Abstract
Approximately half of all human genes have CpG islands (CGIs)around their promoter regions. Although CGIs usually escape methylation, those on Chromosome X in females and those in the vicinity of imprinted genes are exceptions: They have both methylated and unmethylated alleles to display a “composite” pattern in methylation analysis. In addition, aberrant methylation of CGIs is known to often occur in cancer cells. Here we developed a simple HpaII-McrBC PCR method for discrimination of full, null, incomplete, and composite methylation patterns, and applied it to all computationally identified CGIs on human Chromosome 21q. This comprehensive analysis revealed that, although most CGIs (103 out of 149)escape methylation, a sizable fraction (31 out of 149)are fully methylated even in normal peripheral blood cells. Furthermore, we identified seven CGIs showing the composite methylation, and demonstrated that three of them are indeed methylated monoallelically. Further analyses using informative pedigrees revealed that two of the three are subject to maternal allele-specific methylation. Intriguingly, the other CGI is methylated in an allele-specific but parental-origin-independent manner. Thus, the cell seems to have a broader repertoire of methylating CGIs than previously thought, and our approach may contribute to uncover novel modes of allelic methylation.
Mammalian genomes contain CpG dinucleotides much less frequently than expected from their GC contents (i.e., CpG suppression), and most of them are modified by methylation at the 5-position of cytosine (Ponger et al. 2001). However, CpG suppression is not observed or much less evident in characteristic regions termed CpG islands (CGIs) despite their high GC contents (Gardiner-Garden and Frommer 1987; Antequera and Bird 1993). CGIs are generally found near promoter regions of genes, including most housekeeping and many tissue-specific ones, and intriguingly escape methylation, often regardless of the expression of flanking genes (Macleod et al. 1998; Grunau et al. 2000; Ioshikhes and Zhang 2000).
Although aberrant methylation of CGIs is frequently observed in cancer cells, some exceptional CGIs are physiologically methylated in an allele-specific manner. It is well known that one of the two X-chromosomes in females is inactivated. The CGIs on the inactivated X-chromosome are heavily methylated, similar to other regions on this chromosome (Norris et al. 1991). On autosomes, a small number of imprinted genes that display exclusive or highly skewed expression of specific allele depending on their parental origins (Morison and Reeve 1998) have been demonstrated to accompany regions subject to parental-origin-dependent methylation. These regions are termed allelic differentially methylated regions (DMRs), and have been demonstrated to play pivotal roles in genomic imprinting (Wutz et al. 1997; Yoon et al. 2002). Although allelic DMRs show base composition similar to CGIs and often contain tandem repeat sequences (Neumann et al. 1995), they share no apparent sequence similarity.
Allelic DMRs have been extensively searched around imprinted genes but not in other regions. In other words, their distribution has not been analyzed in an unbiased, hypothesis-free manner. Although several methods have been developed for the purpose, they are not truly comprehensive and have missed many DMRs (Plass et al. 1996). We thus intended to thoroughly examine the methylation status of CGIs based on the established genome sequence data, which allows one to identify all CGIs in silico. The experimental method to be used for the evaluation of methylation status should not only be rapid and simple but also be capable of detecting the coexistence of methylated and unmethylated alleles (i.e., composite methylation).
As a method to fulfill the requirement, we developed a simple method called HpaII-McrBC PCR, which is based on the complementary sensitivity of the two enzymes HpaII and McrBC to DNA methylation. We applied it for the analysis of 149 CGIs computationally identified on human Chromosome 21q, one of the most completely sequenced chromosomes. The analysis, which is the very first thorough analysis of CGIs on a chromosome-wide scale, revealed an unexpectedly high incidence of normally methylated CGIs and, furthermore, three allelic DMRs, including one subject to a novel mode of allelic methylation.
RESULTS
HpaII-McrBC PCR for Rapid Evaluation of Allelic Methylation Status
A comprehensive methylation analysis requires a rapid and simple method to examine methylation status. Although the so-called HpaII-PCR has been widely used, it cannot distinguish between fully methylated and compositely methylated sequences, the latter of which include CGIs on X-chromosomes in female and allelic DMRs in the vicinity of imprinted genes. To overcome this drawback, we developed a novel method termed HpaII-McrBC PCR by exploiting two enzymes with complementary methylation sensitivity. The method can readily distinguish regions subject to full, null, composite, and incomplete methylation.
In HpaII-McrBC PCR, genomic DNA is divided into two portions, each of which is subsequently digested with HpaII (or other methylation-sensitive enzymes such as HhaI) or McrBC, and used as templates for PCR (Fig. 1). Whereas HpaII cuts unmethylated alleles at CCGG sites, McrBC digests methylated alleles at RmCN40∼80RmC (Fig. 1; Sutherland et al. 1992; Stewart and Raleigh 1998). In the case of a fully methylated sequence, HpaII totally fails to digest the target, whereas McrBC cuts it completely (Fig. 1A). Amplification would be thus achieved only from the HpaII-digested template. On the other hand, an unmethylated region is digested only with HpaII but not with McrBC, and hence amplification would be successful only from the McrBC-digested DNA. Accordingly, amplification from both HpaII- and McrBC-digested DNAs indicates the presence of both methylated and unmethylated alleles in the sample or the “composite” methylation. If the target region is incompletely methylated, amplification will be obtained from neither HpaII- nor McrBC-digested DNAs, because both enzymes digest the template. Therefore, HpaII-McrBC PCR can, in principle, distinguish four different statuses of allelic methylation (Fig. 1A).
Figure 1.
HpaII-McrBC PCR. (A) Principle of HpaII-McrBC PCR that distinguishes four different patterns in allelic methylation. (Left) The open and closed circles indicate unmethylated CCGG and methylated CmCGG sites, respectively. Similarly, the open and closed squares indicate unmethylated RC and methylated RmC sites, respectively. Each line with two circlesand four squaresindicateseach allele of genomic DNA. (Middle) The +, +/-, and - mean complete, incomplete, and no digestion with HpaII or McrBC, respectively. (Right) A schematic gel pattern of HpaII-McrBC PCR products in individual cases. (B) Proof of principle for HpaII-McrBC PCR. HpaII-McrBC PCR wasapplied to the intronic DMR of mouse Impact, a paternally expressed gene. JF, B6, and (B6 × JF) F1 indicate Mus musculus molossinus JF1, Mus musculus domesticus C57BL/6, and F1 hybrid generated between JF and B6, respectively. The PCR products from mock-treated (-), HpaII-digested, or McrBC-digested DNAs from JF, B6, or (B6 × JF) F1 were electrophoresed, stained with ethidium bromide, and visualized by UV illumination.
As a proof-of-principle experiment, we applied the method to the allelic DMR of mouse Impact, which we identified as a gene expressed exclusively from the paternal allele bearing a maternally methylated CGI or allelic DMR in its first intron (Hagiwara et al. 1997; Okamura et al. 2000). We analyzed an F1 hybrid between Mus musculus domesticus C57BL/6 (B6) and Mus musculus molossinus JF1 (JF), because the CGI displays an obvious length polymorphism between the two species, allowing one to distinguish the two alleles by simple gel electrophoresis (Okamura et al. 2000).
As shown in Figure 1B, both the maternally derived B6 allele (1426 bp) and the paternally derived JF one (1245 bp) were detected from the mock-treated DNA prepared from a (B6 × JF) F1 mouse. Note that only the maternally derived B6 allele was amplified from the HpaII-digested DNA, which serves as a template for methylated portions. On the other hand, only the paternally derived JF allele was amplified from the McrBC-treated DNA, in which methylated alleles were digested. These results unequivocally indicate maternal methylation of this CGI, consistent with the previous observation (Okamura et al 2000).
We also examined the CGI spanning the promoter region of human IMPACT, which we had previously shown to escape methylation biallelically and hence represent a conventional nonmethylated CGI (Okamura et al. 2000). As expected, amplification was achieved only from the McrBC-digested DNA but not from the HpaII-digested one, indicative of unmethylated pattern (data not shown).
These results demonstrated that HpaII-McrBC PCR serves as a rapid and simple method to evaluate allelic methylation status.
Strategy for a Comprehensive HpaII-McrBC PCR Analysis of CGIs on Human Chromosome 21q
Having a versatile method in hand to examine allelic methylation status, we planned a comprehensive methylation analysis of CGIs on human Chromosome 21q. Because this chromosome provides the most complete and accurate sequence data ever generated, one can identify CGIs most thoroughly in silico and examine their methylation status in a truly comprehensive manner.
According to the original definition of CGI, it should be longer than 200 bp, have a GC content higher than 50%, and display an expected CpG frequency (ECF) larger than 0.6 (Gardiner-Garden and Frommer 1987). This definition has, however, turned out to allow contamination of repetitive DNA elements as well as exons. Thus, more stringent criteria are generally used in recent studies. For instance, in the initial annotation of human Chromosome 21q, we used a criterion requiring that the length, GC content, and ECF be larger than 400 bp, 55%, and 0.6, respectively, to identify 137 CGIs, most of which were found linked to the 5′-portions of genes (Hattori et al. 2000). Here we used a slightly relaxed condition (length >400 bp, GC content >50%, ECF >0.6) with the masking of Alu and LINE-1 sequences to extract 149 CGIs in total (Table 1).
Table 1.
