Abstract
The reported human genome sequence includes about 400 gaps of unknown sequence that were not found in the bacterial artificial chromosome (BAC) and cosmid libraries used for sequencing of the genome. These missing sequences correspond to ∼1% of euchromatic regions of the human genome. Gap filling is a laborious process because it relies on analysis of random clones of numerous genomic BAC or cosmid libraries. In this work we demonstrate that closing the gaps can be accelerated by a selective recombinational capture of missing chromosomal segments in yeast. The use of both methodologies allowed us to close the four remaining gaps on the human chromosome 19. Analysis of the gap sequences revealed that they contain several abnormalities that could result in instability of the sequences in microbe hosts, including large blocks of micro- and minisatellites and a high density of Alu repeats. Sequencing of the gap regions, in both BAC and YAC forms, allowed us to generate a complete sequence of four genes, including the neuronal cell signaling gene SCK1/SLI. The SCK1/SLI gene contains a record number of minisatellites, most of which are polymorphic and transmitted through meiosis following a Mendelian inheritance. In conclusion, the use of the alternative recombinational cloning system in yeast may greatly accelerate work on closing the remaining gaps in the human genome (as well as in other complex genomes) to achieve the goal of annotation of all human genes.
The International Human Genome Sequencing Consortium recently reported that ∼99% of the gene-rich euchromatic portion of the genome has been sequenced and assembled. Each base pair of this 99% was sequenced five times on average, ensuring an error rate of less than one base in 50,000. The finished sequence nowhas an N50 contig size of 27 Mb, and the number of gaps has been reduced from 80,000 in the draft to ∼400. These 400 gaps represent genome sequences not found in the screened genomic bacterial artificial chromosome (BAC), P1-derived artificial chromosome (PAC), or other fosmid and cosmid libraries. Most of these gaps represent the type 3 gaps that are the most difficult to evaluate, because the genome sequence flanking these gaps is often not precisely aligned with the fingerprinted clones. The gaps represent ∼30 Mb or 1.0% of the human genome (Grimwood and Schmutz 2003). Although almost the whole genome is considered “finished” and offers a wealth of information, cloning and sequencing the tough leftovers of the human genome is essential. Without these sequences, we will not know what we are missing. Each missed sequence can potentially contain a gene, and each missed gene is potentially a missed drug target. Even gene-poor areas might be critical for gene regulation.
A traditional method of filling gaps includes screening additional BAC and cosmid libraries. However, this approach is time-consuming and may be not applicable to some gap regions with unusual DNA structures. For example, it is well documented that long inverted repeats, AT-rich sequences, and sequences with structures such as Z-DNA are extremely unstable in Escherichia coli (Hagan and Warren 1982; Schroth and Ho 1995; Kang and Cox 1996; Razin et al. 2001). These sequences may be underrepresented or even lost when cloned in E. coli.
The introduction of alternative cloning systems and hosts, allowing isolation of genomic segments that are poorly clonable in E. coli cells, may assist the effort to close the gaps. Such a system is yeast artificial chromosome (YAC) cloning in yeast. Several recent reports demonstrate that genomic segments that are unstable in E. coli vectors can be accurately recovered as YACs in yeast (Bigger et al. 2000; Gardner et al. 2002; Kouprina et al. 2003). In some cases (Dictyostelium discoideum), YACs may represent the only viable method for the construction of large insert libraries (Glockner et al. 2002). An additional advantage of using yeast is the opportunity to directly isolate a desired genomic region by transformation-associated recombination (TAR) cloning (Kouprina and Larionov 2003). Two main TAR cloning schemes were developed and applied for isolation of dozens of full-size mammalian genes. When DNA sequence information is available from the 3′- and 5′-flanking regions of the gene of interest, the region is isolated using a vector with two unique targeting sequences (hooks; Larionov et al. 1997). Another version of TAR cloning, called radial TAR cloning, uses a vector with one hook that is a unique sequence from the chromosomal region of interest and the other hook that is a repeated sequence occurring frequently and randomly in the genomic DNA (i.e., Alu repeats; Kouprina et al. 1998a).
For the purpose of gap closure, the radial TAR cloning is the most suitable, because sequences of the flanking clones may be deleted or rearranged, making the development of two specific targeting hooks difficult. In the present study, the TAR cloning approach and screening of additional genomic libraries were used to close gaps on human chromosome 19. A subsequent analysis of the gap sequences allowed us to annotate four human genes and shed light on the nature of poorly clonable chromosome segments.
RESULTS
Closing the Gaps on Human Chromosome 19
The first phase of the chromosome 19 mapping and sequencing was based on chromosome 19-specific cosmid libraries constructed from flow-sorted chromosomes isolated from human-hamster hybrid cell lines containing chromosome 19 as the only human chromosome (Carrano et al. 1989). Cosmid contigs were extended and merged by BAC libraries, that is, CTC, CTD, CTB, and CIT from Caltech and RPC-11 (Osoegawa et al. 2001). This approach has been extremely successful. However, as the Human Genome Project drewto a close, there were four regions of chromosome 19 that were not spanned by sequenced BAC or cosmid clones. These regions, on 19p13.1, 19p13.2, 19p13.3, and 19q13.4, were referred to as clone gaps of type 3 (GAP1, GAP2, GAP3, and GAP6) that required the collection of additional information for their closure.
