Background: We report the cell death mechanism of KD548-Fc, an Fc-fused DR4/DR5 dual-specific Kringle domain variant.
Results: KD548-Fc-stimulated DR4/DR5 activate Nox1 NADPH oxidase to generate superoxide anion, leading to reactive oxygen species-mediated apoptotic cell death.
Conclusion: DR4/DR5 have the capability to activate Nox1.
Significance: Our results provide a new signaling pathway of DR4/DR5, distinct from the conventional apoptosis pathway.
Keywords: Apoptosis, Cell Death, Death Domain, Reactive Oxygen Species (ROS), TRAIL, Agonists, Signaling Complex, Superoxide Anion, TRADD, TRAIL Receptors
Abstract
Stimulation of the proapoptotic tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) receptors, death receptors 4 (DR4) and 5 (DR5), conventionally induces caspase-dependent apoptosis in tumor cells. Here we report that stimulation of DR4 and/or DR5 by the agonistic protein KD548-Fc, an Fc-fused DR4/DR5 dual-specific Kringle domain variant, activates plasma membrane-associated Nox1 NADPH oxidase to generate superoxide anion and subsequently accumulates intracellular reactive oxygen species (ROS), leading to sustained c-Jun N-terminal kinase activation and eventual apoptotic cell death in human HeLa and Jurkat tumor cells. KD548-Fc treatment induces the formation of a DR4/DR5 signaling complex containing riboflavin kinase (RFK), Nox1, the Nox1 subunits (Rac1, Noxo1, and Noxa1), TNF receptor-associated death domain (TRADD), and TNF receptor-associated factor 2 (TRAF2). Depletion of RFK, but not the Nox1 subunits, TRADD and TRAF2, failed to recruit Nox1 and Rac1 to DR4 and DR5, demonstrating that RFK plays an essential role in linking DR4/DR5 with Nox1. Knockdown studies also reveal that RFK, TRADD, and TRAF2 play critical, intermediate, and negligible roles, respectively, in the KD548-Fc-mediated ROS accumulation and downstream signaling. Binding assays using recombinantly expressed proteins suggest that DR4/DR5 directly interact with cytosolic RFK through RFK-binding regions within the intracellular death domains, and TRADD stabilizes the DR4/DR5-RFK complex. Our results suggest that DR4 and DR5 have a capability to activate Nox1 by recruiting RFK, resulting in ROS-mediated apoptotic cell death in tumor cells.
Introduction
The proapoptotic tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL,2 Apo2L) receptors, death receptors 4 (DR4, TRAIL-R1) and 5 (DR5, TRAIL-R2), are members of the DR family and are attractive anti-cancer targets because their stimulation with the cognate ligand TRAIL induces apoptosis in various tumor cells without significant cytotoxicity on normal cells (1, 2). The additional TRAIL membrane receptors, death decoy receptor 1 (DcR1, TRAIL-R3) and DcR2 (TRAIL-R4), also bind to TRAIL but cannot induce apoptosis due to a lack of fully functional intracellular death domain (DD) (1). Currently, many agonists against DR4 and/or DR5, including recombinant TRAIL and receptor-specific monoclonal antibodies (mAbs), are in various stages of clinical trials for cancer therapy (1).
Like other DRs, such as TNF receptor 1 (TNFR1) and CD95 (Fas), the cytoplasmic DD of DR4 and DR5 (DR4/DR5) acts as a docking site for intracellular adaptor proteins for downstream signaling (2, 3). Activation of DR4/DR5 by homotrimeric TRAIL results in the oligomerization of the intracellular DD to recruit the primary adaptor protein Fas-associated DD (FADD) via homotypic interactions between the DDs and then caspase-8/-10, forming the so-called death-inducing signaling complex (DISC) (4, 5). Activated caspase-8/10 in DISC directly or indirectly activate effector caspases, such as caspase-3/6/7, triggering caspase-dependent apoptotic cell death (1). Activated CD95 also forms the DISC at the intracellular DD to trigger mainly caspase-dependent apoptotic cell death (2). However, TNFα-bound TNFR1 primarily recruits TNFR-associated death domain (TRADD), rather than FADD, via homotypic DD interactions and then other second adaptor molecules, mediating mainly proinflammatory and necrotic cell death signaling (2). Thus, depending on which adaptor protein binds to the DDs of activated DRs, the outcome of the downstream signaling varies. However, the structural and molecular mechanism(s) by which the DDs of DRs distinguish the respective primary adaptor protein is not clear yet.
Reactive oxygen species (ROS), such as superoxide anion (O2˙̄) and hydrogen peroxide (H2O2), are known to induce a wide range of responses, depending on the cell types and ROS levels within the cell, including apoptotic and necrotic cell death (6). Stimulation of TNFR1, CD95, and DR4/DR5 generate intracellular ROS, the major source of which is mitochondria (6–8). The external cell death stimuli mediated by the DRs cause uncoupling of the mitochondrial electron transport chain and/or collapse of the mitochondrial outer membrane potential, producing intracellular ROS (6). For an additional source of ROS, however, recent studies have shown that TNFR1 and CD95 can activate plasma membrane-associated NADPH oxidases (Nox enzymes) to generate superoxide anion and eventually accumulate intracellular ROS (9–11).
NADPH oxidases are integral membrane proteins that transfer electrons from NADPH to oxygen across biological membranes, generating superoxide anion (O2˙̄) and its downstream ROS, such as H2O2 (12). The Nox family encompasses seven different enzymes, Nox1 to -5, DUOX1, and DUOX2, with distinct cellular distribution and functions in human cells (12). NADPH oxidase activity is controlled by regulatory subunits, including the cytosolic subunits of Rac1, Noxo1, and Noxa1 and membrane-bound p22phox for Nox1 to -3 (12, 13). TNFα-induced activation of Nox1 is mediated by recruiting riboflavin kinase (RFK) to TNFR1 (14). RFK is a cytosolic enzyme that catalyzes the phosphorylation of riboflavin to form flavin mononucleotide (FMN), a precursor to FAD (15). FAD is an essential prosthetic group of Nox1 to -4 for electron transport (15). Thus, RFK activation enhances FAD incorporation into Nox1, facilitating the assembly and activation of Nox1 NADPH oxidase (14). CD95L-bound CD95 also produces superoxide anion and ROS by activating Nox enzymes, such as Nox1, Nox3, and Nox4 (10, 11), although the molecular mechanism(s) remain obscure. There have been no reports yet for DR4/DR5-mediated RFK recruiting or Nox activation.
Recently, we have developed the Kringle domain (KD) scaffold based on the KD2 of human plasminogen as an antibody surrogate (16). From a KD synthetic library on the yeast cell surface, we isolated a variant, KD548, with DR4/DR5 dual-specific binding activity (16, 17). The Fc-fused form in its C terminus, KD548-Fc (Fig. 1A), induced cell death in several cancer cell lines in vitro and suppressed tumor growth in xenograft mouse models even for TRAIL-resistant cells (16). However, the detailed cell death mechanism of KD548-Fc has not been reported yet.
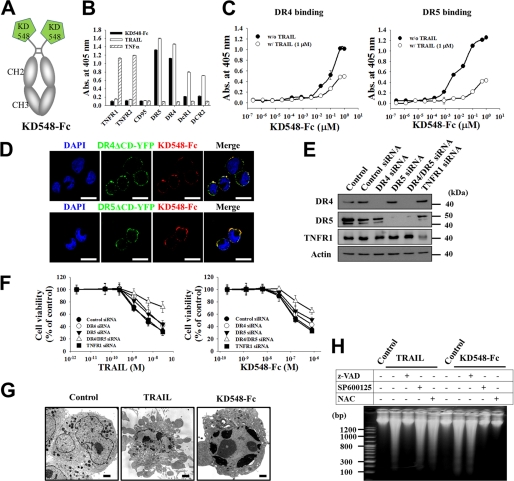
FIGURE 1.