Methylation and Other Features of CpG Islands on Human Chromosome 21q
|
Nucleotide positione
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CpG islandsa | Methylation | Repeatb | Locus | GC %c | Obs/expd | Size (bp) | Start | End | Location in the linked genef | CGI-linked genes |
| #1 (NT_002836.4 740746-742525) | Complete methylation | — | 21q11.1 | 72.0 | 0.88 | 780 | 13998888 | 13999667 | CDS | Similarity to feminization 1 homolog a (C. elegans) |
| #2 (NT_002836.4 798428-799837) | Complete methylation | — | 21q11.1 | 72.2 | 0.80 | 410 | 14056570 | 14056979 | CDS | Similarity to feminization 1 homolog a (C. elegans) |
| #3 (NT_002836.4 1099894-1101335) | Complete methylation | — | 21q11.1 | 68.1 | 0.80 | 442 | 14358036 | 14358477 | 5′-UTR and CDS | Similarity to hypothetical protein DKFZp434A171; KIAA0565; ankyrin repeat-containing proteins |
| #4 (NT_002836.4 2100365-2102278) | Unmethylation | — | 21q11.2 | 76.2 | 0.98 | 914 | 15358453 | 15359366 | 5′-UTR | Nuclear receptor interacting protein 1 (NRIP1) |
| #5 (NT_002836.4 2765740-2767861) | Unmethylation | — | 21q11.2 | 74.7 | 0.88 | 1320 | 16023802 | 16025122 | 5′-UTR and CDS | Ubiquitin specific protease 25 (USP25) |
| #6 (NT_002836.4 4548804-4550994) | Unmethylation | — | 21q21.1 | 71.1 | 0.83 | 1229 | 17806771 | 17807999 | 5′-UTR and CDS | Coxsackie virus and adenovirus receptor (CXADR) |
| #7 (NT_002836.4 4648389-4650557) | Unmethylation | — | 21q21.1 | 73.4 | 0.96 | 1169 | 17906356 | 17907524 | 5′-UTR | BTG family member 3 (BTG3) |
| #8 (NT_002836.4 4854926-4857042) | Unmethylation | — | 21q21.1 | 68.0 | 0.97 | 1117 | 18112871 | 18113987 | 5′-UTR and CDS | Chromosome 21 open reading frame 91 (C21 orf91), hypothetical protein LOC54149 (YG81) |
| #9 (NT_002836.4 12511562-12513060) | Unmethylation | — | 21q21.2 | 70.3 | 0.88 | 499 | 25856202 | 25856700 | — | |
| #10 (NT_002836.4 12684911-12686522) | Unmethylation | — | 21q21.2 | 71.7 | 0.79 | 612 | 26029550 | 26030161 | 5′-UTR | ATP synthase H+ transporting mitochondrial F0 complex subunit F6 |
| #11 (NT_002836.4 13119202-13121651) | Unmethylation | — | 21q21.3 | 70.8 | 0.83 | 1450 | 26463778 | 26465227 | 5′-UTR and CDS | Amyloid beta precursor protein (APP) |
| #12 (NT_002836.4 13793868-13795756) | Unmethylation | — | 21q21.3 | 67.2 | 0.98 | 889 | 27138374 | 27139262 | 5′-UTR and CDS | Matrix metalloprotease (ADAMTS1) |
| #13 (NT_002836.4 13915193-13916938) | Unmethylation | — | 21q21.3 | 70.1 | 0.87 | 746 | 27259699 | 27260444 | 5′-UTR and CDS | A disintegrin-like and metalloprotease with thrombospondin type 1 motif5 (ADAMTS5) |
| #14 (NT_002836.4 13917154-13918663) | Unmethylation | — | 21q21.3 | 61.7 | 0.83 | 510 | 27261660 | 27262169 | — | |
| #15 (NT_002836.4 15834745-15836237) | Unmethylation | — | 21q22.11 | 62.4 | 0.93 | 493 | 29179194 | 29179686 | 5′-UTR and CDS | Putative N6-DNA-methyltransferase (N6AMT1) |
| #16 (NT_002836.4 16246692-16248676) | Unmethylation | — | 21q22.11 | 76.5 | 0.90 | 985 | 29591133 | 29592117 | 5′-UTR | BTB and CNC homology 1, basic leucine zipper transcription factor 1 (BACH1) |
| #17 (NT_002836.4 18506100-18509038) | Incomplete methylation | — | 21q22.11 | 71.7 | 0.94 | 1939 | 31850340 | 31852278 | 5′-UTR | Similarity to T-lymphoma invasion and metastasis-inducing protein 1 (TIAM1) |
| #18 (NT_002836.4 18607855-18609856) | Unmethylation | — | 21q22.11 | 69.0 | 0.94 | 1002 | 31952095 | 31953096 | 5′-UTR and CDS | Superoxide dismutase (SOD-1) |
| #19 (NT_002836.4 18679791-18682057) | Unmethylation | — | 21q22.11 | 74.3 | 0.90 | 1267 | 32024044 | 32025310 | Similarity to CTD-binding SR-like protein rA4 | |
| #20 (NT_002836.4 18821198-18823391) | Unmethylation | — | 21q22.11 | 72.7 | 0.87 | 1374 | 32165461 | 32166834 | 5′-UTR and CDS | Hormonally up-regulated Neu-associated kinase (HUNK) |
| #21 (NT_002836.4 19227134-19228707) | Unmethylation | — | 21q22.11 | 65.1 | 0.85 | 574 | 32571397 | 32571970 | 5′-UTR and CDS | FAPP1-associated protein 1 (FASP1); chromosome 21 open reading frame 45 (C21orf45) |
| #22 (NT_002836.4 19248734-19250388) | Complete methylation | + | 21q22.11 | 54.0 | 1.21 | 655 | 32592997 | 32593651 | — | |
| #23 (NT_002836.4 19359982-19362161) | Unmethylation | — | 21q22.11 | 70.2 | 0.86 | 1180 | 32704245 | 32705424 | — | |
| #24 (NT_002836.4 19561116-19562518) | Unmethylation | — | 21q22.11 | 62.0 | 0.82 | 403 | 32905379 | 32905781 | 5′-UTR | Chromosome 21 open reading frame 59 (C21orf59) |
| #25 (NT_002836.4 19675812-19677620) | Unmethylation | — | 21q22.11 | 71.4 | 0.83 | 809 | 33020075 | 33020883 | 5′-UTR and CDS | Synaptojanin 1 (SYNJ1) |
| #26 (NT_002836.4 19719511-19721351) | Unmethylation | — | 21q22.11 | 70.2 | 0.98 | 841 | 33063774 | 33064614 | 5′-UTR and CDS | Chromosome 21 open reading frame 66 (C21orf66); Putative transcription factor (ORF1) |
| #27 (NT_002836.4 19972011-19973417) | Unmethylation | — | 21q22.11 | 62.9 | 0.83 | 407 | 33316261 | 33316667 | — | |
| #28 (NT_002836.4 19975198-19977216) | Unmethylation | — | 21q22.11 | 68.6 | 0.90 | 1019 | 33319448 | 33320466 | 5′-UTR, CDS and 3′-UTR | Oligodendrocyte lineage transcription factor 2 (OLIG2) |
| #29 (NT_002836.4 20018303-20020533) | Unmethylation | — | 21q22.11 | 73.9 | 0.84 | 1283 | 33362553 | 33363835 | 5′-UTR, CDS and 3′-UTR | Oligodendrocyte transcription factor 1 (Olig1) |
| #30 (NT_002836.4 20178164-20179883) | Unmethylation | — | 21q22.11 | 72.0 | 0.99 | 720 | 33522413 | 33523132 | 5′-UTR | Interferon (alpha, beta and omega) receptor 2(IFNAR2) |
| #31 (NT_002836.4 20273122-20274794) | Unmethylation | — | 21q22.11 | 64.7 | 0.93 | 673 | 33617371 | 33618043 | 5′-UTR and CDS | Interferon (alpha, beta and omega) receptor 1(IFNAR1) |
| #32 (NT_002836.4 20351550-20353202) | Unmethylation | — | 21q22.11 | 79.1 | 0.87 | 653 | 33695798 | 33696450 | 5′-UTR and CDS | Interferon gamma receptor 2 (interferon gamma transducer 1) (IFNGR2) |
| #33 (NT_002836.4 20427755-20429718) | Unmethylation | — | 21q22.11 | 73.0 | 0.93 | 964 | 33772003 | 33772966 | 5′-UTR | Chromosome 21 open reading frame 4 (C21orf4) |
| #34 (NT_002836.4 20536702-20538471) | Unmethylation | — | 21q22.11 | 72.7 | 0.92 | 770 | 33880950 | 33881719 | 5′-UTR and CDS; 3′-UTR | Downstream neighbor of SON (DONSON); Crystallin, zeta (quinone reductase)-like 1 (CRYZL1) |
| #35 (NT_002836.4 20590829-20592722) | Unmethylation | — | 21q22.12 | 75.8 | 0.88 | 894 | 33935077 | 33935970 | 5′-UTR | Intersectin 1 (SH3 domain protein) (ITSN1) |
| #36 (NT_002836.4 21021661-21023784) | Unmethylation | — | 21q22.12 | 77.3 | 1.02 | 1124 | 34365908 | 34367031 | 5′-UTR and CDS | Mitochondrial ribosomal protein S6 (MRPS6) |
| #37 (NT_002836.4 21323771-21325344) | Unmethylation | — | 21q22.12 | 68.1 | 0.84 | 574 | 34668018 | 34668591 | — | |