Radial TAR cloning was successfully used to isolate genomic fragments containing the GAP1, GAP2, GAP3, and GAP6 sequences in yeast. Figure 1 illustrates the scheme of the GAP6 closure between two flanking clones. All four gap regions were selectively cloned as circular YACs using vectors carrying a GAP-specific targeting hook and an Alu repeat as a second targeting sequence (see Methods). Transformation experiments were carried out with freshly prepared yeast spheroplasts and a linearized TAR GAP-specific vector as described (see Methods). For each GAP, from one to five clones positive for either one or both flanking clones were identified (Table 1). The size of the positive YACs was determined (see Methods). The results are summarized in Table 1. Two approaches were taken to verify the integrity of the YACs and their stability during propagation in yeast. In the first approach, YAC DNA was isolated from four subclones of each original GAP1, GAP2, GAP3, and GAP6 clones in plugs, digested by NotI, separated by clamped homogeneous electrical field (CHEF), and then hybridized with an Alu probe. Subclones of each gap carried YACs of the same size, indicating that these clones do not have detectable deletions in yeast. For the second approach, the Alu profiles of four subclones of each clone were determined and shown to be identical, indicating no detectable rearrangements during propagation in yeast cells (Fig. 2A,B). Thus, the YAC clones are relatively stable during propagation in yeast. To evaluate the size of each GAP, the Alu ends of the YAC clones positive for both flanking clones were rescued in E. coli and sequenced (see Methods). The sequences were compared to the draft sequence of chromosome 19 at NCBI and UCSC (build 29, April, 2002) using BLAST. Positions of YAC ends for GAP1, GAP2, GAP3, and GAP6 TAR clones are shown in Table 1. With knowledge of the size of the YACs, the positions of the YAC end sequences, and the hooks in the clones towards the gaps, we estimated their sizes: GAP1, ∼15 kb; GAP2, ∼20 kb; GAP3, ∼15 kb; GAP6 ∼25 kb.
Figure 1.
Scheme of the GAP6 closing between two flanking clones on chromosome 19 by TAR cloning. Radial TAR cloning with a TAR vector containing a unique targeting sequence and an Alu repeat was used to close the gap between contigs AC020922 and AC008735.7. Yeast spheroplasts were transformed with genomic DNA containing human chromosome 19 along with a linearized TAR cloning vector (see Methods). Recombination between the sequences in the vector and genomic fragment containing the GAP6 sequence led to the establishment of circular YACs that extend from the unique sequence to various Alu positions. In the present scheme, only YACs that are positive by diagnostic primers for both contigs (DG1 and DG2) are shown. Orientation of contigs towards centromere and telomere are indicated.
Table 1.
TAR Isolates Containing Gap Sequences
| GAP | Size of YACs | Positions of rescued YAC end in flanking clone | Accession number | |
|---|---|---|---|---|
| GAP1 | ||||
| clone 1 | 90 kb | NC | AY345879 | |
| GAP2 | ||||
| clone 1a | 180 kb | nd | ||
| clone 2 | 100 kb | 11500–11970 | (AC092300.2)b | |
| clone 3 | 50 kb | NC | ||
| clone 4 | 80 kb | NC | ||
| GAP3 | ||||
| clone 1 | 40 kb | 24749–25023 | (AC022149) | |
| clone 2 | 120 kb | 53173–52505 | (AC090427) | AC140008 |
| clone 3 | 180 k | NC | ||
| GAP6 | ||||
| clone 1 | 70 kb | nd | ||
| clone 2 | 70 kb | 14233–14865 | (AC016625.6) | |
| clone 3 | 90 kb | 98504–98946 | (AC020922) | AY207046 |
| clone 4 | 90 kb | 98504–99035 | (AC020922) | |
| clone 5 | 100 kb | 91140–91657 | (AC020922) | |
NC-end positions are uncertain because the end contains a repeat sequence.
In bold are the TAR clones that were sequenced.
TAR clone obtained from total genomic DNA
Flanking clone
Figure 2.
Integrity of TAR YAC isolates in yeast. (A) YAC DNA was isolated from the original GAP6-#1 clone (lane 1), five of its subclones (lanes 2-6) and five transformants with YAC/BACs (lane 7-11), digested by NotI, separated by CHEF, and then hybridized with an Alu probe. Clones have the same size, indicating that these clones do not have detectable deletions in yeast. (B) Alu profiles of the original TAR YAC clones containing the GAP6 sequence and their subclones. Total yeast DNA was isolated from four independent gap-positive transformants and from four subclones of each independent TAR isolate and digested to completion with TaqI. Fragments were separated by gel electrophoresis, transferred to a nylon membrane, and hybridized with an Alu probe. (Alu profile of isolates differs slightly due to differences in the size of TAR isolates.) (a) Four independent TAR isolates of GAP6-#1, -#2, -#3 and -#4. (b-e) correspond to four subclones of each independent isolate, respectively.
For further analysis, circular YACs were retrofitted into BACs by homologous recombination in yeast, and transformed into E. coli. Retrofitted YAC/BACs usually transform E. coli with high efficiency: a 1-μL sample of a melted agarose plug usually produces 100-500 transformants. In contrast, the YAC/BACs with GAP inserts transformed E. coli with an efficiency ∼10 times lower. Most of the BACs rescued in E. coli underwent deletions, suggesting that the inserts are intrinsically unstable in bacterial cells. This observation was consistent with the absence of these sequences in genomic libraries observed when this work was begun. After screening the E. coli transformants, we succeeded in finding BAC clones with no detectable rearrangements for two gaps, GAP1 and GAP6, when transformation and subsequent growth of E. coli cells was performed at 30°C (Fig. 3). Lack of rearrangements in the BAC inserts was confirmed by Alu profile comparison of original YAC isolates, retrofitted YAC/BACs, and BACs with the size of insert not changed (data not shown). The GAP1 and GAP6 BACs were chosen for further sequencing analysis. For GAP2 and GAP3, circular YAC DNAs were isolated from the yeast cells and used for sequencing. In addition, GAP2- and GAP3-deleted BACs were also sequenced to determine whether the deletion(s) occurred at the same region, which might suggest the reason for the instability of these regions in bacterial cells.
Figure 3.
Propagation of BACs with GAP6 sequence in E. coli at 37°C and 30°C. BAC DNA was isolated from three E. coli subclones of the clones #1, #2, #3, and #4 carrying GAP6, digested with NotI, and separated by gel electrophoresis. Deleted/rearranged isolates of BACs grown at 37°C were partially stabilized by growing at 30°C. M1, 1-kb ladder marker; M2, 48-kb ladder marker.
In addition to the TAR cloning strategy, clones for three gaps (GAP1, GAP2, and GAP6) were identified by screening two newlibraries (a BAC library, RP13 and an LLNL fosmid library, XXfos), as well as by additional screening of RP11. No bacterial clones linking the contigs that flank GAP3 were identified.