KD548-Fc induces the apoptotic cell death of tumor cells by specifically binding to DR4 and/or DR5. A, schematic diagram of KD548-Fc, the homodimeric Fc-fused form of KD548, generated by its C-terminal fusion to Fc of human IgG1. B, ELISA to analyze the binding specificity of KD548-Fc for the extracellular domains of the indicated TNF family receptors, in comparison with TRAIL and TNFα. C, competition ELISA. Shown is binding activity of KD548-Fc for the plate-coated extracellular domain of DR4 or DR5 in the absence or presence of 1 μm TRAIL. D, colocalization of KD548-Fc with cell surface-expressed DR4 and DR5. TRITC-labeled KD548-Fc was incubated for 1 h at 4 °C with HeLa cells transiently transfected with DR4ΔCD-YFP or DR5ΔCD-YFP fusion protein, respectively, and then visualized by confocal fluorescence microscopy. Nuclei were costained with DAPI. Bar, 20 μm. E and F, knockdown of DR4, DR5, or TNFR1 by siRNA transfection (E) and the effects on TRAIL- or KD548-Fc-mediated cell death (F) in HeLa cells. Cells untransfected (control) or transfected with the indicated siRNA for 36 h (E) were incubated with the indicated concentrations of TRAIL or KD548-Fc for 40 h prior to the MTT assay (F). DR4/DR5 siRNA indicates the cotransfection of DR4 and DR5 siRNAs. G, representative transmission electron microscopy images of HeLa cells, which were left untreated (control) or treated with either TRAIL (30 nm) or KD548-Fc (0.8 μm) for 20 h. Bar, 2 μm. H, oligonucleosomal DNA fragmentation assay for HeLa cells, pretreated for 1 h with Z-VAD-fmk, SP600125, or NAC and further left untreated (control) or treated with TRAIL (30 nm) for 15 h or KD548-Fc (0.8 μm) for 40 h. B, C, and F, data represent mean ± S.E. (error bars) of three independent experiments carried out in triplicate.
In the present work, we demonstrate that DR4/DR5-specific KD548-Fc induces apoptotic cell death via the ROS-dependent sustained c-Jun N-terminal kinase (JNK) activation pathway without caspase activation in HeLa and Jurkat cells. We found that KD548-Fc-induced ROS accumulation resulted from superoxide anion generation from Nox1 NADPH oxidase, which was mediated by the formation of a KD548-Fc-induced DR4/DR5-signaling complex containing RFK, Nox1, Rac1, TRADD, and TNF receptor-associated factor 2 (TRAF2). RFK is an essential component in linking DR4/DR5 with Nox1. We have identified RFK-binding regions within the DDs of DR4/DR5 by mapping the intracellular domains. Our results provide a new signaling pathway of DR4/DR5, directly activating Nox1 via RFK, which is distinct from conventional DISC-mediated apoptosis.
EXPERIMENTAL PROCEDURES
Cell Lines and Reagents
Human cervical carcinoma HeLa cells were purchased from the ATCC (Manassas, VA) and grown in DMEM supplemented with 10% (v/v) FCS, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen) as described previously (18, 19). RFK-deficient HeLashRFK cells that stably express RFK-specific shRNA (14) were a kind gift from Prof. Martin Krönke (University of Cologne) and were grown in the same conditions with wild type HeLa cells. Reagents, described in detail in the supplemental Experimental Procedures, were of analytical grade.
Protein Expression and Purification
KD548-Fc, where KD548 was formatted into the homodimeric Fc-fused form by C-terminal fusion to crystallizable fragment (Fc) of human IgG1, was expressed and purified using the EasySelectTM Pichia expression kit (Invitrogen), essentially as described before (16). The preparations of the other proteins are described in the supplemental Experimental Procedures. The endotoxin level of the purified proteins was monitored by Pyrosate® (Associates of Cape Cod, Inc.), and, if necessary, endotoxin was removed using an endotoxin removal kit (ProteoSpinTM; Norgen Biotek Corp.) (17). The purified proteins were sterilized by filtration using a cellulose acetate membrane filter (0.2 μm) (Nalgene Co.) before use in cell assays.
Binding Analysis by ELISA
Direct and competitive ELISAs were performed as described previously (16–18). All of the details are described in the supplemental Experimental Procedures.
Cellular Treatments and Viability Assay
In all experiments performed with a single concentration of protein, cells were treated with 0.8 μm KD548-Fc or 30 nm TRAIL for the indicated periods with or without a 1-h pretreatment with 10 μm Z-VAD-fmk (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), 10 μm SP60015 (Calbiochem), 100 μm 3-methyladenine (Sigma), 50 μm chloroquine (Sigma), 10 mm N-acetyl-l-cysteine (NAC; Sigma), 50 μm necrostatin-1 (Sigma), or 10 μm diphenyleneiodonium (Sigma), unless otherwise specified. Cell viability was analyzed using an MTT assay kit (Sigma) and presented as the percentage of viable cells compared with untreated control cells (16).
Oligonucleosomal DNA Fragmentation Assay
Cells (2 × 106 cells/well) were plated overnight in 6-well plates and then treated under the conditions specified in the legend to Fig. 1H. Genomic DNA extraction was done using the G-DEXTM IIc kit (Intron Biotechnology) following the manufacturer's instructions and then analyzed by electrophoresis on a 2% (w/v) agarose gel prior to staining with 0.2 μg/ml ethidium bromide.
Light Microscopy and Transmission Electron Microscopy
To analyze the morphology of dying cells, images were taken by a phase-contrast microscope or transmission electron microscope, as described previously (18, 19).
Western Blots
Cells (5 × 105 cells/well) were seeded in 6-well plates, grown overnight, and then treated under given conditions as specified in the figure legends. The standard procedure for Western blots was then performed as described previously (18, 19). Proteins were detected by the primary antibody and secondary antibody of horseradish peroxidase-conjugated anti-rabbit-IgG (Zymed Laboratories Inc.) or anti-mouse-IgG1 (Cell Signaling) and then developed using a Power Opti-ECL reagent (Animal Genetics Inc.). The chemiluminescent signal was detected by an ImageQuant LAS 4000 mini (GE Healthcare). Actin was used for the equal protein loading control.
RNA Interference
In siRNA knockdown experiments, cells (5 × 105 cells/well in 6-well plates or 1 × 104 cells/well in 96-well plates) were transfected with a 2 μm concentration of the indicated siRNA by electroporation in a 0.1-cm diameter capillary (Digital Biotech) and then further incubated 36 h prior to the following experiments (18, 19). Details of the siRNA sequence for each gene are described in the supplemental Experimental Procedures.
Measurement of ROS Production
The ROS level was detected by using 2′,7′-dichlorofluorescein diacetate (Invitrogen), following the manufacturer's protocol (20). Cells (3 × 105 cells) were treated with KD548-Fc or TRAIL for different time periods, collected, and washed with PBS. After incubation with 2′,7′-dichlorofluorescein diacetate (15 μm) for 30 min at 37 °C, the cells were washed once with cold PBS and immediately analyzed by flow cytometry. The ROS production levels of samples were represented as the -fold increase of DCF fluorescence compared with untreated control cells.
Superoxide Anion Generation Assay
Superoxide anion generation was monitored using the Superoxide Anion Assay Kit (Sigma, catalog no. A6105) following the manufacturer's protocol. Approximately 5 × 105 cells grown in the growth medium were washed and resuspended with 100 μl of assay medium in 96-well plates (SPL). KD548-Fc or TRAIL was added to the samples with NADPH (400 μm) and luminol (200 μm). The samples were mixed and placed immediately into a luminometer (BMG Labtech), and then chemiluminescence was measured every 10 min for 2 h. Cells were maintained at 37 °C during the experiment.
Immunoprecipitation
Immunoprecipitation of the DR4/DR5 complex stimulated by TRAIL or KD548-Fc was performed as described previously (18). Briefly, cells (1 × 107 cells/time point) were left untreated or treated with FLAG-tagged TRAIL (2 μg/ml) or KD548-Fc (1 μm) for the indicated periods at 37 °C. The cells were collected and lysed for 30 min on ice in 1 ml of a lysis buffer (20 mm Tris, pH 7.5, 150 mm NaCl, 10% (v/v) glycerol, 1% (v/v) Triton X-100, 2 mm EDTA) with protease inhibitor mixture (Roche Applied Science). After removing cell debris by centrifugation, the cell lysates were incubated overnight at 4 °C with EZviewTM Red anti-FLAG M2-agarose beads (Sigma) to pull down FLAG-tagged TRAIL, or anti-human IgG (Fc-specific)-agarose beads (Sigma) to pull down KD548-Fc. For immunoprecipitation of RFK, DR4, and DR5, the cell lysates were incubated overnight at 4 °C with mouse anti-RFK (Santa Cruz Biotechnology, Inc.), anti-DR4 (Santa Cruz Biotechnology, Inc.), and anti-DR5 (Santa Cruz Biotechnology, Inc.) antibody, respectively. The complexes were then precipitated by agarose-conjugated goat anti-mouse IgG antibody (Sigma), which does not cross-react with human Fc in KD548-Fc. Beads were washed three times with the lysis buffer before the bound proteins were resuspended by boiling in SDS sample buffer. Equal amounts of precipitates were analyzed by Western blots as described above.
Cell Lines and Reagents, Protein Expression and Purification, ELISA, Cell Viability Assay, RNA Interference, Confocal Fluorescence Microscopy, TUNEL Assay, and Surface Plasmon Resonance Experiments
Please see the supplemental Experimental Procedures.
