA deletion. This process is very important for yeast in nature, because the cells frequently shift their metabolism between fermentation and respiration and are always at risk of starvation.
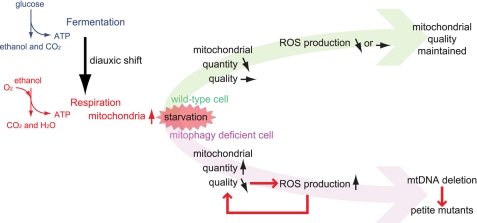
Our current scheme based on this study is summarized in Fig. 4. When yeast cells grow in glucose-rich conditions, such as on the surface of grapes, they preferentially ferment glucose and release ethanol. When cells absorb glucose, they switch their metabolism from fermentation to respiration, the aerobic usage of ethanol. This conversion (called the diauxic shift) is frequently observed in nature. During the diauxic shift, mitochondria proliferate to increase cellular respiration. If the cells then encounter starvation stress, for example, if the nutrients are washed away by rain, mitophagy is initiated and the cells minimize the number of mitochondria to a basal level to fulfill cellular energy requirements, thus suppressing the production of unwanted ROS and mitochondrial damage.
FIGURE 4.
Summary of the importance of mitophagy in yeast. When yeast cells encounter starvation stress in respiratory growth, wild-type cells initiate mitophagy, degrade excess mitochondria, and suppress the production of ROS from mitochondria; as a result, the mitochondria escape oxidative damage. In mitophagy-deficient cells, the mitochondria suffer oxidative damage and produce more ROS that ultimately cause mtDNA mutation.
It has been recently reported that bulk autophagy-deficient cells accumulate high levels of ROS and that these ROS cause the loss of mtDNA during nitrogen starvation (24). Although these phenotypes of bulk autophagy-deficient cells are similar to those of mitophagy-deficient cells, the process of ROS accumulation is totally different. In bulk autophagy-deficient cells, cellular ROS accumulate during nitrogen starvation because the cellular amino acid pool is reduced and the expression of mitochondrial respiratory proteins and the ROS scavenger proteins is suppressed (24). Moreover, the level of ROS accumulated in bulk autophagy-deficient cells during nitrogen starvation was much higher than that in mitophagy-deficient or wild-type cells (data not shown).
It is believed that mitophagy in mammalian cells functions in mitochondrial quality control. There are, however, limited experimental data showing the physiological importance of mitophagy for quality control of mitochondria in mammalian cells, except for PINK1/Parkin-dependent mitophagy. One reason for this is that mitophagy-specific genes such as ATG32 in yeast have not been identified in mammals. Furthermore, although it is clear that PINK1/Parkin-dependent mitophagy, especially when Parkin is overexpressed, eliminates dysfunctional mitochondria in cultured cells, the physiological phenotypes of mitophagy deficient (Parkin knock-out mice or Parkin knockdown cells, for example) have been poorly characterized (25–28). In this article, we clearly demonstrate the physiological importance of mitophagy to maintain mitochondrial quality in yeast. Because mitophagy is conserved from yeast to mammals, the fundamental processes and roles of mitophagy are expected to be similar between yeast and mammals, although they may be highly diverse among mammalian cells. Further studies are required to elucidate the details of these processes in mammals, and to assess their impact on human diseases such as Parkinson disease.
Supplementary Material
Acknowledgment
We thank Daniel J. Klionsky (University of Michigan) for providing the yeast strains and plasmids.
This work was supported by Grants-in-aid for Scientific Research 23689032 (to T. K.), 22020028 (to T. K.), and 22249018 (to D. K.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, a grant from the Takeda Science Foundation (to T. K.), the Naito Foundation (to T. K.), the Mochida Memorial Foundation for Medical and Pharmaceutical Research (to T. K.), the Kowa Life Science Foundation (to T. K.), and the Kyushu University Interdisciplinary Programs in Education and Projects in Research Development (to T. K.).

This article contains supplemental Figs. S1–S5.
- mtDNA
- mitochondrial DNA
- DHE
- dihydroethidium
- ROS
- reactive oxygen species
- SD-N
- synthetic minimal medium lacking nitrogen
- YPD
- yeast extract, peptone, glucose
- YPEG
- yeast extract, peptone, ethanol, glycerol
- YPL
- yeast extract, peptone, lactate.