Analysis of GAP1, GAP2, GAP3, and GAP6 Sequences
To fill the gaps, 11 clones were sequenced. Because TAR isolates and clones from additional libraries overlapped, only six sequences corresponding to the gaps were deposited into GenBank.
For GAP6, the sequence information obtained from the bacterial clone found in the BAC library (AC135592) was confirmed by the sequence information obtained from the TAR isolate (AY207046). Analysis of the sequences revealed the presence of two blocks of telomeric repeats and minisatellites that are known to be unstable in E coli. Such a sequence is presumably a cause of inefficient recovery of the gap in E. coli.
For other gaps, discrepancies were observed between inserts propagated in E. coli and yeast cells. For GAP1, a fosmid clone and the TAR isolate in a BAC form were sequenced. Comparison of the two sequences revealed a great difference in one of the minisatellite regions. This minisatellite is located in intron 8 of the gene SCK1/SLI, spanning GAP1. The size of the minisatellite in the fosmid clone is ∼500 bp (positions 19218-19716 in AC138433), versus 1.2 kb in the TAR BAC clone. PCR analysis of the minisatellite in the original TAR YAC isolate, using the primers flanking this region (TR6, Suppl. Table A1), revealed a fragment 4.2 kb in size, indicating that this region is unstable in E. coli. Analysis of 103 unrelated individuals showed that the minisatellite is nonpolymorphic (see below). This block of minisatellites was completely sequenced using a PCR product generated from a YAC TAR isolate, and a complete sequence of GAP1 was deposited into GenBank (AY345879). Other minor differences between the fosmid clone and the TAR isolate are due to the presence of variable repeat sequences (VNTRs) in the region. Detailed analysis of the VNTR polymorphism within the SCK1/SLI gene is described below.
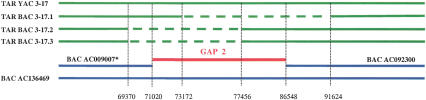
For GAP2, two clones were also sequenced. One of the clones was identified in a BAC library. Another clone was isolated by TAR and retrofitted into BAC. Because transfer of the retrofitted YAC/BAC into E. coli resulted in deletions, three subclones of the BAC were shotgun-sequenced. Comparison of sequences of these subclones revealed that each clone has overlapping deletions in the same region (Fig. 4) and is highly enriched by Alu repeats (33 Alu copies per 11-kb sequence). The gap sequence was reconstructed using the sequences of the deleted BACs. The sequence obtained matched the sequence present in the BAC clone AC136469, identified by screening the BAC library.
Figure 4.
Alignment of the RP11-886P16 BAC clone and three deleted BAC subclones obtained during transferring of the GAP2 YAC/BAC TAR isolate from yeast into E. coli cells. BAC RP11-886P16 (AC136469) was identified by screening the genomic library; sequences of the deleted YAC/BACs, 3-17.1, 3-17.2, and 3.17.3, are in the Supplemental Material. Comparison of sequences revealed that each clone has overlapping deletions in the same region that is highly enriched by Alu repeats. The gap sequence was reconstructed using the sequences of the deleted BACs and additional PCR amplification of the sequence from YAC TAR isolate. The sequence obtained matched the sequence present in the BAC clone AC136469.
Because no clones with GAP3 sequence were identified by screening the additional BAC libraries, the only clones used for closing the gap were the yeast YAC clones obtained by TAR cloning (AC140008). Similar to clones with GAP1 and GAP2 sequences, GAP3 TAR isolates revealed instability during transfer to E. coli cells. For this reason, three BAC subclones of the GAP3 isolate were shotgun-sequenced. The sequencing showed that the GAP3 BAC clones were rearranged in multiple configurations during growth, prohibiting the development of a sequence contig. Most rearrangements are presumably due to a large block of TGG repeats that is known to be unstable in E. coli cells (Pan and Leach 2000). Failure to complete the gap sequence using BAC sequence information encouraged us to sequence it in a YAC form. Our analysis of gap sequences is summarized in Table 2.
Table 2.
Analysis of Gap Chromosome 19 Sequences
| GAP | Size | Position in chromosome 19 sequencea | Reason of instability in E. coli cells |
|---|---|---|---|
| GAP1 | 12.2 kb | 364968–374586 | High density of minisatellites |
| GAP2 | 16 kb | 8370894–8386410 | Dense in Alu repeats (58%) |
| GAP3 | 13.7 kb | 14549278–14563445 | TGG repeat block |
| GAP6 | 23.5 kb | 60613898–60639940 | High density of minisatellites and telomeric repeats |
Build 33, April 2003
All of the Gap Sequences Are a Part of Gene-Encoding Regions
It is noteworthy that all gap sequences analyzed span gene-encoding regions. Analysis of the GAP2 region revealed the presence of EST (BG705726) encoding the hypothetical protein (HSPC240) expressed in CD34+ hematopoietic stem/progenitor cells. The EMR3 gene, encoding human EFG-like module-containing mucin-like receptor (Stacey et al. 2001), has sequence within GAP3. The first seven exons and part of exon 8 of this gene match sequence of the flanking clone AC0022149. Another part of exon 8 lies within the gap sequence. Analysis of the GAP6 sequence revealed the presence of EST (AK094959) corresponding to a hypothetical isochorismatase hydrolase family-containing protein. Analysis of the GAP1 sequence revealed the presence of exon 9 and exon 10 of the SCK1/SLI gene (Kojima et al. 2001). Eleven other exons are located within two flanking clones, AC006124 and AC008988. The gene, the genomic copy of which is 46 kb in size, was originally identified by low-stringency hybridization of brain cDNA libraries. We analyzed this gene more carefully because of the significance of this gene in neuronal cell signaling (Kojima et al. 2001) and the presence of an unusual polymorphism that may affect gene expression.