Statistical Analysis
Data are reported as mean ± S.E. of at least three independent experiments carried out in triplicate. Statistical significance was analyzed by a two-tailed Student's t test on Sigma Plot 8.0 software (SPSS Inc.), and a p value of less than 0.05 was considered significant (16).
RESULTS
KD548-Fc Induces Cell Death of Tumor Cells by Specific Binding to DR4 and DR5
The homodimeric KD548-Fc maintained the DR4/DR5 dual binding specificity of KD548 with very weak cross-reactivity for DcR1 and DcR2, determined by ELISA (Fig. 1B) (16, 17). However, KD548-Fc did not cross-react with the soluble extracellular domains of other TNF family receptors, such as TNFR1, TNFR2, and CD95 (Fig. 1B). Surface plasmon resonance analysis demonstrated that KD548-Fc bound to DR5 with a dissociation equilibrium constant (KD) of ∼11 nm and DR4 with a KD of ∼16 nm (supplemental Fig. S1A). Competition ELISA revealed that KD548-Fc partially competed with TRAIL for the binding to the extracellular domains of DR4 or DR5 (Fig. 1C), like KD548 (16), suggesting that it recognizes regions partially overlapping with but distinct from those of TRAIL on DR4 and DR5. When the specific binding activity of KD548-Fc for cell surface-expressed DR4 and DR5 was analyzed by confocal fluorescence microscopy in HeLa cells, KD548-Fc colocalized with DR4 or DR5 (Fig. 1D), like the staining with the positive control of anti-DR4 or anti-DR5 mAb (supplemental Fig. S1B). Knockdown of DR4 and DR5 alone or of both by the transfection of DR4- and DR5-specific siRNA into HeLa cells (Fig. 1E) reduced the cell surface binding levels of KD548-Fc (supplemental Fig. S1C) and KD548-Fc-mediated cell death (Fig. 1F), showing more pronounced effects on the simultaneous knockdown of DR4 and DR5, most likely due to the DR4/DR5 dual-specific binding property of KD548-Fc, as in the case of TRAIL. The partial cell death at the higher concentration of KD548-Fc and TRAIL, even after knockdown of both DR5 and DR4, could be attributed to the incomplete depletion of DR5 and DR4 throughout the incubation period of 40 h after siRNA transfection. TNFR1 knockdown did not affect the cell death mediated by KD548-Fc or TRAIL (Fig. 1F). Taken together, these results demonstrated that KD548-Fc induces tumor cell death by specific engagement of either the proapoptotic DR4 or DR5, or of both expressed on the cell surface.
KD548-Fc Induces Apoptotic Cell Death of Tumor Cells
To determine the cell death mode of KD548-Fc, we investigated the morphological and biochemical features of dying cells by exposure to KD548-Fc, in comparison with TRAIL. The anchorage-dependent HeLa cells dying by KD548-Fc treatment became shrunk and rounded prior to detachment from the culture plates, similar to those undergoing apoptotic cell death by TRAIL (supplemental Fig. S1D). Transmission electron microscopy analysis revealed that, compared with untreated cells, KD548-Fc-treated HeLa and Jurkat cells exhibited the typical characteristics of apoptotic cell death, such as nuclear fragmentation and chromatin condensation lying against the nuclear envelope, similar to those of TRAIL-treated cells (Fig. 1G and supplemental Fig. S2A). However, the characteristics of necrotic cell death, such as cytoplasmic swelling and the rupture of the plasma membrane (21), were not observed in the cells. Both TRAIL and KD548-Fc induced the typical discrete ladder of oligonucleosomal DNA fragmentation in HeLa and Jurkat cells (Fig. 1H and supplemental Fig. S2B), signifying the apoptotic cell death mechanism (21). The TUNEL assay in HeLa cells (supplemental Fig. S1E) and Annexin-V-FITC labeling in Jurkat cells (supplemental Fig. S2C) also indicated that KD548-Fc induces apoptotic cell death of tumor cells.
KD548-Fc-induced Apoptosis Is Mediated by ROS-dependent Sustained JNK Activation
The KD548-Fc-mediated apoptotic cell death pathway was characterized by pretreatments with pharmacological inhibitors and by monitoring the activation of some proteins involved in cell death by Western blots in HeLa and Jurkat cells. The pretreatments of HeLa and Jurkat cells with autophagy inhibitors, 3-methyladenine and chloroquine, or the necrosis inhibitor, necrostatin-1, negligibly influenced the cytotoxicity of KD548-Fc and TRAIL (Fig. 2A and supplemental Fig. S2D), indicating that neither autophagy nor necrosis was involved in the cell death. The pancaspase inhibitor Z-VAD-fmk completely blocked the death of HeLa and Jurkat cells by TRAIL but not by KD548-Fc (Fig. 2A and supplemental Fig. S2D). Further TRAIL, but not KD548-Fc, induced significant cleavage of caspase-8, caspase-3, and poly(ADP-ribose) polymerase (a substrate of caspase-3) from their pro-forms in the cells (Fig. 2B and supplemental Fig. S2, E and F). These results indicated that caspase activation is essential for TRAIL-induced cell death (1) but not for KD548-Fc-mediated cell death. Interestingly, the JNK inhibitor SP600125 and antioxidant NAC effectively suppressed the KD548-Fc-induced oligonucleosomal DNA fragmentation and cell death, but such effects were not observed in the TRAIL-treated cells (Figs. 1H and 2A and supplemental Fig. S2, B and D). Also, KD548-Fc induced prolonged JNK activation that started within 10 h and then persisted through a 30-h incubation in the cells (Fig. 2B and supplemental Fig. S2E). These results suggested that JNK activation and/or oxidative stress, but not caspase activation, play critical roles in KD548-Fc-induced cell death.
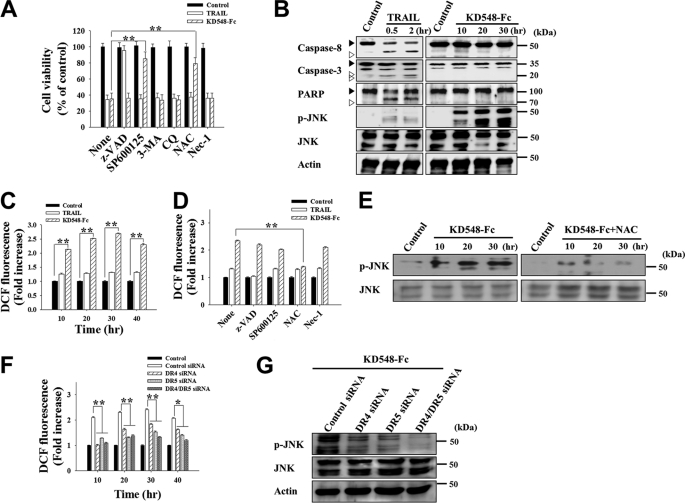
FIGURE 2.
KD548-Fc-induced apoptotic cell death is dependent on ROS generation and JNK activation, but not caspase activation, in HeLa cells. A, cell viability of the cells, pretreated for 1 h with the indicated pharmacological inhibitor and further untreated (control) or treated with TRAIL (30 nm) or KD548-Fc (0.8 μm) for 40 h prior to the MTT assay. B, Western blot analysis of caspase-8 and -3, poly(ADP-ribose) polymerase (PARP), and JNK activation by treatment of TRAIL (30 nm) or KD548-Fc (0.8 μm). The cells were left untreated (control) or treated with TRAIL or KD548-Fc for the indicated periods. C, ROS production in the cells left untreated (control) or treated with TRAIL (30 nm) or KD548-Fc (0.8 μm) for the indicated periods. D, effects of pharmacological inhibitors on the KD548-Fc-induced ROS production in the cells, pretreated for 1 h with the indicated pharmacological inhibitor, and then left untreated (control) or treated with TRAIL (30 nm) or KD548-Fc (0.8 μm) for 20 h. E, effect of NAC on JNK phosphorylation in the cells, pretreated for 1 h with or without NAC and then left untreated (control) or treated with KD548-Fc (0.8 μm) for the indicated periods prior to Western blot analysis. F and G, effects of knockdown of DR4, DR5, or both (DR4/DR5) on ROS production (F) and JNK phosphorylation (G) induced by KD548-Fc (0.8 μm) treatment. Cells transfected with the indicated siRNA for 36 h were left untreated (control) or treated with KD548-Fc for the additional indicated periods (F) or for 20 h (G). A, C, D, and F, data represent mean ± S.E. (error bars) of three independent experiments carried out in triplicate. *, p < 0.05; **, p < 0.001 compared with the corresponding control. F, the indicated p values correspond to each sample transfected with DR4, DR5, or DR4/DR5 siRNA versus the control siRNA-transfected cells.