| #38 (NT_002836.4 21562903-21564940) | Unmethylation | — | 21q22.12 | 75.3 | 0.95 | 1038 | 34907150 | 34908187 | 5′-UTR and CDS | Down syndrome critical region gene 1 (DSCR1) |
| #39 (NT_002836.4 21617860-21620045) | Unmethylation | + | 21q22.12 | 75.8 | 0.88 | 1186 | 34962107 | 34963292 | — | |
| #40 (NT_002836.4 21740221-21742136) | Complete methylation | — | 21q22.12 | 71.8 | 0.84 | 1064 | 35084468 | 35085531 | CDS and 3′-UTR | Runt-related transcription factor 1 (RUNX1); acute myeloid leukemia 1 (AML1) |
| #41 (NT_002836.4 21835266-21836717) | Unmethylation | — | 21q22.12 | 67.2 | 0.79 | 452 | 35179513 | 35179964 | 5′-UTR and CDS | Runt-related transcription factor 1 (RUNX1); acute myeloid leukemia 1 (AML1) |
| #42 (NT_002836.4 21837000-21839961) | Incomplete methylation | — | 21q22.12 | 73.9 | 0.90 | 1961 | 35181247 | 35183208 | 5′-UTR | Runt-related transcription factor 1 (RUNX1); acute myeloid leukemia 1 (AML1) |
| #43 (NT_002836.4 22835347-22836774) | Complete methylation | — | 21q22.13 | 60.5 | 0.94 | 428 | 36179519 | 36179946 | 5′-UTR and CDS | Homo sapiens protein phosphatase 1, regulatory (inhibitor) subunit 2 pseudogene 2 (PPP1R2P2) |
| #44 (NT_002836.4 23008324-23010418) | Unmethylation | — | 21q22.13 | 62.5 | 0.86 | 1095 | 36352469 | 36353563 | 5′-UTR | Chromosome 21 open reading frame 18 (C21orf18) |
| #45 (NT_002836.4 23018419-23020042) | Unmethylation | — | 21q22.13 | 70.0 | 0.87 | 624 | 36362564 | 36363187 | 5′-UTR and CDS | Carbonyl reductase 1 (CBR1) |
| $46 (NT_002836.4 23083543-23085110) | Unmethylation | — | 21q22.13 | 69.8 | 0.83 | 568 | 36427688 | 36428255 | 5′-UTR and CDS | Carbonyl reductase 3 (CBR3) |
| #47 (NT_002836.4 23104500-23106945) | Unmethylation | — | 21q22.13 | 73.0 | 0.78 | 1446 | 36448645 | 36450090 | 5′-UTR | mRNA expressed in placenta |
| #48 (NT_002836.4 23268373-23270063) | Unmethylation | — | 21q22.13 | 73.6 | 0.86 | 691 | 36612817 | 36613507 | 5′-UTR and CDS | Nuclear matrix protein NXP-2 (NXP-2) |
| #49 (NT_002836.4 23333456-23335039) | Unmethylation | — | 21q22.13 | 76.8 | 0.79 | 584 | 36677900 | 36678483 | 5′-UTR and CDS | Similarity to chromatin assembly factor 1 subunit B (p60) |
| #50 (NT_002836.4 23646833-23649367) | Unmethylation | — | 21q22.13 | 74.0 | 0.83 | 1535 | 36991300 | 36992834 | 5′-UTR and CDS | Single-minded (Drosophila) homolog 2 (SIM2) transcript variant SIM2s |
| #51 (NT_002836.4 23648887-23650365) | Unmethylation | — | 21q22.13 | 65.9 | 0.75 | 479 | 36993354 | 36993832 | Intron | Single-minded (Drosophila) homolog 2 (SIM2) |
| #52 (NT_002836.4 23657131-23658548) | Unmethylation | — | 21q22.13 | 71.7 | 0.80 | 418 | 37001598 | 37002015 | CDS | Single-minded (Drosophila) homolog 2 (SIM2) transcript variant SIM2s |
| #53 (NT_002836.4 23695691-23697656) | Unmethylation | — | 21q22.13 | 73.8 | 0.99 | 966 | 37040158 | 37041123 | CDS and 3′-UTR | Single-minded (Drosophila) homolog 2 (SIM2) |
| #54 (NT_002836.4 23914335-23916288) | Unmethylation | — | 21q22.13 | 78.5 | 0.82 | 954 | 37258805 | 37259758 | — | |
| #55 (NT_002836.4 23928709-23930164) | Composite methylation | — | 21q22.13 | 69.3 | 0.75 | 456 | 37273178 | 37273633 | 5′-UTR | Holocarboxylase synthetase |
| #56 (NT_002836.4 23938177-23939758) | Unmethylation | — | 21q22.13 | 74.2 | 0.85 | 582 | 37282645 | 37283226 | 5′-UTR | Holocarboxylase synthetase |
| #57 (NT_002836.4 23953977-23956357) | Incomplete methylation | — | 21q22.13 | 70.2 | 0.82 | 1381 | 37298442 | 37299822 | 5′-UTR and CDS | Down syndrome critical region gene 6 (DSCR6) |
| #58 (NT_002836.4 24021123-24022718) | Unmethylation | — | 21q22.13 | 73.6 | 0.90 | 596 | 37365583 | 37366178 | 5′-UTR | Down syndrome critical region gene 5 (DSCR5) |
| #59 (NT_002836.4 24206418-24207868) | Composite methylation | — | 21q22.13 | 58.3 | 0.90 | 451 | 37550792 | 37551242 | Intron | Down syndrome critical region gene 3 (DSCR3) |
| #60 (NT_002836.4 24215237-24217598) | Unmethylation | — | 21q22.13 | 67.5 | 0.78 | 1362 | 37559611 | 37560972 | 5′-UTR and CDS | Down syndrome critical region gene 3 (DSCR3) |
| #61 (NT_002836.4 24314183-24317285) | Unmethylation | — | 21q22.13 | 75.0 | 0.95 | 2103 | 37658554 | 37660656 | 5′-UTR | Dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A (DYRK1A) |
| #62 (NT_002836.4 24511883-24513479) | Unmethylation | — | 21q22.13 | 67.0 | 0.94 | 597 | 37856254 | 37856850 | — | |
| #63 (NT_002836.4 25608278-25609973) | Unmethylation | — | 21q22.2 | 74.5 | 1.04 | 696 | 38952732 | 38953427 | 5′-UTR | Transcriptional regulator ERG2 gene |
| #64 (NT_002836.4 25753617-25755681) | Unmethylation | — | 21q22.2 | 71.1 | 0.78 | 1259 | 39098071 | 39099329 | 5′-UTR | Erythroblastosis virus oncogene homolog 2 (ets-2) |
| #65 (NT_002836.4 26130818-26133115) | Unmethylation | — | 21q22.2 | 66.7 | 0.86 | 1298 | 39475251 | 39476548 | 5′-UTR and CDS | Down syndrome critical region gene 2 (DSCR2) |
| #66 (NT_002836.4 26259167-26262261) | Unmethylation | — | 21q22.2 | 71.5 | 0.91 | 2095 | 39604704 | 39606798 | 5′-UTR and CDS | WD repeat domain 9 (WDR9) |
| #67 (NT_002836.4 26295243-26297124) | Unmethylation | — | 21q22.2 | 76.7 | 0.90 | 882 | 39640780 | 39641661 | 5′-UTR and CDS | High-mobility group (nonhistone chromosomal) protein 14 (HMG14) |
| #68 (NT_002836.4 26325240-26326648) | Incomplete methylation | — | 21q22.2 | 61.8 | 1.02 | 409 | 39672415 | 39672823 | 5′-UTR and CDS | Tryptophan rich basic protein (WRB) |
| #69 (NT_002836.4 26390768-26392267) | Unmethylation | — | 21q22.2 | 67.8 | 0.91 | 500 | 39737943 | 39738442 | 5′-UTR | Chromosome 21 open reading frame 13 (C21 orf13) |
| #70 (NT_002836.4 26556991-26558660) | Unmethylation | — | 21q22.2 | 68.9 | 0.95 | 670 | 39904918 | 39905587 | 5′-UTR | C21 orf88 protein form A (C21 orf88) |
| #71 (NT_002836.4 27791317-27792866) | Unmethylation | — | 21q22.2 | 75.2 | 0.99 | 550 | 41138898 | 41139447 | 5′-UTR and CDS | Down syndrome cell adhesion molecule (DSCAM) |
| #72 (NT_002836.4 28112154-28114765) | Unmethylation | — | 21q22.3 | 73.4 | 0.81 | 1612 | 41459726 | 41461337 | 5′-UTR and CDS | Beta-site APP-cleaving enzyme 2 (BACE2) |
| #73 (NT_002836.4 28452242-28454382) | Unmethylation | — | 21q22.3 | 72.2 | 0.73 | 1141 | 41799550 | 41800690 | — | |
| #74 (NT_001035.5 179474-181456) | Composite methylation | — | 21q22.3 | 70.9 | 0.87 | 983 | 42072090 | 42073072 | 5′-UTR and CDS | Ankyrin repeat domain 3 (ANKRD3) |
| #75 (NT_003545.2 47617-49632) | Complete methylation | — | 21q22.3 | 78.3 | 1.00 | 1016 | 42192927 | 42193942 | CDS | PR-domain zinc finger protein 15 (PRDM15) |
| #76 (NT_003545.2 122339-124366) | Unmethylation | — | 21q22.3 | 77.2 | 0.87 | 1028 | 42267649 | 42268676 | 5′-UTR and CDS | Similarity to human cDNA DKFZp586F0422 |