Sequence analysis of the entire SCK1/SLI gene allowed the determination of 12 blocks of tandem repeats (Fig. 5). The degree of polymorphism of these minisatellites was examined using diagnostic PCR primers (Suppl. Table A1) in human DNA samples isolated from 103 unrelated individuals as well as in the TAR YAC clone and DNA isolated from the hybrid cell line containing a single human chromosome 19. The results revealed 10 blocks of variable minisatellites (VNTRs) and two blocks that contained nonpolymorphic minisatellites (TR6 and TR7; Table 3; Suppl. Fig. A1). For the VNTR1 minisatellite in intron 10 of SCK1/SLI, nine alleles ranging in size from 130 bp to 615 bp in length, corresponding to two to 11 copies of the repeat, and a degree of heterozygosity of 0.746 were recovered. The most common allele had 10 repeats. VNTR2, VNTR3, and VNTR4 are located within intron 9 of SCK1/SLI. For the VNTR2 minisatellite, seven alleles ranging in size from 400 bp to 680 bp in length, corresponding to 10-18 copies of the repeat and a degree of heterozygosity of 0.717, were recovered. The most common allele had 16 repeats. Five alleles of VNTR3 range from 13 to 19 repeats. The most common allele had 19 repeats and a degree of heterozygosity of 0.671. Four alleles of VNTR4 ranging from 50 to 63 repeats, with 51 repeats for the most common allele and a corresponding degree of heterozygosity of 0.186, were found. SCK1/SLI includes six additional variable minisatellites, that is, VNTR5 located in intron 8 and VNTR8, VNTR9, VNTR10, VNTR11, and VNTR12 located in intron 1. VNTR5 has 27 alleles with 51 repeats for the most common 3.0 kb allele and a degree of heterozygosity of 0.94. VNTR8 has two alleles with 17 repeats most common. VNTR9 has three alleles with 8 repeats most common, VNTR10 has four alleles with 12 repeats most common. VNTR11 has four alleles with 45 repeats most common, and VNTR12 has three alleles with 17 repeats most common (Table 3; Suppl. Fig. A1). The degree of heterozygosity was 0.1, 0.279, 0.351, 0.076, and 0.039, respectively (Suppl. Table A2). The repeats within one VNTR diverge by ∼20%. No polymorphism was found for the minisatellite TR6 located in intron 8 or TR7 located in intron 4. For TR6, the number of repeats was 293, giving a PCR product of 4.2 kb, and for TR7 the number of repeats was 24, giving PCR products of 410 bp for all 103 individuals (data not shown).
Figure 5.
Minisatellites in SCK1/SLI. (A) A schematic diagram of the sequence spanning the SCK1/SLI gene. Exons are represented by boxes either above (top strand) or below (bottom strand) the line. Thirteen putative exons, coding for the Sck protein, were identified by BLAST analysis. The approximate positions of minisatellites, detected by the Tandem Repeats Finder Program (Benson 1999), are indicated by *. At the top, a blown-up portion of the insert shows the relative positions within introns of the 12 minisatellites. (B) The sequences of 12 minisatellite repeat units. Minisatellites 1, 2, 3, 4, 5, 7, 8, 9, 10, 11, and 12 are polymorphic, whereas minisatellites 6 and 7 are monomorphic in the population sample studies.
Table 3.
Polymorphic Alleles in SCK1/SL1 VNTRs
| # Repeats | Size (bp) | N = 206 | Frequency | # Repeats | Size (kb) | N = 206 | Frequency | ||
|---|---|---|---|---|---|---|---|---|---|
| VNTR1 | 1.7 | 130 bp | 2 | 0.010 | VNTR5 | 29.0 | 1.65 kb | 6 | 0.029 |
| 2.7 | 190 bp | 34 | 0.165* | 30.0 | 1.70 kb | 9 | 0.044 | ||
| 3.7 | 250 bp | 59 | 0.286 | 31.0 | 1.75 kb | 5 | 0.024 | ||
| 4.7 | 310 bp | 35 | 0.170 | 33.0 | 1.95 kb | 4 | 0.019 | ||
| 5.7 | 370 bp | 2 | 0.010 | 35.5 | 2.10 kb | 2 | 0.010 | ||
| 6.7 | 420 bp | 1 | 0.005 | 37.3 | 2.20 kb* | 16 | 0.078 | ||
| 8.7 | 540 bp | 2 | 0.010 | 38.3 | 2.25 kb | 4 | 0.019 | ||
| 9.7 | 615 bp | 70 | 0.340 | 39.3 | 2.30 kb | 4 | 0.019 | ||
| 11.0 | 720 bp | 1 | 0.005 | 41.0 | 2.40 kb | 4 | 0.019 | ||
| 42.0 | 2.45 kb | 3 | 0.015 | ||||||
| 43.0 | 2.50 kb | 4 | 0.019 | ||||||
| VNTR2 | 9.7 | 400 bp | 4 | 0.019 | 44.9 | 2.55 kb | 5 | 0.024 | |
| 12.7 | 500 bp | 43 | 0.209 | 46.7 | 2.65 kb | 21 | 0.102 | ||
| 13.7 | 540 bp | 12 | 0.058 | 47.7 | 2.80 kb | 7 | 0.034 | ||
| 14.7 | 570 bp | 60 | 0.291 | 49.5 | 2.90 kb | 5 | 0.024 | ||
| 15.7 | 600 bp | 80 | 0.388* | 51.3 | 3.00 kb | 17 | 0.083 | ||
| 16.7 | 640 bp | 4 | 0.019 | 53.0 | 3.10 kb | 10 | 0.049 | ||
| 17.7 | 680 bp | 3 | 0.015 | 55.0 | 3.20 kb | 4 | 0.019 | ||
| 57.0 | 3.30 kb | 6 | 0.029 | ||||||
| 58.0 | 3.35 kb | 4 | 0.019 | ||||||
| VNTR3 | 12.9 | 370 bp | 3 | 0.015 | 65.0 | 3.75 kb | 2 | 0.010 | |
| 13.9 | 400 bp | 59 | 0.286 | 69.0 | 4.00 kb | 5 | 0.024 | ||
| 14.9 | 430 bp | 54 | 0.262 | 75.0 | 4.30 kb | 25 | 0.121 | ||
| 15.9 | 460 bp | 3 | 0.015 | 77.0 | 4.40 kb | 17 | 0.083 | ||
| 18.9 | 560 bp | 87 | 0.422* | 83.0 | 4.70 kb | 11 | 0.053 | ||
| 85.0 | 4.80 kb | 4 | 0.019 | ||||||
| 87.0 | 4.90 kb | 2 | 0.010 | ||||||
| VNTR4 | 50.3 | 1.79 kb | 3 | 0.015 | |||||
| 51.3 | 1.85 bp | 185 | 0.898* | ||||||
| 60.3 | 2.30 kb | 17 | 0.083 | VNTR8 | 15.7 | 680 bp | 11 | 0.053* | |
| 63.3 | 2.45 kb | 1 | 0.005 | 16.7 | 720 bp | 195 | 0.947 | ||
| VNTR9 | 4.7 | 420 bp | 23 | 0.112 | VNTR11 | 41.7 | 1.03 kb | 2 | 0.010 |
| 6.7 | 550 bp | 10 | 0.049* | 42.7 | 1.06 kb | 2 | 0.010 | ||
| 7.7 | 620 bp | 173 | 0.840 | 43.7 | 1.08 kb | 4 | 0.019 | ||
| 44.7 | 1.10 kb | 198 | 0.961* | ||||||
| VNTR10 | 11.0 | 750 bp | 6 | 0.029 | |||||
| 11.7 | 800 bp | 162 | 0.786* | VNTR12 | 16.8 | 670 bp | 202 | 0.980* | |
| 12.7 | 860 bp | 2 | 0.010 | 18.8 | 730 bp | 3 | 0.015 | ||
| 16.0 | 1070 bp | 36 | 0.175 | 19.8 | 760 bp | 1 | 0.005 |
In bold are minisatellite alleles identified in a fosmid clone.