Because the antioxidant NAC significantly inhibited KD548-Fc-mediated cell death, we measured the time course of intracellular accumulation of ROS by 2′,7′-dichlorofluorescein diacetate staining and then flow cytometry (20). KD548-Fc-treated HeLa and Jurkat cells displayed substantially amplified ROS levels, exhibiting more than 2-fold higher ROS levels over the time course up to 40 h compared with those of untreated control cells (Fig. 2C and supplemental Fig. S2G). However, TRAIL treatment exhibited marginal ROS accumulation in the cells. The antioxidant NAC significantly abolished ROS production and JNK activation induced by KD548-Fc (Fig. 2, D and E, and supplemental Fig. S2H). In contrast, the JNK inhibitor SP600125 slightly decreased the KD548-Fc-induced ROS accumulation (Fig. 2D). These results indicated that ROS act as upstream signal causing JNK activation and its prolonged activation. Z-VAD-fmk and necrostatin-1 negligibly affected ROS accumulation in the KD548-Fc-treated cells (Fig. 2D and supplemental Fig. S2H), in line with their insignificant effects on the cell death (Fig. 2A and supplemental Fig. S2D).
The depletion of DR4 and DR5 alone or of both by siRNA in HeLa cells substantially decreased both ROS accumulation and sustained JNK activation mediated by KD548-Fc, with the simultaneous knockdown of the two receptors showing more pronounced effects than either one of the two receptors (Fig. 2F and supplemental Fig. 2G). This result confirmed that KD548-Fc induces ROS generation and sustained JNK activation via the specific engagement of DR5 and/or DR4.
KD548-Fc Activates Nox1 NADPH Oxidase to Generate Superoxide Anion and Intracellular ROS
Recent studies have shown that TNFR1 and CD95 have a capability to activate plasma membrane-associated NADPH oxidases to produce superoxide anion, leading to intracellular ROS accumulation (9, 11, 14). To address whether ROS accumulation caused by KD548-Fc-bound DR4/DR5 stimulation is mediated by activating NADPH oxidase(s), we monitored the generation of superoxide anion, O2˙̄, in real time using a probe of luminol that specifically reacts with O2˙̄ to generate chemiluminescence (11) in HeLa cells after KD548-Fc treatment. Surprisingly enough, luminol chemiluminescence started to increase dramatically at ∼30–40 min and reached a maximal level at ∼80 min after KD548-Fc stimulation (Fig. 3A), which was similar to the kinetic profiles of TNFα-mediated O2˙̄ production via Nox1 activation in mouse fibroblasts (9). However, TRAIL failed to produce O2˙̄. In the settings of HCT116 and Jurkat cells, KD548-Fc also generated O2˙̄ showing similar kinetics to that of HeLa cells (supplemental Figs. S3B and S4B). The O2˙̄ production in response to KD548-Fc was dramatically attenuated by the addition of the antioxidant NAC or the NADPH oxidase inhibitor diphenyleneiodonium (11) (Fig. 3A), suggesting the involvement of NADPH oxidase(s) in the KD548-Fc-mediated O2˙̄ production.
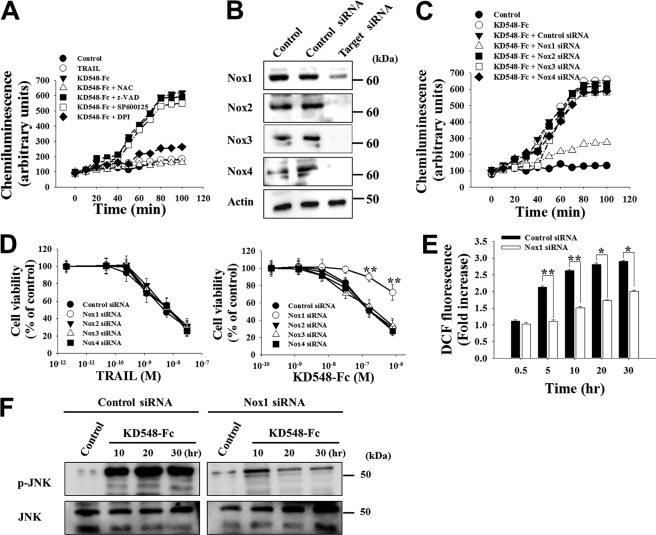
FIGURE 3.
KD548-Fc activates Nox1 NADPH oxidase to produce superoxide anion, leading to intracellular ROS accumulation, sustained JNK activation, and eventual apoptotic cell death in HeLa cells. A, superoxide anion generation assay of the cells left untreated (control) or treated with TRAIL (30 nm) or KD548-Fc (0.8 μm) in the absence or presence of the indicated chemicals. The data are representative of three separate experiments with the same results. B, Western blots showing the knockdown of Nox1, Nox2, Nox3, or Nox4 expression by siRNA transfection into HeLa cells. Control, untransfected cells. C and D, effects of knockdown of Nox1, Nox2, Nox3, or Nox4 on superoxide anion generation (C) and cell viability (D) in the cells, left untreated (control) or treated with KD548-Fc (C and D) or TRAIL (D). C, cells were treated with 0.8 μm of KD548-Fc. D, cells transfected with siRNA for 36 h were further incubated with the indicated concentrations of KD548-Fc or TRAIL for 40 h. E and F, effect of Nox1 knockdown on ROS production (E) or JNK phosphorylation (F) in the cells, untreated (control) or treated with KD548-Fc (0.8 μm) for the indicated periods. D and E, data represent mean ± S.E. (error bars) of three independent experiments carried out in triplicate. *, p < 0.05; **, p < 0.001 compared with the cells transfected with control siRNA.
Among the seven NADPH oxidases in Nox family proteins, Nox1, Nox2, Nox3, and/or Nox4 have been activated by TNFR1 or CD95 stimulation (7). Western blot analysis confirmed the expression of Nox1 to Nox4 in HeLa and HCT116 cells (Fig. 3B and supplemental Fig. S3A), consistent with previous results (14, 22). To address which Nox enzyme is involved in the O2˙̄ production by KD548-Fc, each Nox enzyme was knocked down by siRNA transfection into HeLa and HCT116 cells prior to the O2˙̄ production assay. Only Nox1 knockdown dramatically blocked KD548-Fc-induced O2˙̄ production, whereas the knockdown of Nox2, Nox3, or Nox4 did not show such an effect in the cells (Fig. 3C and supplemental Fig. S3B). In a similar context, the knockdown of Nox1, but not Nox2, Nox3, and Nox4, significantly protected HeLa and HCT116 cells from the KD548-Fc-induced cell death (Fig. 3D and supplemental Fig. S3C). In Jurkat cells, Nox1 knockdown also substantially reduced the O2˙̄ production and cell death triggered by KD548-Fc (supplemental Fig. S4, B and C). Furthermore, Nox1 knockdown significantly abolished the intracellular ROS accumulation and sustained JNK activation mediated by KD548-Fc in the three cell types (Fig. 3, E and F, and supplemental Figs. S3, D and E, and S4, D and E). These results suggested that Nox1 is mainly responsible for O2˙̄ generation by KD548-Fc stimulation, leading to the intracellular ROS accumulation, sustained JNK activation, and eventual apoptotic cell death.
KD548-Fc Stimulation Induces the DR4/DR5-associated Signaling Complex Composed of RFK, Nox1, Nox1 Cytosolic Subunits, TRADD, and TRAF2
Recent data have shown that TNFα-stimulated TNFR1 activates Nox1 by inducing the formation of a signaling complex containing Nox1, RFK, and TRADD in mouse and human cells (14). To elucidate how DR4/DR5-specific KD548-Fc activates Nox1, we analyzed the KD548-Fc-induced DR4/DR5 signaling complex by immunoprecipitation from the lysates of KD548-Fc-treated HeLa and Jurkat cells through KD548-Fc itself, in comparison with TRAIL. TRAIL induced the conventional DR4/DR5 DISC assembled with FADD and caspase-8, but not with RIP1, TRADD, or TRAF2, in both cells (Fig. 4A and supplemental Fig. S5A), which is consistent with earlier works (1). Interestingly enough, KD548-Fc coprecipitated DR5 and DR4 with RFK, Nox1, the Nox1 cytosolic subunits (Rac1, Noxo1, and/or Noxa1), TRADD, and TRAF2 within 30 min of treatment of the two cells (Fig. 4A and supplemental Fig. S5A), demonstrating that KD548-Fc induced a physical interaction between DR4/DR5 and Nox1. The cytosolic subunits are essential for Nox1 activation (12, 13). However, the signaling complex did not contain FADD and caspase-8, which are two critical upstream activators of the caspase-dependent apoptotic cell death mediated by TRAIL (1). RIP1, an essential component in TNFα-induced necrotic cell death signaling (9), was not coprecipitated with KD548-Fc-stimulated DR4/DR5. Consistent with the signaling complex, KD548-Fc showed cytotoxicity in Jurkat cells deficient in FADD, caspase-8, or RIP1 comparable with that of the wild-type cells (supplemental Fig. S5B). KD548-Fc did not pull down CD95 and TNFR1 (Fig. 4A and supplemental Fig. S5A), eliminating the possibility of CD95- or TNFR1-mediated signaling complex formation. Down-regulation of DR5 and DR4 alone or together by siRNA transfection significantly reduced the amount of the recruited molecules to the KD548-Fc-induced signaling complex (Fig. 4B), signifying the specificity of the signaling complex formation via DR4 and DR5.