| #77 (NT_00354.2 178197-181181) | Unmethylation | — | 21q22.3 | 72.0 | 0.96 | 1985 | 42323507 | 42325491 | 5′-UTR | Zinc finger protein 295 (ZNF295) |
| #78 (NT_003545.2 387867-389371) | Unmethylation | — | 21q22.3 | 73.0 | 0.89 | 505 | 42533177 | 42533681 | 5′-UTR | ABCG1 gene for ABC transporter (ATP-binding cassette, sub-family G (WHITE) member 1 |
| #79 (NT_003545.2 403601-405007) | Unmethylation | — | 21q22.3 | 72.4 | 1.01 | 407 | 42548911 | 42549317 | Intron | ABCG1 gene for ABC transporter (ATP-binding cassette, sub-family G (WHITE) member 1 |
| #80 (NT_003545.2 665089-666509) | Unmethylation | — | 21q22.3 | 69.8 | 0.78 | 421 | 42810399 | 42810819 | 5′-UTR and CDS; 5′-UTR | TSGA2 mRNA for testis specific protein A2; putative glycerol 3-phosphate permease (SLC37A1) |
| #81 (NT_003545.2 682430-684731) | Unmethylation | — | 21q22.3 | 72.8 | 0.83 | 1354 | 42827740 | 42829093 | 5′-UTR (exons -2, -1 and 1) | Putative glycerol 3-phosphate permease (SLC37A1) |
| #82 (NT_003545.2 756256-760387) | Complete methylation | + | 21q22.3 | 61.2 | 1.12 | 3132 | 42901566 | 42904697 | — | |
| #83 (NT_003545.2 822395-823837) | Unmethylation | — | 21q22.3 | 74.7 | 0.81 | 443 | 42967705 | 42968147 | 5′-UTR and CDS | Phosphodiesterase 9A (PDE9A) |
| #84 (NT_003545.2 854975-856401) | Complete methylation | — | 21q22.3 | 62.3 | 0.86 | 427 | 43000285 | 43000711 | CDS | Phosphodiesterase 9A (PDE9A) |
| #85 (NT_003545.2 1017136-1019217) | Complete methylation | + | 21q22.3 | 62.9 | 1.34 | 1082 | 43162446 | 43163527 | 3′-UTR | WD repeat domain 4 (WDR4) |
| #86 (NT_003545.2 1047902-1049365) | Unmethylation | — | 21q22.3 | 70.6 | 0.87 | 464 | 43193212 | 43193675 | 5′-UTR and CDS | WD repeat domain 4 (WDR4) |
| #87 (NT_003545.2 1142956-1145447) | Unmethylation | — | 21q22.3 | 76.2 | 0.96 | 1492 | 43288266 | 43289757 | 5′-UTR | Similarity to PBX/knotted 1 homeobox 1 |
| #88 (NT_003545.2 1243354-1246655) | Incomplete methylation | — | 21q22.3 | 69.9 | 0.84 | 2302 | 43388664 | 43390965 | 5′-UTR | Cystathionine beta-synthase (CBS) |
| #89 (NT_003545.2 1275925-1277810) | Unmethylation | — | 21q22.3 | 72.1 | 0.99 | 886 | 43421235 | 43422120 | 5′-UTR and CDS | U2(RNU2) small nuclear RNA auxillary factor 1 |
| #90 (NT_002835.3 30733-32180) | Complete methylation | + | 21q22.3 | 60.4 | 1.07 | 448 | 43574697 | 43575144 | CDS | PR-domain zinc finger protein 15 (PRDM15) |
| #91 (NT_002835.3 158046-161231) | Unmethylation | — | 21q22.3 | 76.1 | 0.86 | 2526 | 43702010 | 43704535 | 5′-UTR and CDS | Similarity to Mus musculus SNF1-like kinase (Snf1lk) |
| #92 (NT_002835.3 297286-298768) | Complete methylation | — | 21q22.3 | 58.1 | 0.96 | 483 | 43841250 | 43841732 | 5′-UTR, CDS and 3′-UTR | H2B histone family member S (LOC115376) |
| #93 (NT_002835.3 389981-391414) | Complete methylation | — | 21q22.3 | 69.5 | 0.91 | 434 | 43933945 | 43934378 | 5′-UTR and CDS | Heatschock transcription factor 2 binding protein (HSF2BP) |
| #94 (NT_002835.3 391442-393133) | Unmethylation | — | 21q22.3 | 68.0 | 0.98 | 692 | 43935406 | 43936097 | 5′-UTR; 5′-UTR and CDS | Heatshock transcription factor 2 binding protein (HSF2BP); KIAA0179 protein |
| #95 (NT_002835.3 450630-453100) | Incomplete methylation | — | 21q22.3 | 74.4 | 0.92 | 1471 | 43994594 | 43996064 | 5′-UTR and CDS | Pyridoxal (pyridoxine, vitamin B6) kinase (PDXK) |
| #96 (NT_002835.3 473197-474680) | Complete methylation | + | 21q22.3 | 82.2 | 0.86 | 484 | 44017161 | 44017644 | CDS | Pyridoxal (pyridoxine, vitamin B6) kinase (PDXK) |
| #97 (NT_002835.3 508085-509942) | Unmethylation | — | 21q22.3 | 72.9 | 0.94 | 858 | 44052049 | 44052906 | 5′-UTR and CDS | Cystatin B (stefin B) (CSTB) |
| #98 (NT_002835.3 521399-523051) | Unmethylation | — | 21q22.3 | 71.6 | 0.90 | 653 | 44065363 | 44066015 | 5′-UTR and CDS | Novel nuclear protein 1 (NNP-1/Nop52) |
| #99 (NT_002835.3 596448-599173) | Unmethylation | — | 21q22.3 | 80.4 | 0.84 | 1844 | 44140412 | 44142255 | 5′-UTR | Similarity to 1-acylglycerol-3-phosphate O-acyltransferase 3 |
| #100 (NT_002835.3 743918-746243) | Unmethylation | — | 21q22.3 | 73.7 | 0.91 | 1442 | 44287766 | 44289207 | 5′-UTR and CDS | GT334 protein (GT334); transmembrane protein 1 (TMEM1) |
| #101 (NT_002835.3 839100-840858) | Unmethylation | — | 21q22.3 | 70.4 | 0.99 | 759 | 44383064 | 44383822 | 5′-UTR and CDS | Periodic tryptophan protein 2 (PWP2) |
| #102 (NT_002835.3 865543-867041) | Unmethylation | — | 21q22.3 | 69.5 | 0.81 | 499 | 44409507 | 44410005 | 5′-UTR and CDS | Chromosome 21 open reading frame 33 (C21orf33), homolog of ES1 protein (zebrafish) |
| #103 (NT_002835.3 900040-901476) | Composite methylation | + | 21q22.3 | 53.0 | 0.92 | 437 | 44444004 | 44444440 | CDS | Chromosome 21 open reading frame 32 (C21orf32) |
| #104 (NT_002835.3 972548-974376) | Unmethylation | — | 21q22.3 | 77.6 | 0.84 | 829 | 44516512 | 44517340 | 5′-UTR and CDS | KIAA0653 protein, Transmembrane protein B7-H2 ICOS ligand (B7-like protein) |
| #105 (NT_002835.3 974194-975798) | Unmethylation | — | 21q22.3 | 71.4 | 0.79 | 605 | 44518158 | 44518762 | — | |
| #106 (NT_002835.3 1017861-1019307) | Incomplete methylation | — | 21q22.3 | 75.1 | 0.83 | 447 | 44561825 | 44562271 | 5′-UTR and CDS | Autoimmune regulator (autoimmune polyendocrinopathy candidiasis ectodermal dystrophy) (AIRE) |
| #107 (NT_002835.3 1031958-1033740) | Unmethylation | — | 21q22.3 | 79.3 | 0.89 | 783 | 44575922 | 44576704 | 5′-UTR | PFKL gene for liver-type 6-phosphofructokinase (EC 2.7.1.11) |
| #108 (NT_002835.3 1070778-1072793) | Unmethylation | + | 21q22.3 | 76.2 | 0.85 | 1016 | 44614742 | 44615758 | 5′-UTR and CDS | Chromosome 21 open reading frame 2 (C21orf2) |
| #109 (NT_002835.3 1082093-1084166) | Unmethylation | + | 21q22.3 | 76.6 | 0.87 | 1074 | 44626057 | 44627130 | — | |
| #110 (NT_002835.3 1101487-1103181) | Complete methylation | + | 21q22.3 | 71.9 | 0.81 | 695 | 44645451 | 44646145 | CDS | Transient receptor potential channel 7 (TRPC7) |
| #111 (NT_002835.3 1187382-1188869) | Unmethylation | — | 21q22.3 | 75.8 | 0.79 | 488 | 44731346 | 44731833 | — | |
| #112 (NT_002835.3 1438369-1439902) | Composite methylation | + | 21q22.3 | 64.9 | 0.85 | 534 | 44982314 | 44982847 | Intron | Chromosome 21 open reading frame 29 (C21orf29) |
| #113 (NT_002835.3 1441617-1443042) | Unmethylation | — | 21q22.3 | 62.4 | 0.79 | 426 | 44985562 | 44985987 | — | |
| #114 (NT_002835.3 1533742-1535201) | Unmethylation | — | 21q22.3 | 73.2 | 0.91 | 460 | 45077687 | 45078146 | 5′-UTR and CDS | Ubiquitin-conjugating enzyme E2G2 (UBE2G2); homologous to yeast UBC7 |
| #115 (NT_002835.3 1549573-1551858) | Unmethylation | — | 21q22.3 | 74.6 | 0.93 | 1286 | 45093518 | 45094803 | 5′-UTR | SMT3 (suppressor of mif two 3, yeast) homolog 1 (SMT3H1) |