Asterisks show minisatellite alleles in a TAR isolate.
Eleven families were selected for segregation analysis of VNTRs in the SCK1/SLI gene. Blood was collected from grandparents, parents, and one to three children from each family. Hereditary segregation of 10 VNTRs was traced for two generations in nine families and three generations in two families. In most cases, alleles of VNTR1, VNTR2, VNTR3, VNTR4, VNTR5, VNTR8, VNTR9, VNTR10, VNTR11, and VNTR12 could be identified and their transmission traced from parent to child. The results showed that these VNTRs are subject to Mendelian inheritance (i.e., children carried one VNTR allele from each parent). New VNTRs were not observed during this analysis (Suppl. Table A2). Thus, these 10 VNTRs in the SCK1/SLI gene are meiotically stable and could potentially be used as DNA typing markers to follow meiotic segregation of SCK1/SLI alleles.
The individual differences in minisatellite lengths of the SCK1/SLI gene may result in differences in expression pattern. Sequence analysis of the SCK1/SLI gene revealed that the minisatellites contain specific cis-regulatory elements/domains that may interact with transcription factor proteins such as HRE1, ZF87, XPB1, GATA3, KE1, REX, NF-KB, uE4, and ETS, which are involved in region-specific expression. It is also possible that changes in DNA conformation due to the repetitive nature of the minisatellites might influence gene transcription. It should be also noted that, although the most striking individual differences are in the length of minisatellites, there may be also differences in their sequence, such as single-base mutations, which could also contribute to the variability in expression.
DISCUSSION
Chromosome 19 is among the smallest and gene-dense human chromosomes, spanning 64 Mb and estimated to contain ∼1760 genes. Sequencing and assessment of the chromosome sequence were performed by the Joint Genome Institute (JGI) and the Stanford Human Genome Center and relied almost exclusively on cosmid and BAC libraries. In general, this approach has been extremely successful. However, as the Human Genome Project drewto a close, there were four regions of the chromosome 19 that were not spanned by sequenced BAC clones. Because these regions were not identified in five different BAC and cosmid libraries, they were referred to as type 3 gaps.
In this work, we demonstrate that closing the gaps can be achieved by a combination of two strategies, that is, screening of newBAC and fosmid libraries and selective TAR cloning in yeast. The opportunity to compare the clones isolated in different hosts allowed us to determine the structure of the missing genomic segments. Sequence analysis of the chromosome 19 gap isolates revealed at least three types of sequences that could destabilize the corresponding inserts during cloning in microbe hosts. Two gap regions contained large blocks of micro- and/or minisatellite repeats. Another gap region was highly enriched by Alu repeats. In the fourth clone, a large block of TGG trinucleotide repeat was detected. We showed previously that regions containing AT-rich blocks are also unstable in BAC vectors (Kouprina et al. 2003). Thus in each case the cause of instability of the genomic segment in host cells may be different. This means that gap closure can be most efficiently achieved by using both E. coli and yeast cloning systems. For practical reasons, the use of direct TAR cloning in yeast versus a library of random clones has some advantages. In addition to a high selectivity, typically TAR cloning produces multiple independent isolates, each of which is flanked by the targeting sequences included in the TAR vector. Therefore, even if a clone is deleted or rearranged, the comparison of independent isolates generates a discontinuous DNA sequence. Sometimes sequencing of the gap region may require nonconventional approaches. Here, a TAR clone, containing a DNA insert that was unstable in E. coli, was sequenced in a YAC form. Another clone containing numerous similar repeated sequences was retrofitted to a BAC form and sequenced using a BAC direct-sequencing strategy (Polushin et al. 2001).
The fact that some human DNA sequences, including unique genes, are unstable and even unclonable in E. coli (Kouprina et al. 2003 and results therein) raises an important question. To what extent might the true sequence of chromosome 19 and other chromosomes be altered or lost in the sequence generated from BAC, PAC, cosmid, or fosmid clones? To address this question, TAR cloning may be one of the best available approaches. Human DNA can be selectively cloned from human-rodent monochromosomal hybrid cell lines by a TAR vector with Alu repeats as targeting sequences (Kouprina et al. 1998b). This strategy has been used to construct a chromosome 19-specific YAC/BAC library (N. Kouprina, unpubl.). Comparison of the insert ends for ∼400 clones with the sequence of chromosome 19 confirms a high-quality assembly. In the majority of the clones (>98%), the predicted and determined distances between insert ends coincided, and orientation of the ends was correct. The same strategy can be applied for verification of other chromosome sequences.