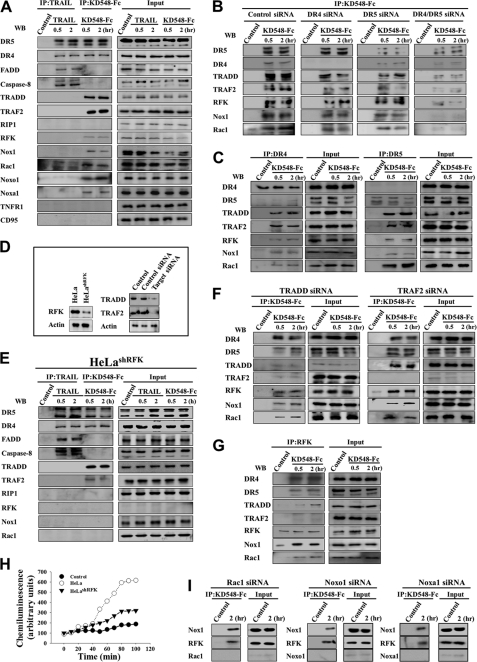
FIGURE 4.
KD548-Fc induces a DR4/DR5-associated signaling complex composed of RFK, Nox1, Rac1, TRADD, and TRAF2 in HeLa cells, where RFK plays an essential role in recruiting Nox1 to DR4/DR5. A, Western blots (WB) showing immunoprecipitation (IP) of the lysates of TRAIL- and KD548-Fc-treated cells using anti-FLAG antibody (for TRAIL) and anti-human Fc antibody (for KD548-Fc), respectively. Cells were left untreated (control) or treated with FLAG-tagged TRAIL (30 nm) or KD548-Fc (0.8 μm) for 0.5 or 2 h and then subjected to immunoprecipitation and/or Western blot. B, Western blots showing KD548-Fc immunoprecipitation of the lysates of DR4-, DR5-, or DR4/DR5-depleted cells using anti-human Fc antibody after KD548-Fc (0.8 μm) treatment for 0.5 or 2 h or no treatment (control). C, Western blots showing DR4 and DR5 immunoprecipitation of the cell lysates using anti-DR4 and anti-DR5 antibody, respectively, after KD548-Fc (0.8 μm) treatment for 0.5 or 2 h or no treatment (control). D, Western blot analysis of the expression levels of RFK, TRADD, and TRAF2 in wild type, RFK-deficient HeLashRFK, TRADD, and TRAF2 siRNA-transfected HeLa cells, respectively. Control, untransfected cells. E, Western blots showing immunoprecipitation of the lysates of TRAIL- and KD548-Fc-treated RFK-deficient HeLashRFK cells using anti-FLAG antibody (for TRAIL) and anti-human Fc antibody (for KD548-Fc), respectively. Experiments were performed as described in A. F, Western blots showing KD548-Fc immunoprecipitation of the lysates of TRADD- or TRAF2-depleted cells using anti-human Fc antibody after KD548-Fc (0.8 μm) treatment for 0.5 or 2 h or no treatment (control). G, Western blots showing RFK immunoprecipitation of the cell lysates using anti-RFK antibody after KD548-Fc (0.8 μm) treatment for 0.5 or 2 h or no treatment (control). H, superoxide anion generation assay in wild-type and RFK-deficient HeLashRFK cells by KD548-Fc (0.8 μm) treatment. Control, untreated wild type HeLa cells. I, Western blots showing immunoprecipitation of the lysates of Rac1-, Noxo1-, or Noxa1-depleted cells using anti-human Fc antibody after KD548-Fc (0.8 μm) treatment for 2 h or no treatment (control).
KD548-Fc possesses DR4/DR5 dual binding specificity. To determine which receptor is involved in the signaling complex formation, we immunoprecipitated DR5 or DR4 using the respective specific mAb from the lysates of KD548-Fc-treated HeLa cells. Both DR5 and DR4 independently coimmunoprecipitated RFK, Nox1, Rac1, TRADD, and TRAF2 (Fig. 4C), suggesting that DR5 and DR4 have the independent and redundant ability to form the signaling complex by KD548-Fc.
RFK, but Not TRADD, TRAF2, and Nox1 Subunits, Is Essential in Recruiting Nox1 to DR4/DR5
To determine the individual role of RFK, TRADD, and TRAF2 in the recruiting of Nox1 to DR4/DR5, we analyzed the KD548-Fc-stimulated DR4/DR5 signaling complex by pulling down KD548-Fc after the down-regulation of each protein in HeLa cells (Fig. 4D). In RFK-deficient HeLashRFK cells that stably express RFK-specific shRNA (14), immunoprecipitation of KD548-Fc coprecipitated DR4/DR5, TRADD, and TRAF2 but failed to pull down RFK, Nox1, and Rac1 (Fig. 4E), indicating that RFK plays a critical role in linking DR4/DR5 with Nox1 and Rac1. In the lysates of TRADD-depleted HeLa cells, KD548-Fc coprecipitated DR4/DR5 with RFK, Nox1, and Rac1 but only negligibly with TRAF2 (Fig. 4F), indicating that TRADD is dispensable in recruiting RFK, Nox1, and Rac1 to DR4/DR5 but is an essential component for TRAF2 recruiting to the complex. TRAF2 down-regulation abrogated only TRAF2 recruitment without significant influence on the recruitment of the other components to the signaling complex (Fig. 4F). RFK immunoprecipitation from the lysates of KD548-Fc-treated HeLa cells coprecipitated DR4/DR5, Nox1, Rac1, and TRADD, but not TRAF2, suggesting that RFK has strong interactions with the coprecipitated proteins but not with TRAF2 (Fig. 4G). RFK-deficient HeLashRFK cells exhibited minimal production of O2˙̄ by KD548-Fc treatment, compared with wild-type HeLa cells (Fig. 4H).
KD548-Fc immunoprecipitation also coprecipitated the Nox1 cytosolic subunits, such as Rac1, Noxo1, and Noxa1 (12, 13). Knockdown of each cytosolic subunit (Rac1, Noxo1, and Noxa1) did not abolish the recruitment of both Nox1 and RFK to the KD548-Fc-induced signaling complex (Fig. 4I), suggesting that the cytosolic subunits are not required for recruiting Nox1 to DR4/DR5. Taken together, the above results suggested that, in the formation of the KD548-Fc-induced signaling complex, RFK is a critical component in recruiting Nox1 to DR4/DR5, but TRADD, TRAF2, and the cytosolic Nox1 subunits are dispensable.
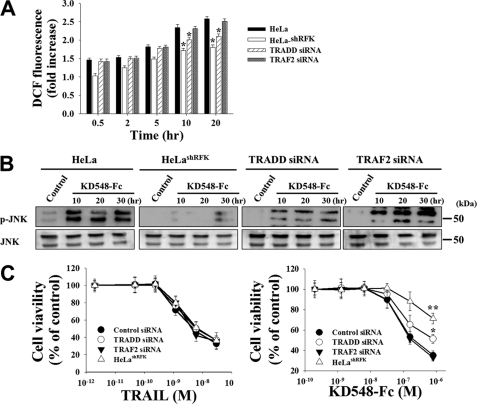
RFK and TRADD, but Not TRAF2, Are Important in ROS Accumulation and Downstream Signaling Induced by KD548-Fc
We further investigated any specific role(s) of TRADD, RFK, and TRAF2 in the KD548-Fc-induced ROS accumulation after the down-regulation of each protein in HeLa cells. In HeLashRFK cells, ROS production was almost completely reduced in response to KD548-Fc (Fig. 5A), in line with a reduction in O2˙̄ generation (Fig. 4H). TRADD knockdown resulted in substantial attenuation of ROS accumulation, but much less strongly than that caused by RFK deficiency. TRAF2 knockdown did not show a significant reduction in ROS production, compared with that of the wild type cells. In a similar context, knockdown of RFK or TRADD with RFK knockdown more substantial effect, but not TRAF2, attenuated the sustained JNK activation and decreased the susceptibility to cell death induced by KD548-Fc (Fig. 5, B and C). In contrast, TRAIL treatment exhibited significant cytotoxicity, regardless of the knockdown of TRADD, RFK, or TRAF2 (Fig. 5C). Taken together, the above results suggested that, in KD548-Fc-induced ROS generation and subsequent downstream signaling cascades, RFK, TRADD, and TRAF2 play critical, intermediate, and negligible roles, respectively.