| #116 (NT_002835.3 1604933-1607527) | Unmethylation | — | 21q22.3 | 70.5 | 0.97 | 1595 | 45148878 | 45150472 | 5′-UTR and CDS | Pituitary tumor-transforming 1 interacting protein (PTTG1IP) |
| #117 (NT_002835.3 1664660-1666157) | Unmethylation | — | 21q22.3 | 78.9 | 0.74 | 498 | 45208605 | 45209102 | Intron | Prediction gene 54 (PRED54) |
| #118 (NT_002835.3 1671866-1673671) | Unmethylation | — | 21q22.3 | 70.3 | 1.00 | 806 | 45215811 | 45216616 | 5′-UTR | Chromosome 21 open reading frame 67 (C21orf67) |
| #119 (NT_002835.3 1680257-1681681) | Complete methylation | + | 21q22.3 | 65.8 | 0.90 | 425 | 45224202 | 45224626 | Intron | Prediction gene 55 (PRED55) |
| #120 (NT_002835.3 1710450-1712127) | Complete methylation | + | 21q22.3 | 72.7 | 1.07 | 678 | 45254395 | 45255072 | — | |
| #121 (NT_002835.3 1750441-1752578) | Unmethylation | — | 21q22.3 | 75.4 | 0.81 | 1138 | 45294386 | 45295523 | — | |
| #122 (NT_002835.3 1806394-1808604) | Unmethylation | — | 21q22.3 | 77.5 | 1.03 | 1211 | 45350339 | 45351549 | 5′-UTR | Double-stranded RNA-specific adenosine deaminase 2 (ADAR2) |
| #123 (NT_002835.3 1865795-1867783) | Complete methylation | + | 21q22.3 | 67.2 | 0.85 | 989 | 45409731 | 45410719 | 5′-UTR and CDS | Double-stranded RNA-specific adenosine deaminase 2 (ADAR2) |
| #124 (NT_002835.3 2020037-2021768) | Unmethylation | — | 21q22.3 | 75.5 | 0.79 | 732 | 45563914 | 45564645 | KIAA0958; chromosome 21 open reading frame 80 (C21orf80) | |
| #125 (NT_002835.3 2077997-2079517) | Complete methylation | + | 21q22.3 | 54.7 | 0.77 | 521 | 45621843 | 45622363 | Intron | Prediction gene 59 (PRED59) |
| #126 (NT_002835.3 2112166-2114090) | Complete methylation | + | 21q22.3 | 63.8 | 1.15 | 925 | 45656012 | 45656936 | — | |
| #127 (NT_002835.3 2136861-2139459) | Unmethylation | — | 21q22.3 | 79.1 | 0.88 | 1599 | 45680707 | 45682305 | 5′-UTR and CDS | Collagen type XVIII alpha 1 (COL18A1) |
| #128 (NT_002835.3 2274532-2276182) | Incomplete methylation | — | 21q22.3 | 77.8 | 0.84 | 651 | 45818349 | 45818999 | 5′-UTR (exons 1a, 1c, and 1b) | Reduced folate carrier (RFC1; SLC19A1) |
| #129 (NT_002835.3 2286418-2288635) | Complete methylation | + | 21q22.3 | 75.0 | 0.79 | 1218 | 45830961 | 45832178 | — | |
| #130 (NT_002835.3 2322402-2323845) | Composite methylation | — | 21q22.3 | 72.5 | 0.78 | 444 | 45866945 | 45867388 | — | |
| #131 (NT_002835.3 2364595-2366958) | Complete methylation | + | 21q22.3 | 66.7 | 0.98 | 1364 | 45909214 | 45910577 | — | |
| #132 (NT_002835.3 2371534-2374424) | Unmethylation | — | 21q22.3 | 76.3 | 0.89 | 1891 | 45918451 | 45920341 | — | |
| #133 (NT_002835.32475023-2477222) | Complete methylation | + | 21q22.3 | 57.9 | 1.15 | 1200 | 46021940 | 46023139 | — | |
| #134 (NT_002835.3 2653007-2655508) | Complete methylation | + | 21q22.3 | 54.8 | 1.25 | 1502 | 46199895 | 46201396 | Intron | Poly (rC)-binding protein 3 (PCBP3) |
| #135 (NT_002835.3 2716963-2718791) | Complete methylation | + | 21q22.3 | 72.0 | 0.79 | 829 | 46263850 | 46264678 | CDS | Alpha-1 collagen type VI (COL6A1) |
| #136 (NT_002835.3 2732562-2734239) | Complete methylation | — | 21q22.3 | 67.5 | 0.84 | 678 | 46279449 | 46280126 | CDS | Alpha-1 collagen type VI (COL6A1) |
| #137 (NT_002835.3 2735023-2736682) | Complete methylation | + | 21q22.3 | 76.8 | 0.73 | 660 | 46281910 | 46282569 | — | |
| #138 (NT_002835.3 2827157-2828726) | Unmethylation | — | 21q22.3 | 79.6 | 0.75 | 570 | 46374044 | 46374613 | 5′-UTR | Alpha-2 collagen type VI (COL6A2) |
| #139 (NT_002835.3 2841396-2842933) | Complete methylation | — | 21q22.3 | 68.5 | 0.81 | 538 | 46388283 | 46388820 | CDS | Alpha-2 collagen type VI (COL6A2) |
| #140 (NT_002835.3 2855101-2856552) | Complete methylation | — | 21q22.3 | 63.7 | 0.91 | 452 | 46401988 | 46402439 | CDS | Alpha-2 collagen type VI (COL6A2) |
| #141 (NT_002835.3 2861362-2862966) | Complete methylation | — | 21q22.3 | 69.4 | 0.86 | 605 | 46408249 | 46408854 | CDS | Alpha-2 collagen type VI-a′ |
| #142 (NT_002835.3 2890249-2891697) | Composite methylation | — | 21q22.3 | 70.8 | 0.77 | 449 | 46437137 | 46437585 | CDS and 3′-UTR | Similarity to Mus musculus adult male testis cDNA |
| #143 (NT_002835.3 2957438-2959877) | Unmethylation | — | 21q22.3 | 73.6 | 0.75 | 1576 | 46504326 | 46505901 | 5′-UTR and CDS | Lanosterol synthase (2,3-oxidosqualene-lanosterolcyclase) (LSS) |
| #144 (NT_002835.3 3014936-3016783) | Unmethylation | — | 21q22.3 | 69.5 | 0.95 | 848 | 46561824 | 46562671 | 5′-UTR and CDS | Minichromosome maintenance deficient 3 (S. cerevisiae)- associated protein (MCM3AP) |
| #145 (NT_002835.3 3052859-3055543) | Unmethylation | — | 21q22.3 | 71.7 | 0.88 | 1955 | 46599747 | 46601701 | 5′-UTR and CDS | Pericentrin 2 (kendrin) (PCNT2) |
| #146 (NT_002835.3 3132299-3134479) | Complete methylation | + | 21q22.3 | 58.0 | 1.07 | 1181 | 46679187 | 46680367 | Intron | Pericentrin, kendrin |
| #147 (NT_002835.3 3187879-3189962) | Unmethylation | + | 21q22.3 | 75.3 | 0.77 | 1280 | 46734767 | 46736046 | — | |
| #148 (NT_002835.3 3364581-3366420) | Unmethylation | — | 21q22.3 | 70.8 | 0.83 | 840 | 46911470 | 46912309 | 5′-UTR | HMT 1 (hnRNP methyltransferase, S. cerevisiae)-like 1 |
| #149 (NT_002835.3 3396443-3398312) | Unmethylation | — | 21q22.3 | 67.3 | 0.81 | 934 | 46943268 | 46944201 | — | |
The name in parenthesis for each CpG island indicates its symbol in URL (http://hgp.gsc.riken.go.jp/CGI/)
The plus (+) and minus (–) indicate the presence and absence of tandem repeat sequences, respectively
The GC % indicates the GC-content of each CpG island
The Obs/exp indicates the ratio of observed and expected CpG frequency
The start and end point of each CpG island are shown using nucleotide positions in the sequence of human chromosome 21q in UCSC genome browser (http://genome.ucsc.edu/)
The 5′-UTR (5′-untranslated region), CDS (coding region) and 3′-UTR (3′-untranslated region) indicate the location of the CpG island in its flanking gene
For these CGIs, we designed PCR primers using a free program called prima. We selected each primer pair so that the amplicon keeps its GC content as low as possible and contains more than two recognition sites for HpaII or HhaI, thereby avoiding the difficulty in amplification and minimizing the effect of incomplete digestion, and at least one site that would be recognized by McrBC when methylated. The program designed 101 primer pairs that work in amplification, and 47 other pairs were designed manually (Supplemental Table S1 available online at www.genome.org). Unfortunately, we failed to find any suitable amplicons containing methylation-sensitive enzyme sites for a particular CGI, which we analyzed directly by the bisulfite genomic sequencing method.