All four chromosome 19 gaps are mapped to regions corresponding to genes. The GAP1 region corresponds to the neuronally expressed Shc adaptor homolog SCK1/SLI (Kojima et al. 2001). GAP3 sequence overlaps the gene EMR3, encoding an EFG-like module-containing mucin-like receptor (Stacey et al. 2001). Two other gaps, GAP2 and GAP6, contain EST sequences for two hypothetical proteins. Poorly clonable DNAs are present in the intronic regions of these genes. The SCK1/SLI gene contains a cluster of sequences (blocks of minisatellites) that are at high risk for deletions/rearrangements in BACs. Twelve different minisatellite sequences are located in 12 introns of the gene; 10 of them are highly polymorphic. Such a high density of minisatellite repeats is presumably due to the location of the gene near a telomere, as was previously shown for other telomere-linked genes (Leem et al. 2002; Kim et al. 2003). The role of minisatellites in intronic regions is obscure. Recent data indicate that, when present in first introns, minisatellites can affect gene regulation (Lovejoy et al. 2003). It is worth noting that minisatellites in SCK1/SLI contain multiple binding sites for transcription factors, and therefore may be involved in gene regulation.
The fact that gaps in the human genome may correspond to chromosomal regions encoding functional genes emphasizes the importance of the final step of genome sequencing. There are still ∼400 gaps of unknown sequences in the human genome. Because clones for these gaps were not found in the BAC, PAC, fosmid, or cosmid libraries used for the genome sequencing, most of the gap sequences presumably represent poorly clonable DNA segments that cannot be easily covered by newgenomic libraries. Moreover, it is also unlikely that these gaps may be recovered from the sequencing of chimpanzee BAC clone libraries, which is now in progress. Indeed, sequencing of chimpanzee chromosome 21, which is syntenic to human chromosome 22, showed that both chromosomes contain gaps in the same regions (Takamatsu et al. 2002; Y. Sakaki, pers. comm.). Thus, although some of the annotated gaps in the human genome may be closed by long-range PCR and/or by screening newlibraries, a large number of genomic regions should also be isolated in alternative cloning hosts such as yeast, to be sure that sequence information is not lost during cloning in E. coli. In this study, we demonstrate that TAR cloning in yeast can facilitate the rapid collection of additional clones for gap closure. The same strategy can be also applied for closing the gaps in other complex genomes.
METHODS
Construction of the TAR Vectors and Cloning by In Vivo Recombination in Yeast
The TAR circularizing vectors, pVC-GAP1, pVC-GAP2, pVC-GAP3, and pVC-GAP6, containing one unique targeting sequence and an Alu repeat as the second targeting sequence were constructed as follows. Either a 187-bp Alu XbaI-BamHI (for pVC-GAP3) or ApaI-XhoI (for pVC-GAP1, pVC-GAP2, and pVC-GAP6) fragment was inserted into a polylinker site of pVC604 (CEN6-HIS3; Kouprina and Larionov 1999). For pVC-GAP1, a 390-bp SalI-EcoRI fragment corresponding to a unique sequence from the clone AC006124 was inserted into the polylinker of pVC604. The targeting sequence corresponds to positions 2391-2780 in the clone (Suppl. Table A1). For pVC-GAP2, a 163-bp SalI-EcoRI fragment corresponding to a unique sequence from the clone AC009007.4 was inserted into the polylinker of pVC604. The targeting sequence corresponds to positions 26731-26894 in the clone (Suppl. Table A1). The TAR circularizing vector pVC-GAP3 was constructed as follows. A 138-bp BamHI-EcoRI fragment corresponding to a unique sequence of the clone AC010527 was inserted into the polylinker of pVC604. The targeting sequence corresponds to positions 9931-10068 in the clone (Suppl. Table A1). The TAR circularizing vector pVC-GAP6 was constructed as follows. A 160-bp SalI-EcoRI fragment corresponding to a unique sequence from the clone AC008735.7 was inserted into the polylinker of pVC604. The targeting sequence corresponds to positions 9736-9577 in the clone (Suppl. Table A1). The vectors pVC-GAP1, pVC-GAP2, and pVC-GAP6 were cut with SalI, and the vector pVC-GAP3 was cut with BamHI (the sites were located between the Alu and unique sequences) before yeast transformation. Transformation experiments were carried out with freshly prepared yeast spheroplasts and a linearized TAR GAP-specific vector as described (Kouprina and Larionov 1999). In each experiment ∼2 μg of genomic DNA isolated from a human/hamster monochromosomal somatic cell hybrid UV5HL9-5B, 1 μg of vector, and 8 × 108 spheroplasts were used (Leem et al. 2003). Between 500 and 1000 His+ transformants were then screened by diagnostic primers (Suppl. Table A1) to identify transformants positive for one or both flanking clones. The diagnostic primers were developed based on draft sequence of chromosome 19 (build 29, April 2002) and rescued YAC end sequences. Transformants were first combined into pools and examined by diagnostic primers. Then, individual colonies containing the GAP1, GAP2, GAP3, or GAP6 sequences were isolated from each positive pool by a second round of PCR screening. The PCR products were sequenced to verify that they match the predicted sequences of chromosome 19 and map to one of the gap flanking clones. One clone containing the sequence of GAP2 was isolated from total genomic DNA and turned out to be a chimera.
Yeast and Mammalian Cell Culture
The highly transformable S. cerevisiae strain VL6-48 was used for TAR cloning (Kouprina and Larionov 1999). Agarose plugs containing high-molecular-weight genomic DNA and DNA in solution were prepared from normal human/hamster monochromosomal somatic cell hybrid UV5HL9-5B, containing human chromosome 19 (LLNL), and used for TAR cloning experiments.