REFERENCES
- 1. Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y. (2009) Dynamics and diversity in autophagy mechanisms. Lessons from yeast. Nat. Rev. Mol. Cell Biol. 10, 458–467 [DOI] [PubMed] [Google Scholar]
- 2. Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. (2008) Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lemasters J. J. (2005) Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 8, 3–5 [DOI] [PubMed] [Google Scholar]
- 4. Priault M., Salin B., Schaeffer J., Vallette F. M., di Rago J. P., Martinou J. C. (2005) Impairing the bioenergetic status and the biogenesis of mitochondria triggers mitophagy in yeast. Cell Death Differ. 12, 1613–1621 [DOI] [PubMed] [Google Scholar]
- 5. Kanki T., Wang K., Baba M., Bartholomew C. R., Lynch-Day M. A., Du Z., Geng J., Mao K., Yang Z., Yen W. L., Klionsky D. J. (2009) A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol. Biol. Cell 20, 4730–4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Okamoto K., Kondo-Okamoto N., Ohsumi Y. (2009) Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell 17, 87–97 [DOI] [PubMed] [Google Scholar]
- 7. Kanki T., Wang K., Cao Y., Baba M., Klionsky D. J. (2009) Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell 17, 98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kanki T., Klionsky D. J. (2008) Mitophagy in yeast occurs through a selective mechanism. J. Biol. Chem. 283, 32386–32393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aoki Y., Kanki T., Hirota Y., Kurihara Y., Saigusa T., Uchiumi T., Kang D. (2011) Phosphorylation of serine 114 on Atg32 mediates mitophagy. Mol. Biol. Cell 22, 3206–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kanki T., Klionsky D. J., Okamoto K. (2011) Antioxid Redox Signal. 14, 1989–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kanki T., Klionsky D. J. (2010) The molecular mechanism of mitochondria autophagy in yeast. Mol. Microbiol. 75, 795–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Youle R. J., Narendra D. P. (2011) Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12, 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deas E., Wood N. W., Plun-Favreau H. (2011) Mitophagy and Parkinson disease. The PINK1-parkin link. Biochim. Biophys. Acta 1813, 623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vives-Bauza C., Przedborski S. (2011) Mitophagy, the latest problem for Parkinson disease. Trends Mol. Med. 17, 158–165 [DOI] [PubMed] [Google Scholar]
- 15. Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R. L., Kim J., May J., Tocilescu M. A., Liu W., Ko H. S., Magrané J., Moore D. J., Dawson V. L., Grailhe R., Dawson T. M., Li C., Tieu K., Przedborski S. (2010) PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. U.S.A. 107, 378–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Geisler S., Holmström K. M., Skujat D., Fiesel F. C., Rothfuss O. C., Kahle P. J., Springer W. (2010) PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12, 119–131 [DOI] [PubMed] [Google Scholar]
- 17. Ziviani E., Tao R. N., Whitworth A. J. (2010) Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc. Natl. Acad. Sci. U.S.A. 107, 5018–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Narendra D. P., Jin S. M., Tanaka A., Suen D. F., Gautier C. A., Shen J., Cookson M. R., Youle R. J. (2010) PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8, e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Narendra D., Tanaka A., Suen D. F., Youle R. J. (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ferguson L. R., von Borstel R. C. (1992) Induction of the cytoplasmic ”petite“ mutation by chemical and physical agents in Saccharomyces cerevisiae. Mutat. Res. 265, 103–148 [DOI] [PubMed] [Google Scholar]
- 21. Noda T. (2008) Viability assays to monitor yeast autophagy. Methods Enzymol. 451, 27–32 [DOI] [PubMed] [Google Scholar]
- 22. Clark S. L., Jr. (1957) Cellular differentiation in the kidneys of newborn mice studies with the electron microscope. J. Biophys. Biochem. Cytol. 3, 349–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Takeshige K., Baba M., Tsuboi S., Noda T., Ohsumi Y. (1992) Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119, 301–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Suzuki S. W., Onodera J., Ohsumi Y. (2011) Starvation induced cell death in autophagy-defective yeast mutants is caused by mitochondria dysfunction. PLoS One 6, e17412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Perez F. A., Palmiter R. D. (2005) Parkin-deficient mice are not a robust model of parkinsonism. Proc. Natl. Acad. Sci. U.S.A. 102, 2174–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Goldberg M. S., Fleming S. M., Palacino J. J., Cepeda C., Lam H. A., Bhatnagar A., Meloni E. G., Wu N., Ackerson L. C., Klapstein G. J., Gajendiran M., Roth B. L., Chesselet M. F., Maidment N. T., Levine M. S., Shen J. (2003) Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J. Biol. Chem. 278, 43628–43635 [DOI] [PubMed] [Google Scholar]
- 27. Itier J. M., Ibanez P., Mena M. A., Abbas N., Cohen-Salmon C., Bohme G. A., Laville M., Pratt J., Corti O., Pradier L., Ret G., Joubert C., Periquet M., Araujo F., Negroni J., Casarejos M. J., Canals S., Solano R., Serrano A., Gallego E., Sanchez M., Denefle P., Benavides J., Tremp G., Rooney T. A., Brice A., Garcia de Yebenes J. (2003) Parkin gene inactivation alters behavior and dopamine neurotransmission in the mouse. Hum. Mol. Genet. 12, 2277–2291 [DOI] [PubMed] [Google Scholar]
- 28. Von Coelln R., Thomas B., Savitt J. M., Lim K. L., Sasaki M., Hess E. J., Dawson V. L., Dawson T. M. (2004) Loss of locus coeruleus neurons and reduced startle in parkin null mice. Proc. Natl. Acad. Sci. U.S.A. 101, 10744–10749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Robinson J. S., Klionsky D. J., Banta L. M., Emr S. D. (1988) Protein sorting in Saccharomyces cerevisiae. Isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 8, 4936–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shintani T., Huang W. P., Stromhaug P. E., Klionsky D. J. (2002) Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell 3, 825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.