FIGURE 5.
RFK, TRADD, and TRAF2 play critical, intermediate, and negligible roles, respectively, in the KD548-Fc-induced ROS generation, sustained JNK activation, and cell death in HeLa cells. A–C, ROS production (A), Western blots of JNK phosphorylation (B), and cell viability (C) of wild-type, RFK-deficient HeLashRFK, and TRADD- and TRAF2-depleted cells by KD548-Fc treatment. Cells were left untreated (control) or treated with KD548-Fc (0.8 μm) for the indicated periods (A and B) or treated with the indicated concentrations of KD548-Fc or TRAIL for 40 h (C). For siRNA-transfected cells, cells were transfected with the indicated siRNA for 36 h prior to TRAIL or KD548-Fc treatment. A and C, data represent mean ± S.E. (error bars) of three independent experiments carried out in triplicate. *, p < 0.05; **, p < 0.001 compared with wild type HeLa cells (A) or the cells transfected with control siRNA (C).
DR5 and DR4 Have RFK Binding Regions within DDs
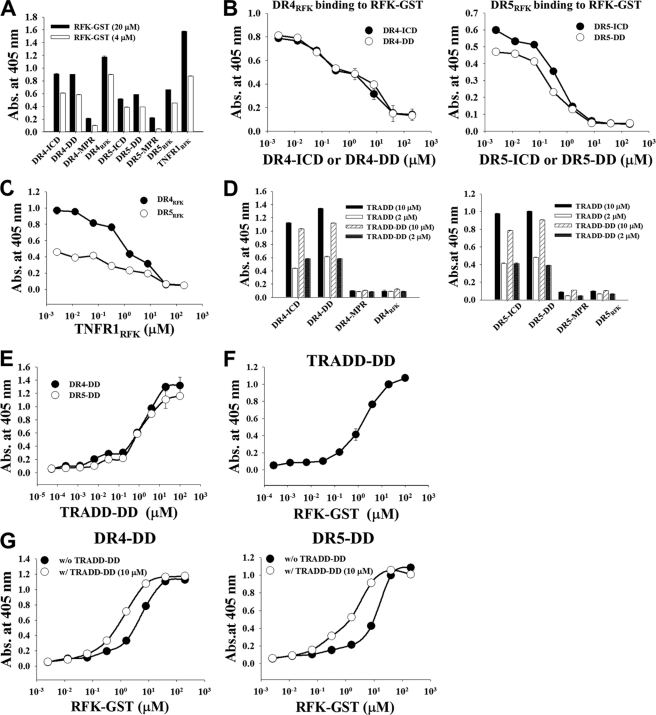
Direct interaction of DR4 and/or DR5 with cytosolic RFK has not yet been demonstrated. To identify which intracellular regions of DR4 and DR5 are responsible for the interaction with RFK, we dissected the intracellular regions of DR4 and DR5 at the domain level and determined the interactions with RFK in the recombinantly purified soluble forms. As shown in supplemental Fig. S6A, the whole intracellular region of DR4 (residues 263–468, DR4-ICR) and DR5 (residues 232–440, DR5-ICR) was divided into two regions. One is the membrane-proximal region (MPR) before starting the DD, dubbed DR4-MPR (residues 263–366) and DR5-MPR (residues 232–341); the other is the C-terminal region containing the DD, dubbed DR4-DD (residues 365–448) and DR5-DD (residues 339–422). All of the proteins were solubly expressed in the cytosol of bacteria and purified to more than 90% purity (supplemental Fig. S6D). Recombinant RFK was prepared as a C-terminal glutathione S-transferase fusion protein (RFK-GST). In the TNFR1-RFK interactions, the N-terminal residues 359–391 within the DD of TNFR1, dubbed TNFR1RFK, are responsible for RFK binding (14, 23). By sequence alignment of the DDs of DR4/DR5 with TNFR1-DD (supplemental Fig. S6B), we assigned the regions encompassing residues 367–400 within DR4-DD and residues 342–374 within DR5-DD as the putative RFK binding regions and synthesized them as peptides, dubbed DR5RFK and DR4RFK.
DR4-ICR/DR5-ICR, DR4-DD/DR5-DD, and DR4RFK/DR5RFK, but not DR4-MPR/DR5-MPR without the DD, displayed significant binding to RFK-GST, with DR4RFK and DR5RFK showing the highest binding level (Fig. 6A). DR4-ICR/DR4-DD and DR5-ICR/DR5-DD efficiently competed with DR4RFK and DR5RFK, respectively, for RFK-GST binding in a concentration-dependent manner (Fig. 6B), suggesting that the regions of DR4RFK and DR5RFK within the DDs are indeed responsible for the interactions of DR4 and DR5 with RFK. The TNFR1RFK peptide showed a concentration-dependent competition binding to RFK-GST with DR4RFK and DR5RFK (Fig. 6C), suggesting that DR4RFK and DR5RFK recognize regions of RFK that are same with or similar to those recognized by TNFR1RFK.
FIGURE 6.
DR4 and DR5 interact with RFK through the RFK-binding regions within the DDs to form the DR4/DR5-RFK complex, which is stabilized by the presence of TRADD. A, binding activity of RFK-GST (4 or 20 μm) for plate-coated intracellular fragments of DR4 and DR5, analyzed by ELISA. For a detailed description of each fragment, see supplemental Fig. S6. B and C, competition ELISA. B, binding activity of DR4RFK (30 μm) and DR5RFK (30 μm) for plate-coated RFK-GST was assessed in the presence of serially diluted DR4-ICD or DR4-DD and DR5-ICD or DR5-DD, respectively. C, binding activity of DR4RFK (30 μm) and DR5RFK (30 μm) for plate-coated RFK-GST was assessed in the presence of serially diluted TNFR1RFK. D, binding activity of TRADD or TRADD-DD (2 or 10 μm) to plate-coated intracellular fragments of DR4 and DR5. E–G, direct ELISA. E, binding activity of TRADD-DD to plate-coated DR4-DD or DR5-DD. F, binding activity of RFK-GST to plate-coated TRADD-DD. G, binding activity of RFK-GST to plate-coated DR4-DD or DR5-DD in the absence or presence of 10 μm TRADD-DD. A–G, data represent mean ± S.E. (error bars) of three independent experiments carried out in triplicate.
DDs of DR5 and DR4 Interact Simultaneously with RFK and TRADD
Because TRADD was recruited to the KD548-Fc-induced DR4/DR5 signaling complex, we also determined which intracellular regions of DR4 and DR5 are responsible for the interactions with whole TRADD or TRADD-DD (residues 179–289) that was prepared from bacterial expression (supplemental Fig. S6D). The strong interactions of DR4 and DR5 with TRADD occurred via their DDs, with apparent KD of ∼2.1 and 2.9 μm, respectively (Fig. 6, D and E). However, no interactions were observed between TRADD and either DR4RFK or DR5RFK (Fig. 6D). Further, TRADD-DD exhibited concentration-dependent interactions with RFK-GST (KD ≈ 2.4 μm) (Fig. 6F). Very interestingly, the presence of TRADD-DD augmented ∼5-fold the interactions between RFK-GST and either DR4-DD (KD of ∼5.5 and 1.3 μm in the absence and presence of TRADD-DD, respectively) or DR5-DD (KD of ∼9.5 and 1.6 μm in the absence and presence of TRADD-DD, respectively) (Fig. 6G). Taken together, these results suggested that TRADD-DD stabilizes the interactions of DR4/DR5 with RFK by simultaneous interactions with the DDs of DR4/DR5 and RFK at regions distinct from those responsible for DR4RFK/DR5RFK and RFK interactions.