Isolation and Physical Analysis of YAC and BAC Clones
Isolation of the circular YAC DNA from yeast for sequencing was carried out as described (Devenish and Newlon 1982). To estimate the size of circular YACs, chromosomal size DNA from yeast transformants was prepared in plugs, then digested with NotI before analysis by CHEF gel electrophoresis and visualized by blot-hybridization with an Alu-probe. YAC ends were rescued as follows. Yeast genomic DNA from YAC-containing clones was digested with a restriction endonuclease, ligated, and transformed into E. coli. For GAP2 and GAP6, either EcoRI or HindIII was used, and for GAP3 EcoRI was used. Transformants were selected on media containing ampicillin (50 μg/mL). Plasmids with rescued YAC ends were isolated using a standard protocol, and the insert was sequenced using T3 and T7 primers. To identify fragments containing Alu sequences (Alu profiles), yeast DNA was digested to completion with TaqI. Samples were separated by gel electrophoresis, transferred to a nylon membrane, and hybridized with an Alu probe. Retrofitting of YACs into BACs, electroporation of YAC/BACs into E. coli cells, and BAC DNA isolation were carried out as described (Kouprina and Larionov 1999), except that electroporation and the bacterial culture growth was carried out at 30°C for 10 h to stabilize the human insert. The size of BAC DNA was determined after digestion with NotI and further analysis by CHEF gel electrophoresis.
Genomic Libraries Used to Recover Gap Sequences
Bacterial clones spanning the gap regions were identified by screening BAC libraries RP13 and RP11 (Osoegawa et al. 2001) and a fosmid XXfos library constructed at the Lawrence Liver-more National Laboratory (LLNL).
Sequencing
The ends of the insert for each YAC/BAC clone were sequenced using standard vector-specific primers M13F/M13R. The sequences were compared to the draft human genome sequence of chromosome 19 at NCBI (http://www.ncbi.nlm.nih.gov/genome/guide/) and UCSC (http://genome.ucsc.edu/goldenPath/apr2001Traks.html; Build 30, April 2003) using BLAST genome analysis software (http://www.ncbi.nlm.nih.gov/BLAST/blast_databases.html). For shotgun sequencing, the DNA from BACs or DNA from circular YACs containing GAP2, GAP3, and GAP6 were purified at 30°C, sonicated to 2-kb or 10-kb fragments, and cloned into an M13 vector. The GAP1 region was directly sequenced from BAC DNA (Polushin et al. 2001) by Fidelity Systems Inc. Unstable minisatellite segments from GAP1 were PCR-amplified from a YAC TAR isolate or genomic DNA, cloned into a TA vector and sequenced by a standard method. Sequencing of the clones containing GAP2, GAP3, and GAP6 were carried out at the JGI. Loci annotation and submission to GenBank were done using Sequin. Accession numbers are AC138433, AC136469, AY345879, AC140008, AC135592, and AY207046. The Tandem Repeats Finder software (Benson 1999) was used to detect VNTRs and other repeated regions.
Analysis of Polymorphism in the SCK1/SLI Gene Minisatellites
The primers used to analyze the gene polymorphism are based on the SCK1/SLI genomic sequence (Table 1). Yeast genomic DNA isolated from the transformants of GAP1, containing the SCK1/SLI gene, was amplified using these primers under standard PCR conditions: 50 mM KCl, 10 mM Tris-HCl, pH 9.0, 3.0 mM MgCl2, 0.2 mM dTTP, dCTP, dGTP, and dATP in a final volume of 50 μL. Thermocycling conditions were as follows: one cycle of 2 min of initial denaturation at 94°C, 30 cycles of 30 sec at 94°C, and 1 min at 68°C (1 min per 1 kb of DNA), followed by a 10-min extension at 72°C in a 9700 Thermocycler (Perkin-Elmer). To assess the degree of polymorphism of SCK1/SLI minisatellites, DNA was analyzed from 103 healthy unrelated individuals. DNA was isolated from peripheral blood lymphocytes using standard methods. PCR analysis of human DNA samples was performed using Takara Ex Taq polymerase (Takara) with 100 ng genomic DNA. PCR products were analyzed by gel electrophoresis (1 volt/cm) in 1×TAE buffer through a 1.2% agarose gel.
Acknowledgments
This research was partially supported by the Biological and Environmental Research Program (BER), U.S. Department of Energy, Interagency Agreement No. DE-AI02-01ER63079.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1929904. Article published online before print in January 2004.
Footnotes
References
- Benson, G. 1999. Tandem repeats finder: A program to analyze DNA sequences. Nucl. Acids Res. 27: 573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigger, B., Tolmachov, O., Collombet, J.M., and Coutelle, C. 2000. Introduction of chloramphenicol resistance into the modified mouse mitochondrial genome: Cloning of unstable sequences by passage through yeast. Anal Biochem. 277: 236-242. [DOI] [PubMed] [Google Scholar]
- Carrano, A.V., de Jong, P.J., Branscomb, E., Slezak, T., and Watkins, B.W. 1989. Constructing chromosome- and region-specific cosmid maps of the human genome. Genome 31: 1059-1065. [DOI] [PubMed] [Google Scholar]
- Devenish, R.J. and Newlon, C.S. 1982. Isolation and characterization of yeast ring chromosome III by a method applicable to other circular DNAs. Gene 3: 277-288. [DOI] [PubMed] [Google Scholar]
- Gardner, M.J., Shallom, S.J., Carlton, J.M., Salzberg, S.L., Nene, V., Shoaibi, A., Ciecko, A., Lynn, J., Rizzo, M., Weaver, B., et al. 2002. Sequence of Plasmodium falciparum chromosomes 2, 10, 11, and 14. Nature 419: 531-534. [DOI] [PubMed] [Google Scholar]
- Glockner, G., Eichinger, L., Szafranski, K., Pachebat, J.A., Bankier, A.T., Dear, P.H., Lehmann, R., Baumgart. C., Parra, G., Abril, J.F., et al. 2002. Sequence and analysis of chromosome 2 of Dictyostelium discoideum. Nature 418: 79-85. [DOI] [PubMed] [Google Scholar]