Methylation Status of 149 CGIs on Human Chromosome 21q
Using genomic DNAs isolated from peripheral blood leukocytes (PBLs) donated by four healthy individuals, we analyzed the 149 CGIs (i.e., 148 by HpaII-McrBC PCR and one by bisulfite genomic sequencing). Consequently, 31, 103, 8, and 7 CGIs were found to display full, null, incomplete, and composite methylation patterns, respectively (Fig. 2; Tables 1 and 2).
Figure 2.
Examples of HpaII-McrBC PCR assays for CpG islands on human Chromosome 21. The results of self-Harr plot and HpaII-McrBC PCR were depicted for (A) a completely methylated CGI #123, (B) an unmethylated CGI #114, (C) a compositely methylated CGI #112, and (D) an incompletely methylated CGI #106. In the self-Harr-plot, each CGI is indicated asan open square and the regionsflanking the island (500 bp on both sides) are shaded. Oblique lines running in parallel with the diagonal line indicate the tandem repeat sequence. The PCR products from mock-treated (lane 1), HpaII-digested (lane 2), MspI-digested (lane 3), and McrBC-digested DNAs (lane 4) were electrophoresed, stained with ethidium bromide, and visualized by UV illumination. The part where the diagonal line isnot shown indicatesthe position of interspersed repeats, which were masked by RepeatMasker.
Table 2.
Methylation Status and Characteristics of the 149 CGIs Analyzed
| Complete methylation | 31 | |
| Promoter or 5′-UTR | 5 | |
| CDS or 3′-UTR | 14 | |
| Tandem repeat sequence | 18 | |
| Both tandem repeat sequence and Promoter or 5′-UTR | 1 | |
| Both tandem repeat sequence and CDS or 3′-UTR | 5 | |
| Null methylation | 103 | |
| Promoter or 5′-UTR | 78 | |
| CDS or 3′-UTR | 2 | |
| Tandem repeat sequence | 4 | |
| Both tandem repeat sequence and CDS or 3′-UTR | 0 | |
| Composite methylation | 7 | |
| Promoter or 5′-UTR | 2 | |
| CDS or 3′-UTR | 2 | |
| Tandem repeat sequence | 2 | |
| Both tandem repeat sequence and CDS or 3′-UTR | 1 | |
| Incomplete methylation | 8 | |
| Promoter or 5′-UTR | 8 | |
| CDS or 3′-UTR | 0 | |
| Tandem repeat sequence | 0 | |
| Both tandem repeat sequence and CDS or 3′-UTR |
Each gene-associated CGI was classified by its location in the gene, namely, promoter, 5′-untranslated region (UTR), coding sequence (CDS), and 3′-UTR. The presence of tandem repeat sequence examined by self-Herr plot is also indicated.
Although the highest incidence of unmethylated pattern was consistent with conventional observations on CGIs, a considerable incidence (31/149; ∼21%) of fully methylated ones was rather unexpected. Of the 31 CGIs displaying the complete methylation pattern, 14 overlap with the coding sequence (CDS) or 3′-untranslated regions (UTRs). Of the 14 CGIs, seven are entirely included within exons so that GC-rich codon sequences seem to contribute to fulfill the requirements for CGIs, whereas the others have only partial overlap with exons and nonexonic regions with CGI-like base compositions. We also found that 18 bear tandem repeat sequences. Although previous studies pointed out that CG-rich tandem repeat sequences often associate with DMRs and are subject to monoallelic methylation, the results indicate that they are rather methylated on both alleles (Table 1). These CGIs tend to be excluded from the vicinity of promoters and 5′-UTRs and may represent an unconventional class of CGIs, although their distribution is, similar to that of nonmethylated or conventional ones, biased toward the subtelomeric, gene-rich region (Hattori et al. 2000).
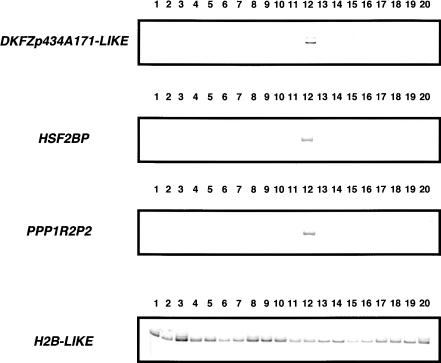
Our analysis revealed five methylated CGIs associated with promoters or 5′-UTRs of genes. We thus examined the expression of these genes by RT-PCR (Fig. 3). PPP1R2P2 (protein phosphatase 1 regulatory inhibitor subunit 2 pseudogene 2) and HSF2BP (heat-shock transcription factor 2 binding protein) were expressed in testis but not in PBLs, in which their CGIs are methylated. H2B-LIKE (similar to H2B histone family member S) was expressed ubiquitously including PBLs, and the CGI #92 linked to this gene includes not only its 5′-UTR but also its CDS and 3′-UTR. ADAR2 (double-stranded RNA-specific adenosine deaminase) was previously reported to be expressed ubiquitously (Chen et al. 2000). Whereas the CGI #123 associated with ADAR2 spans its second exon and is methylated, the other CGI of this gene corresponding to the first exon (i.e., CGI #122) escapes methylation (Table 1). Thus, methylation of CGI #123 and CGII #92 does not affect expression of these genes in PBLs. The DKFZp434A171-LIKE gene was expressed in testis but not in PBLs.
Figure 3.
Expression of genes methylated in their promoters or 5′-UTRs. RT-PCR was performed using total RNA from various human tissues (1, bone marrow; 2, adrenal gland; 3, thymus; 4, prostate; 5, trachea; 6, thyroid; 7, spleen; 8, small intestine; 9, colon; 10, uterus; 11, placenta; 12, testis; 13, heart; 14, skeletal muscle; 15, liver; 16, lung; 17, kidney; 18, whole brain; 19, salivary gland; 20, peripheral blood cells). The products were resolved by polyacrylamide gel electrophoresis, stained with SYBR Green, and visualized by UV illumination.
The HpaII-McrBC screening revealed 14 CGIs showing the composite methylation pattern, although some of them displayed uneven amplification from HpaII- and McrBC-digested DNAs. We further analyzed the 14 CGIs using the bisulfite genomic sequencing method. Treatment of denatured DNA with sodium bisulfite leads to the conversion of unmethylated cytosine, but not 5-methyl cytosine, to uracil. Following PCR amplification of each CGI from bisulfite-treated genomic DNA, the products were cloned and individually sequenced (Supplemental Fig. S1). The analysis revealed that clones for six out of the 14 CGIs were composed of two distinct classes: one totally lacking cytosine and the other maintaining a considerable fraction of CpG dinucleotides. The remaining eight CGIs showed complete, null, or incomplete methylation patterns, presumably because of incomplete digestion by either or both enzymes leading to an apparent composite methylation pattern in HpaII-McrBC PCR. Nevertheless, the results demonstrate that the HpaII-McrBC PCR can effectively enrich CGIs composed of methylated and unmethylated alleles.
In addition to the six CGIs described above, we analyzed CGI #103, which lacks any appropriate enzyme sites for HpaII-McrBC PCR, directly by the bisulfite sequencing to find that it consists of both methylated and unmethylated alleles (Supplemental Fig. S1). We thus identified seven CGIs with composite methylation patterns. Two of these CGIs (#55 and #74) are found in 5′-UTRs, two (#103 and #142) in CDS, and two (#59 and #112) in introns, whereas the remaining one (#130) is not included in any gene. Tandem repeats were found in two CGIs, namely, #103 and #112(Table 1).
Identification of Three CGIs Subject to Allele-Specific Methylation
Next we intended to examine whether or not the seven CGIs identified as above are methylated in an allele-specific manner. For this purpose, nucleotide sequence polymorphisms have to be identified. We found single-nucleotide polymorphisms (SNPs) for four of the seven CGIs, but failed to find any for the other three.
We analyzed individuals heterozygous for these SNPs by directly sequencing the HpaII-McrBC PCR products: Amplification product from HpaII- or McrBC-digested DNAs represents methylated or unmethylated allele, respectively. For CGI #142, both alleles were detected from either HpaII- or McrBC-digested DNA. This CGI may be completely methylated in some cells but unmethylated in other cells. Alternatively, the CGI is subject to random monoallelic methylation. The other three CGIs, namely, #59, #112, and #130 (Fig. 4A), were found methylated in an allele-specific manner as described in detail below.
Figure 4.
Monoallelically methylated CGIs and their nearest neighbor genes. (A) The positions of CGI #112, #59, and #130 are depicted with their nearest neighbor genes. (B) The allelic expression status of DSCR3 wasexamined by direct sequencing of RT-PCR productsderived from PBLsof an A/T heterozygote. (C) An RT-PCR-RFLP assay was developed for SLC19A1 using HhaI, and applied to cDNAsfrom PBLsof three individuals(left, A/G heterozygote; middle, A/A homozygote; right, G/G homozygote). Note that the primers used in this assay simultaneously amplify two splicing isoforms, which are detected astwo distinct bandsin the gel.