DISCUSSION
Conventionally activated DR4 and DR5, in response to the cognate ligand TRAIL or agonistic mAbs, form the canonical DISC, which activates initiator caspase-8/10 and then effector caspase-3/6/7, triggering apoptosis in various tumor cells (1). In this study, we have shown that KD548-Fc-stimulated DR4/DR5 activate plasma membrane-associated Nox1 NADPH oxidase to generate superoxide anion by forming a unique signaling complex, including RFK, Nox1, the Nox1 cytosolic subunits (Rac1, Noxo1, and Noxa1), TRADD, and TRAF2. The signaling complex was formed within 30 min after cells were treated with KD548-Fc, coincident with the O2˙̄ generation. Although other Nox enzymes, such as Nox2, Nox3, and Nox4, are expressed in HeLa and HCT116 cells, our data suggest that only Nox1 is activated by KD548-Fc-mediated DR4/DR5 stimulation in the cells. Nox1 knockdown substantially abolished intracellular ROS accumulation, suggesting that O2˙̄ generated outside the cells is the main source of ROS by conversion into plasma membrane-permeable ROS species, such as H2O2 (9). Our data also suggest that the Nox1-mediated ROS accumulation is responsible for sustained JNK activation-dependent apoptotic cell death in response to KD548-Fc.
Nox1 recruitment to the intracellular DDs of DR4/DR5 seems to be directly mediated by the adaptor protein RFK rather than the Nox1 cytosolic subunits, like in the case of RFK-mediated linking of TNFR1 with Nox1 (14). In the RFK-deficient HeLashRFK cells, KD548-Fc-induced ROS accumulation, sustained JNK activation, and apoptotic cell death were dramatically inhibited, supporting the essential role of RFK in the DR4/DR5-mediated Nox1 activation. TRADD knockdown did not prevent the recruitment of RFK, Nox1, and Rac1 to KD548-Fc-stimulated DR4/DR5, but it substantially attenuated ROS accumulation, sustained JNK activation, and apoptotic cell death in HeLa cells. These results are similar to the role of TRADD in TNFα-induced Nox1 activation in mouse and human cells, where TRADD was dispensable in the formation of the TNFα-induced signaling complex of TNFR1-RFK-Nox1 (14) but essential for TNFα-induced ROS accumulation mediated by Nox1 activation (9). In the study with recombinantly purified proteins, TRADD improved ∼5-fold the interactions of RFK with the DDs of DR4/DR5, suggesting that the ternary complex of DR4/DR5-DD·TRADD·RFK is more stable than binary complexes consisting of two of the three molecules. These data suggest that RFK can physically and functionally couple KD548-Fc-stimulated DR4/DR5 to Nox1, the complex of which is stabilized by TRADD, enhancing the activity of RFK and/or Nox1. TRAF2 played a negligible role in the formation of the KD548-Fc-induced signaling complex and subsequent downstream signaling. TRADD contains an N-terminal TRAF2-binding domain and a C-terminal DD interacting with other DDs (24) (supplemental Fig. S6C). Thus, it seems that TRAF2 is recruited to the activated complex indirectly via the adaptor protein TRADD, but its effect on the downstream signaling is not clear at this point.
Our results suggest that KD548-Fc-stimulated DR4/DR5 simultaneously interacts with RFK and TRADD via the DDs. The DD is a homotypic protein interaction module composed of a bundle of six α-helices, the tertiary structures of which are closely superimposed despite the low sequence homology below 30% (3). The DDs of DR5 and DR4 share ∼63% identity with each other (supplemental Fig. S6B), indicative of their redundant functions. Our data have shown that the DR4RFK and DR5RFK regions, structurally equivalent to the RFK-binding regions of the DD of TNFR1 (TNFR1RFK) (14), are indeed responsible for the interactions of DR4 and DR5 with RFK. The RFK has 6-stranded antiparallel β-barrel structures (15), which is much different from the DD structure. Accordingly, the DDs of DR4/DR5 have a capability to interact heterotypically with RFK, like the interactions of TNFR1-DD with RFK (14) and CD95-DD with the caspase recruitment domain (25). Although DR4/DR5 activated by TRAIL primarily recruit FADD via the DD interactions (2), KD548-Fc-stimulated DR4/DR5 interact with TRADD through the homotypic DD interactions. Previous reports have also shown that DR4 and/or DR5 interact directly with TRADD rather than FADD (18, 26, 27).
The question is how TRAIL and KD548-Fc recruit different signaling adaptors to the same DDs of DR4/DR5. We hypothesize two possible reasons: different binding regions and valency between TRAIL and KD548-Fc. KD548-Fc recognizes partially overlapping but different extracellular regions of DR5 and DR4 from those of TRAIL. Recently, we reported that the KD548 single domain without Fc fusion has bivalent binding ability, with each binding site possessing independent DR4/DR5 dual-specific binding ability (17). Thus, homodimeric KD548-Fc has tetravalent binding sites, each site of which has DR4/DR5 dual binding specificity. Tetravalent KD548-Fc might induce higher oligomeric states of DR4/DR5 than those induced by homotrimeric TRAIL. The higher valency and distinct binding sites of KD548-Fc from those of TRAIL raise the possibility that KD548-Fc induces a distinct conformation and/or oligomeric states in the intracellular DDs of DR4/DR5, which may provide preferential binding surfaces to RFK and TRADD over the primary adaptor FADD of TRAIL. Thus, it seems that, depending on the external stimuli, the DD of activated DR4 and/or DR5 may adopt distinct conformations, which provide preferential interaction sites for the adaptor proteins, such as FADD, TRADD, and/or RFK.
ROS play a critical role in apoptotic and necrotic cell death by oxidatively damaging many molecular targets and thus affecting various signaling cascades (6, 7). One of the prominent effects of ROS is the direct activation of JNK and then sustaining the activation by inhibiting mitogen-activated protein kinase phosphatases (28). ROS generated by KD548-Fc-mediated Nox1 activation leads to prolonged JNK activation, as does ROS produced by TNFR1- or CD95-mediated Nox activation (9, 10). Interestingly, in the sustained JNK activation-dependent cell death induced by Nox1-mediated ROS, TNFα induces necrotic cell death (9), whereas DR4/DR5-specific KD548-Fc induces apoptotic cell death, like CD95-mediated apoptosis (10). The apoptotic morphological features of dying cells caused by exposure to KD548-Fc and little effects of the necrosis inhibitor necrostatin-1 and autophagy inhibitors (3-methyladenine and chloroquine) on KD54-Fc-induced cell death exclude necrosis and autophagy as the primary cell death mechanisms of KD548-Fc. In good agreement with our results, ROS-mediated sustained JNK activation was responsible for the caspase-independent apoptotic cell death of Jurkat cells mediated by an agonistic mAb against DR5 (20). The protein RIP1 is required for CD95L-, TNFα-, and TRAIL-induced necrotic cell death in human cells (29) and the ROS-mediated necrotic cell death by TNFα in mouse fibroblasts (9). However, RIP1 was absent in the DR4/DR5-associated signaling complex induced by KD548-Fc in HeLa and Jurkat cells. Thus, the different signaling complexes induced between TNFα and KD548-Fc might explain the distinct cell death mode. However, further study is required to elucidate the exact mechanism by which Nox1-mediated ROS accumulation and JNK activation triggered by KD548-Fc-stimulated DR4/DR5 induce apoptosis.
DR4/DR5 and Nox1 are expressed in a wide range of cells (1, 12). KD548-Fc killed a variety of tumor cells in vitro and efficiently inhibited tumor growth in mouse models, including TRAIL-resistant cells (16), suggesting that the alternative cell death pathway of KD548-Fc to that of TRAIL might overcome some of the TRAIL resistance mechanism. Because there have been no reports of physiological agonists mimicking KD548-Fc, however, the physiological relevance of the KD548-Fc-induced DR4/DR5 signaling pathway remains to be determined. In conclusion, our results propose that DR4 and/or DR5 have the capability to activate Nox1 via RFK recruitment, resulting in intracellular ROS accumulation, sustained JNK activation, and eventually apoptotic cell death in tumor cells.
Supplementary Material
Acknowledgments
We thank Prof. Martin Krönke (University of Cologne) for providing HeLashRFK cells and the bacterial plasmid encoding RFK-GST. We also thank Dr. Avi Ashkenazi (Genentech Inc.) and You-Sun Kim (Ajou University, Korea) for helpful discussions.
This work was supported by grants from the Korean Health Technology R&D Project (A101800) from the Ministry for Health, Welfare & Family Affairs, the Converging Research Center Program (2009-0093653) and the Priority Research Center Program (2011-0022978) from the National Research Foundation of MEST, and the “GRRC” Project of the Gyeonggi Provincial Government, Republic of Korea.

This article contains supplemental Experimental Procedures and Figs. S1–S6.
- TRAIL
- TNF-related apoptosis-inducing ligand
- Nox
- NADPH oxidase
- ROS
- reactive oxygen species
- DR
- death receptor
- DD
- death domain
- DISC
- death-inducing signaling complex
- TNFR
- TNF receptor
- TRADD
- TNFR-associated death domain
- RFK
- riboflavin kinase
- KD
- Kringle domain
- Z
- benzyloxycarbonyl
- fmk
- fluoromethyl ketone
- NAC
- N-acetyl-l-cysteine
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- MPR
- membrane-proximal region.