- Grimwood, J. and Schmutz, J. 2003. Genomics: Six is seventh. Nature 425: 775-776. [DOI] [PubMed] [Google Scholar]
- Hagan, C.E. and Warren, G.J. 1982. Lethality of palindromic DNA and its use in selection of recombinant plasmids. Gene 19: 147-151. [DOI] [PubMed] [Google Scholar]
- Kang, H.K. and Cox, D.W. 1996. Tandem repeats 3′ of the IGHA genes in the human immunoglobulin heavy chain gene cluster. Genomics 35: 189-195. [DOI] [PubMed] [Google Scholar]
- Kim, J.H., Leem, S.H., Sunwoo, Y., and Kouprina, N. 2003. Separation of long-range human TERT gene haplotypes by transformation-associated recombination cloning in yeast. Oncogene 22: 2452-2456. [DOI] [PubMed] [Google Scholar]
- Kojima, T., Yoshikawa, Y., Takada, S., Sato, M., Nakamura, T., Takahashi, N., Copeland, N.G., Gilbert, D.J., Jenkins, N.A., and Mori, N. 2001. Genomic organization of the Shc-related phosphotyrosine adapters and characterization of the full-length Sck/ShcB: Specific association of p68-Sck/ShcB with pp135. Biochem. Biophys. Res. Commun. 284: 1039-1047. [DOI] [PubMed] [Google Scholar]
- Kouprina, N. and Larionov, V. 1999. Selective isolation of mammalian genes by TAR cloning. In Current protocols in human genetics, 1, pp. 1, 5.17.1-5.17.21. Wiley, New York. [DOI] [PubMed] [Google Scholar]
- ———. 2003. Exploiting the yeast Saccharomyces cerevisiae for the study of the organization of complex genomes. FEMS Microbiol. Rev. 27: 629-649. [DOI] [PubMed] [Google Scholar]
- Kouprina, N., Annab, L., Graves, J., Afshari, C., Barrett, J.C., and Larionov, V. 1998a. Functional copies of a human gene can be directly isolated by transformation-associated recombination cloning with a small 3′ end target sequence. Proc. Natl. Acad. Sci. 95: 4469-4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina, N., Campbell, M., Graves, J., Campbell, E., Meincke, L., Tesmer, J., Grady, D.L., Doggett, N.A., Moyzis, R.K., Deaven, L.L., et al. 1998b. Construction of human chromosome 16 and 5-specific circular YAC/BAC libraries in vivo recombination in yeast (TAR cloning). Genomics 53: 21-28. [DOI] [PubMed] [Google Scholar]
- Kouprina, N., Leem, S.-H., Solomon, G., Ly, A., Koriabine, M., Otstot, J., Pak, E., Dutra, A., Zhao, S., Barrett, J.C., et al. 2003. Segments missing from the draft human genome sequence can be isolated by TAR cloning in yeast. EMBO Rep. 4: 257-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larionov, V., Kouprina, N., Solomon, G., Barrett, J.C., and Resnick, M.A. 1997. Direct isolation of human BRCA2 gene by transformation-associated recombination in yeast. Proc. Natl. Acad. Sci. 94: 7384-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leem, S.H., Londono-Vallejo, J.A., Kim, J.H., Bui, H., Tubacher, E., Solomon, G., Park, J.E., Horikawa, I., Kouprina, N., Barrett, J.C., et al. 2002. The human telomerase gene: Complete genomic sequence and analysis of tandem repeat polymorphisms in intronic regions. Oncogene 21: 769-777. [DOI] [PubMed] [Google Scholar]
- Leem, S.H., Noskov, V.N., Park, J.E., Kim, S.I., Larionov, V., and Kouprina, N. 2003. Optimum conditions for selective isolation of genes from complex genomes by transformation-associated recombination cloning. Nucleic Acids Res. 31: e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy, E.A., Scott, A.C., Fiskerstrand, C.E., Bubb, V.J., and Quinn, J.P. 2003. The serotonin transporter intronic VNTR enhancer correlated with a predisposition to affective disorders has distinct regulatory elements within the domain based on the primary DNA sequence of the repeat unit. Eur. J. Neurosci. 17: 417-420. [DOI] [PubMed] [Google Scholar]
- Osoegawa, K., Mammoser, A.G., Wu, C., Frengen, E., Zeng, C., Catanese, J.J., and de Jong, P.J. 2001. A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res. 11: 493-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, X. and Leach, D.R. 2000. The roles of mutS, sbcCD and recA in the propagation of TGG repeats in Escherichia coli. Nucleic Acids Res. 28: 3178-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polushin, N., Malykh, A., Malykh, O., Zenkova, M., Chumakova, N., Vlassov, V., and Kozyavkin, S. 2001. 2′-modified oligonucleotides from methoxyoxalamido and succinimido precursors: Synthesis, properties, and applications. Nucleosides Nucleotides Nucl. Acids 20: 507-511. [DOI] [PubMed] [Google Scholar]
- Razin, S.V., Ioudinkova, E.S., Trifonov. E., and Scherrer, K. 2001. Non-clonability correlates with genome instability: A case of unique DNA region. J. Mol. Biol. 307: 481-486. [DOI] [PubMed] [Google Scholar]
- Schroth, G.P. and Ho, P.S. 1995. Occurrence of potential cruciform and H-DNA forming sequences in genomic DNA. Nucl. Acids Res. 23: 1977-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey, M., Lin, H.H., Hilyard, K.L., Gordon, S., and McKnight, A.J. 2001. Human epidermal growth factor (EGF) module-containing mucin-like hormone receptor 3 is a newmember of the EGF-TM7 family that recognizes a ligand on human macrophages and activated neutrophils. J. Biol. Chem. 276: 18863-18870. [DOI] [PubMed] [Google Scholar]
- Takamatsu, K., Maekawa, K., Togashi, T., Choi, D.K., Suzuki, Y., Taylor, T.D., Toyoda, A., Sugano, S., Fujiyama, A., Hattori, M., et al. 2002. Identification of two novel primate-specific genes in DSCR. DNA Res. 9: 89-97. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://genome.ucsc.edu/goldenPath/apr2001Traks.html; UCSC.
- http://www.ncbi.nlm.nih.gov/BLAST/blast_databases.html; BLAST genome analysis software.
- http://www.ncbi.nlm.nih.gov/genome/guide/; NCBI.