Maternal Allele-Specific Methylation of CGI #112
For the CGI #112, a fragment spanning two SNP sites was PCR-amplified using genomic DNA isolated from PBLs or placental tissue. Direct sequencing of the PCR products from 40 Japanese individuals revealed 20 A/T and 12 C/T heterozygotes for SNP1 (dbSNP ID TSC0115741) and SNP2(dbSNP ID TSC0115740), respectively. As seven individuals were found heterozygous for both SNP1 and SNP2, 25 out of 40 Japanese examined were informative for allelic methylation studies. We analyzed nine out of the 25 individuals heterozygous for either A/T or C/T SNPs by PCR from HhaI- or McrBC-digested DNAs from either PBLs or placenta followed by direct sequencing. In all cases, only a single allele was methylated.
We thus examined six informative pedigrees to reveal the parental origin of the methylated allele (Supplemental Table S2). An example using PBL DNA is shown in Figure 5A. The progeny in this pedigree is a C/T heterozygote, whose father and mother are a C/T heterozygote and a C/C homozygote, respectively. Thus, the progeny bears a paternally transmitted T allele and a maternally derived C allele. HhaI digestion prior to PCR, which cuts the unmethylated allele, eliminated the paternal T peak from the electropherogram, and McrBC digestion, eliminating the methylated allele, resulted in the amplification of the paternal T allele. These results clearly indicated that the methylated allele of this CGI is transmitted from the maternal lineage. Maternal allele-specific methylation of this CGI was also demonstrated in five other cases using placental DNA (Supplemental Table S2). We thus concluded that CGI #112 is maternally methylated in both PBLs and placenta.
Figure 5.
Maternal allele-specific methylation pinpointed to tandem repeats. (A) Maternal allele-specific methylation of CGI #112. A map of CGI #112 (500 bp) isshown on the top with the positionsof A/T and C/T SNPs(i.e., SNP1 and SNP2). The arrowsin the map indicate the tandem repeats. The CGI wasPCR-amplified from untreated (bottom left), HhaI-digested (bottom center), and McrBC-digested (bottom right) genomic DNA isolated from PBLs of a C/T heterozygote at SNP2. The amplified productswere subjected to direct sequencing. The vertical arrowhead in each electropherogram denotes the SNP2 (C/T) sites. (B) Bisulfite genomic sequencing of CGI #112. Each row of circles corresponds to each clone of bisulfite PCR products. Open and closed circles stand for unmethylated and methylated C residues, respectively. The SNP1 (A/T) site is indicated by the arrowhead.
We further analyzed the methylation of CGI #112 using the bisulfite genomic sequencing method. The result using PBL DNA from an A/T heterozygote of SNP1 is depicted in Figure 5B. The 12 clones sequenced were composed of six bearing a maternal T allele and six with a paternal A allele. Intriguingly, the maternal allele-specific methylation occurred mainly in the tandem repeat sequence, which is composed of five 40-bp units mutually showing 82.5% identity. This DMR would thus serve as an interesting model to pursue the relation between tandem repeat sequence and allele-specific methylation.
Mosaicism in Allelic Methylation Status of CGI #59
We used an A/C SNP (dbSNP ID TSC0066520) for the analysis of CGI #59. We identified nine A/C heterozygotes and 15 A/A homozygotes from the 24 Japanese individuals, and analyzed five of the nine heterozygotes by sequencing HpaII-McrBC PCR products from PBLs or placental tissues. Monoallelic methylation was demonstrated in all of the five cases: Only the A allele was methylated in four cases, whereas the C allele was methylated in the other case.
To reveal the parental origin of allele-specific methylation, we examined three informative pedigrees (Supplemental Table S2), an example of which is shown in Figure 6A. As the father is an A/C heterozygote and the mother is an A/A homozygote, the progeny has a maternal A allele and a paternal C allele. The PCR product from HhaI-digested PBL DNA, leaving the methylated allele, contained only the maternal A allele (Fig. 6A). However, amplified product from McrBC-digested DNA reproducibly displayed an A/C doublet peak (Fig. 6A). These results indicate that the maternal A allele is methylated only in a fraction of PBLs but unmethylated in the other fraction, whereas the paternal C allele escapes methylation in all cells. This interpretation was further supported by bisulfite sequencing: All clones derived from the paternal C allele showed unmethylated pattern throughout the island, but those from the maternal A allele were divided into two groups, one almost completely methylated and the other totally escaping methylation (Fig. 6B).
Figure 6.
Mosaicism in maternal allele-specific methylation. (A) Maternal allele-specific methylation of CGI #59. Direct sequencing was performed using the PCR products from mock-treated (bottom left), HhaI- (bottom center), and McrBC-digested (bottom right) DNA isolated from PBLs. The arrowhead indicatesthe A/C SNP site. Note that the maternally inherited A allele wasdetected from both Hha I-treated (or methylated) DNA and McrBC-treated (or unmethylated) DNA. (B) Bisulfite genomic sequencing of CGI #59. Each row of circles corresponds to each clone of bisulfite PCR products. Open and closed circles stand for unmethylated and methylated C residues, respectively. The A/C SNP site is also indicated. Note that the clonesfor the maternally inherited A allele are composed of two populations, one completely methylated and the other completely escaping methylation.
We next analyzed two other pedigrees using placental DNA of the progenies (data not shown). In contrast with the results using PBLs, both progenies showed an A/C doublet peak from HhaI-digested DNA (or methylated allele) and a paternal C peak from McrBC-digested DNA (or unmethylated allele). Consistent with these results, bisulfite sequencing revealed that all clones for the maternal A allele displayed a methylated pattern, whereas those for the paternal C allele were composed of completely methylated and unmethylated ones.
Taken together, CGI #59 is either maternally methylated or biallelically unmethylated in PBLs, but is subject to either maternal methylation or biallelic methylation in placenta. It may be intriguing to note that CGI #59 escapes methylation in PBLs rather than placenta, because the latter tissue has been known to show lower overall methylation than other tissues. Thus, maternal allele-specific methylation of this CGI conceivably occurs in a cell-type-specific manner.
Because CGI #59 is in the first intron of DSCR3 (Fig. 4A), we examined its allelic expression using PBLs from heterozygotes. As shown in Figure 4B, we failed to find evidence for apparent allele-specific expression in PBLs.
Allele-Specific, Parental-Origin-Independent Methylation of CGI #130
We revealed a C/G SNP in CGI #130, and identified 37 C/G heterozygotes, 23 G/G homozygotes, and seven C/C homozygotes. Of the 37 heterozygotes, 14 were analyzed by direct sequencing of HpaII-McrBC PCR products. Strikingly, all of the examined individuals contained a methylated C allele (i.e., one PBL and 13 placental tissues). Eight samples (i.e., one PBL and seven placenta) showed a single peak for C or G at the SNP site from HhaI- or McrBC-digested DNAs, respectively. However, the other six placental DNA samples displayed a C/G doublet peak from the HhaI-digested samples but only a G peak from the McrBC-digested ones. These placental tissues seem to contain a fraction of cells bearing biallelically methylated CGI #130, in addition to the cells in which the CGI is monoallelically methylated.
To reveal the parental origin of allele-specific methylation, we analyzed 11 informative pedigrees by direct sequencing of HpaII-McrBC PCR products (Supplemental Table S2), two examples of which were shown in Figure 7, A and B. The progeny in Figure 7A is a C/G heterozygote with a methylated C allele and an unmethylated G allele. We genotyped the parents and identified the father and the mother as a C/G heterozygote and a G/G homozygote, respectively. Thus, the methylated C allele was paternally inherited in this case. In contrast, the pedigree shown in Figure 7B was composed of a G/G-homozygous father, a C/G-heterozygous mother, and C/G-heterozygous progeny, whose maternally transmitted C allele is methylated. In total, we found that four and seven of the 11 heterozygotes inherited the methylated C allele from paternal and maternal lineages, respectively (Supplemental Table S2). Thus, in C/G-heterozygous individuals, this CGI is methylated in a C-allele-specific manner regardless of its parental origin.
Figure 7.
Allele-specific, parental-origin-independent methylation of CGI #130. Direct sequencing was performed using the PCR products from mock-treated (bottom left), HhaI- (bottom center), and McrBC-digested (bottom right) DNA. In the pedigree shown in A, the paternally inherited C allele ismethylated. On the other hand, the maternally inherited C allele ismethylated in the pedigree shown in B.
We next wondered whether this CGI is subject to monoallelic methylation also in G/G or C/C homozygotes. Successful amplification of this CGI from both HhaI-digested and McrBC-digested DNAs strongly indicated that the allele-specific methylation occurs also in G/G and C/C homozygotes (data not shown). This notion was further reinforced by the results of bisulfite sequencing, wherein both completely methylated and unmethylated clones were identified from both G/G and C/C homozygotes.
Based on these findings, we concluded that CGI #130 is methylated in an allele-specific but parental-origin-independent manner. Intrigued by this unique methylation pattern, we examined the allelic expression status of SLC19A1, which presently serves as the nearest neighbor gene of the CGI #130 (Fig. 4A). As shown in Figure 4C, an RT-PCR-RFLP (restriction fragment length polymorphism) assay indicated its biallelic expression in PBLs.