REFERENCES
- 1. Pennarun B., Meijer A., de Vries E. G., Kleibeuker J. H., Kruyt F., de Jong S. (2010) Playing the DISC. Turning on TRAIL death receptor-mediated apoptosis in cancer. Biochim. Biophys. Acta 1805, 123–140 [DOI] [PubMed] [Google Scholar]
- 2. Wilson N. S., Dixit V., Ashkenazi A. (2009) Death receptor signal transducers. Nodes of coordination in immune signaling networks. Nat. Immunol. 10, 348–355 [DOI] [PubMed] [Google Scholar]
- 3. Park H. H. (2011) Structural analyses of death domains and their interactions. Apoptosis 16, 209–220 [DOI] [PubMed] [Google Scholar]
- 4. Kischkel F. C., Lawrence D. A., Chuntharapai A., Schow P., Kim K. J., Ashkenazi A. (2000) Apo2L/TRAIL-dependent recruitment of endogenous FADD and caspase-8 to death receptors 4 and 5. Immunity. 12, 611–620 [DOI] [PubMed] [Google Scholar]
- 5. Sprick M. R., Weigand M. A., Rieser E., Rauch C. T., Juo P., Blenis J., Krammer P. H., Walczak H. (2000) FADD/MORT1 and caspase-8 are recruited to TRAIL receptors 1 and 2 and are essential for apoptosis mediated by TRAIL receptor 2. Immunity. 12, 599–609 [DOI] [PubMed] [Google Scholar]
- 6. Shen H. M., Pervaiz S. (2006) TNF receptor superfamily-induced cell death. Redox-dependent execution. FASEB J. 20, 1589–1598 [DOI] [PubMed] [Google Scholar]
- 7. Morgan M. J., Liu Z. G. (2010) Reactive oxygen species in TNFα-induced signaling and cell death. Mol. Cells 30, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee M. W., Park S. C., Kim J. H., Kim I. K., Han K. S., Kim K. Y., Lee W. B., Jung Y. K., Kim S. S. (2002) The involvement of oxidative stress in tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL)-induced apoptosis in HeLa cells. Cancer Lett. 182, 75–82 [DOI] [PubMed] [Google Scholar]
- 9. Kim Y. S., Morgan M. J., Choksi S., Liu Z. G. (2007) TNF-induced activation of the Nox1 NADPH oxidase and its role in the induction of necrotic cell death. Mol. Cell 26, 675–687 [DOI] [PubMed] [Google Scholar]
- 10. Reinehr R., Becker S., Eberle A., Grether-Beck S., Häussinger D. (2005) Involvement of NADPH oxidase isoforms and Src family kinases in CD95-dependent hepatocyte apoptosis. J. Biol. Chem. 280, 27179–27194 [DOI] [PubMed] [Google Scholar]
- 11. Suzuki Y., Ono Y., Hirabayashi Y. (1998) Rapid and specific reactive oxygen species generation via NADPH oxidase activation during Fas-mediated apoptosis. FEBS Lett. 425, 209–212 [DOI] [PubMed] [Google Scholar]
- 12. Petry A., Weitnauer M., Görlach A. (2010) Receptor activation of NADPH oxidases. Antioxid. Redox. Signal. 13, 467–487 [DOI] [PubMed] [Google Scholar]
- 13. Jiang F., Zhang Y., Dusting G. J. (2011) NADPH oxidase-mediated redox signaling. Roles in cellular stress response, stress tolerance, and tissue repair. Pharmacol. Rev. 63, 218–242 [DOI] [PubMed] [Google Scholar]
- 14. Yazdanpanah B., Wiegmann K., Tchikov V., Krut O., Pongratz C., Schramm M., Kleinridders A., Wunderlich T., Kashkar H., Utermöhlen O., Brüning J. C., Schütze S., Krönke M. (2009) Riboflavin kinase couples TNF receptor 1 to NADPH oxidase. Nature 460, 1159–1163 [DOI] [PubMed] [Google Scholar]
- 15. Karthikeyan S., Zhou Q., Mseeh F., Grishin N. V., Osterman A. L., Zhang H. (2003) Crystal structure of human riboflavin kinase reveals a β barrel fold and a novel active site arch. Structure 11, 265–273 [DOI] [PubMed] [Google Scholar]
- 16. Lee C. H., Park K. J., Sung E. S., Kim A., Choi J. D., Kim J. S., Kim S. H., Kwon M. H., Kim Y. S. (2010) Engineering of a human kringle domain into agonistic and antagonistic binding proteins functioning in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 107, 9567–9571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee C. H., Park K. J., Kim S. J., Kwon O., Jeong K. J., Kim A., Kim Y. S. (2011) Generation of bivalent and bispecific kringle single domains by loop grafting as potent agonists against death receptors 4 and 5. J. Mol. Biol. 411, 201–219 [DOI] [PubMed] [Google Scholar]
- 18. Park K. J., Lee S. H., Kim T. I., Lee H. W., Lee C. H., Kim E. H., Jang J. Y., Choi K. S., Kwon M. H., Kim Y. S. (2007) A human scFv antibody against TRAIL receptor 2 induces autophagic cell death in both TRAIL-sensitive and TRAIL-resistant cancer cells. Cancer Res. 67, 7327–7334 [DOI] [PubMed] [Google Scholar]
- 19. Sung E. S., Kim A., Park J. S., Chung J., Kwon M. H., Kim Y. S. (2010) Histone deacetylase inhibitors synergistically potentiate death receptor 4-mediated apoptotic cell death of human T-cell acute lymphoblastic leukemia cells. Apoptosis 15, 1256–1269 [DOI] [PubMed] [Google Scholar]
- 20. Chen C., Liu Y., Zheng D. (2009) An agonistic monoclonal antibody against DR5 induces ROS production, sustained JNK activation and Endo G release in Jurkat leukemia cells. Cell Res. 19, 984–995 [DOI] [PubMed] [Google Scholar]
- 21. Kroemer G., Galluzzi L., Vandenabeele P., Abrams J., Alnemri E. S., Baehrecke E. H., Blagosklonny M. V., El-Deiry W. S., Golstein P., Green D. R., Hengartner M., Knight R. A., Kumar S., Lipton S. A., Malorni W., Nuñez G., Peter M. E., Tschopp J., Yuan J., Piacentini M., Zhivotovsky B., Melino G., Nomenclature Committee on Cell Death 2009 (2009) Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 16, 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cheng G., Cao Z., Xu X., van Meir E. G., Lambeth J. D. (2001) Homologs of gp91phox. Cloning and tissue expression of Nox3, Nox4, and Nox5. Gene. 269, 131–140 [DOI] [PubMed] [Google Scholar]
- 23. Adam-Klages S., Adam D., Wiegmann K., Struve S., Kolanus W., Schneider-Mergener J., Krönke M. (1996) FAN, a novel WD-repeat protein, couples the p55 TNF receptor to neutral sphingomyelinase. Cell 86, 937–947 [DOI] [PubMed] [Google Scholar]
- 24. Kieser A. (2008) Pursuing different “TRADDes”. TRADD signaling induced by TNF receptor 1 and the Epstein-Barr virus oncoprotein LMP1. Biol. Chem. 389, 1261–1271 [DOI] [PubMed] [Google Scholar]
- 25. Nam Y. J., Mani K., Ashton A. W., Peng C. F., Krishnamurthy B., Hayakawa Y., Lee P., Korsmeyer S. J., Kitsis R. N. (2004) Inhibition of both the extrinsic and intrinsic death pathways through nonhomotypic death fold interactions. Mol. Cell 15, 901–912 [DOI] [PubMed] [Google Scholar]
- 26. Chaudhary P. M., Eby M., Jasmin A., Bookwalter A., Murray J., Hood L. (1997) Death receptor 5, a new member of the TNFR family, and DR4 induce FADD-dependent apoptosis and activate the NF-κB pathway. Immunity 7, 821–830 [DOI] [PubMed] [Google Scholar]
- 27. Cao X., Pobezinskaya Y. L., Morgan M. J., Liu Z. G. (2011) The role of TRADD in TRAIL-induced apoptosis and signaling. FASEB J. 25, 1353–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kamata H., Honda S., Maeda S., Chang L., Hirata H., Karin M. (2005) Reactive oxygen species promote TNFα-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell 120, 649–661 [DOI] [PubMed] [Google Scholar]
- 29. Holler N., Zaru R., Micheau O., Thome M., Attinger A., Valitutti S., Bodmer J. L., Schneider P., Seed B., Tschopp J. (2000) Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nat. Immunol. 1, 489–495 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.