DISCUSSION
HpaII-McrBC PCR for a Large-Scale Methylation Analysis
A large-scale methylation analysis requires a simple method for evaluation of methylation status. Although a PCR method using methylation-sensitive restriction endonucleases such as HpaII (Singer-Sam et al. 1990) is simple enough, it cannot distinguish fully methylated status from coexistence of both methylated and unmethylated copies, which we call composite methylation. On the other hand, various methods using the sodium bisulfite treatment (Kubota et al. 1997; Xiong and Laird 1997; Eads et al. 2000) can detect the composite methylation status. However, they are much more tedious than the simple HpaII-PCR, and hence are not suitable for a large-scale analysis. Furthermore, they inevitably degrade genomic DNA down to fragments of 500-1000 bp long, which can serve only as a poor template for PCR to make it impossible to scan longer distances.
Here we developed a novel HpaII-McrBC PCR method by exploiting two restriction enzymes with complementary methylation sensitivities (Fig. 1). This simple method allows one to easily detect composite methylation by scanning much longer stretches than the methods based on the sodium bisulfite treatment.
One drawback of the method is the occasionally encountered, unpredictable behavior of McrBC. We experienced an unexpected PCR amplification from McrBC-treated genomic DNA, even though the completely methylated island bears enough recognition sites for the enzyme. We cannot explain and circumvent such troubles, until the precise mechanism for McrBC action is understood in the future.
Despite this drawback, the unsurpassed simplicity and speed of the HpaII-McrBC PCR method would make it most suitable for a large-scale methylation analysis. Indeed, the comprehensive analysis discussed below has proved it as an effective screen to reduce the number of samples that have to be subjected to tedious bisulfite sequencing. Notably, the screen is free from false negatives for DMRs, because incomplete digestion by either enzyme classifies the target sequence as a potential candidate DMR, which would be examined further by bisulfite sequencing, but not as fully methylated or unmethylated CGIs (Fig. 1). It is thus ideal for the search of allelic DMRs often associated with imprinted genes.
Comprehensive Methylation Analysis of CGIs on Human Chromosome 21q
Using the newly developed HpaII-McrBC PCR method as an initial screening, we investigated the methylation status of 149 CGIs on human Chromosome 21q, whose complete sequence enabled us to exhaustively identify CGIs under a defined criterion in silico. This analysis thus serves as the first comprehensive methylation analysis encompassing an entire chromosome arm to provide a global view of CGI methylation (Tables 1 and 2).
Although most CGIs (103/149, ∼69%) escape methylation, an unexpectedly high incidence (31/149, ∼21%) was observed for full methylation of CGI even in normal peripheral blood cells (Table 2). These normally methylated CGIs often contain tandem repeat sequences composed of CG-rich units. Although it has been pointed out that such iterated structures are often found around imprinted genes, they are not unique to allelic DMRs of imprinted genes but are more frequently found in normally methylated CGIs (Table 2). One may argue that such repeats should not be included in CGIs. Notably, even removing such repeats from analysis, we observed that a substantial fraction (13/125, ∼10%) of CGIs are methylated. Although one may also argue that the lack of evidence for unmethylation in other tissues or developmental stages disqualifies these sequences as CGIs, we would emphasize that the computationally extracted CGIs contain a substantial fraction of CGI-like sequences that are methylated even in normal tissues.
On the other hand, it should be noted that tandem repeats are not always associated with methylation. We found four sequences that escape methylation and contain tandem repeats (Table 2): CGI #108 are located in the 5′-UTR of Chromosome 21 open reading frame 2(C21orf2), whereas the other three (i.e., CGI #39, #109, and #147) are not linked to any gene.
Consistent with our findings, a genome-wide screen using an enrichment cloning procedure was reported during the course of our work to reveal 43 CGIs methylated in normal somatic tissues (Strichman-Almashanu et al. 2002). Because our comprehensive analysis revealed 31 normally methylated CGIs on Chromosome 21q comprising ∼1.2% of the human genome, our genome likely bears >3000 normally methylated CGIs. Relaxation of the criteria for CGI is expected to further increase the number of such CGIs, because shorter CGIs tend to be more often methylated (Strichman-Almashanu et al. 2002). In this context, it is intriguing to note that we used slightly relaxed criteria for CGI than we did in the initial sequence analysis to include additional 12CGIs, which were found to comprise six completely methylated, three unmethylated, two compositely methylated, and one incompletely methylated CGIs.
It is also intriguing to examine the methylation status of these normally methylated CGIs in other tissues in both physiological and pathological conditions, including Down syndrome and various cancers, in which aberrant copy number of this chromosome was demonstrated (Kafri et al. 1992; Kuromitsu et al. 1997; Stephen et al. 2001). Provided with appropriate DNA samples, our system is readily applicable to such studies, which would shed light on the roles for methylation in cellular physiology and pathology.
Allelically Methylated CGIs on Chromosome 21q
Our comprehensive analysis uncovered three CGIs subject to allele-specific methylation, and they may well accompany genes expressed in an allele-specific manner. We thus analyzed the allelic expression status of their nearest neighbor genes (Fig. 4). Although we have not yet obtained an informative sample for C21orf29 because of its testis-specific expression, we successfully examined the allelic expression of DSCR3 and SLC19A1 in PBLs but failed to obtain any evidence for their monoallelic expression (Fig. 4). Allelic expression status of these genes in other tissues would be worth further pursuit. Notably, it becomes increasingly evident that mammalian cells express a larger number of noncoding RNA species than previously expected. Furthermore, monoallelic expression has been demonstrated for such noncoding RNAs derived from various imprinted regions. It is thus conceivable that genes for such noncoding RNAs remain uncovered in the vicinity of these CGIs.
The detailed methylation analysis of CGI #112, one of the maternally methylated CGIs, has revealed a unique pattern of methylation enriched around the tandem repeat sequence (Fig. 5). Because a coincidence has been observed between allelic methylation and tandemly iterated structure, it may provide an interesting example to study their mechanistic relationship.
The analysis of the other maternally methylated CGI, namely, CGI #59, reveals a mosaicism in its allelic methylation. PBLs can be divided into two populations, one in which the CGI is maternally methylated and the other in which it fully escapes methylation (Fig. 6). In contrast, placental tissue is composed of two cell populations, one maternally methylating this CGI and the other methylating it biallelically. It remains to be elucidated whether or not the mosaicism corresponds to cell types and is of physiological significance. It is also interesting to examine the mosaic pattern in other tissues under both physiological and pathological states.
Finally, detailed analysis of the remaining monoallelically methylated CGI termed CGI #130 provided an interesting case for allelic methylation: Its methylation is restricted to a particular allele called the C allele independently of its parental origin. Our analysis of C/G heterozygotes for this SNP clearly demonstrated that some bear a maternally transmitted methylated C allele, whereas others have a paternally derived methylated C allele (Fig. 7). Intriguingly, this CGI is monoallelically methylated even in individuals who are homozygous for a C or G allele. These findings indicate that a particular allele is dominant over the others in its susceptibility to methylation. To the best of our knowledge, this represents a previously unknown mode for allele-specific methylation. The molecular mechanism and biological significance of this phenomenon are of particular interest. Such pursuit would be greatly enhanced by the identification of similar CGIs in more experimentally tractable animals like mouse.
Allele-specific methylation has been investigated mainly using allelic DMRs found around the established imprinted genes, in which differential methylation is dependent on its parental origin, often spans a long range, and is regardless of the expression of adjacent imprinted gene. In contrast, our analysis revealed an example for more pinpointed allele-specific methylation (Fig. 5), variable allelic methylation in cell populations (Fig. 6), and allele-specific parental-origin-independent methylation (Fig. 7).
It is thus likely that the ways for human cells to modify their genomes by allele-specific methylation have more variations than previously expected. To fully uncover the methylation repertoire and its biology, our approach would be powerful, in particular, in the coming age of postgenomic sequence with a wealth of SNPs.
METHODS
In Silico Extraction of CGIs From Human Chromosome 21q Sequence
The CGIs to be analyzed were computationally identified in the human Chromosome 21q sequence with different parameter sets. The parameters used were minimal length 200, 300, 400, and 500 bp; minimal GC content, 0.5 and 0.55; and an expected CpG frequency (ECF), >0.6, where ECF = (the number of CpGs × length of the sequence)/(the number of Cs × the number of Gs). With each of the eight possible parameter sets, we identified CGIs and compared the results with known CGIs. This revealed that the parameter set with minimal length >400, minimal GC content >0.5, and ECF >0.6, is the best, and we decided to use the CGIs that were identified with this parameter set. Because some of the highly repetitive sequences such as Alu and LINE-1 elements contain regions that fulfill the above criteria, these elements were masked using the software RepeatMasker (http://repeatmasker.genome.washington.edu) prior to the identification of CGIs. The sequences of 149 CGIs identified with parameters, minimal length >400, minimal GC content >0.5, and ECF >0.6, can be seen at http://hgp.gsc.riken.go.jp/CGI/.
Primer Design for the CGIs
A free-ware program for primer extraction, prima, was downloaded from http://www.uk.embnet.org/Software/EMBOSS/, and used for designing PCR primers from the extracted CGIs under the following parameters: targetstart 500, targetend 800, minprimertm 53, maxprimertm 63, minprodlen 400, maxprodlen 1000, minpmgccont 40, maxpmgccont 55, minprodgccont X1, maxprodgccont X2, minprimerlen 23, and maxprimerlen 25, where [X1, X2] were [40, 55], [55, 60], [60, 65], or [65, 70]. If the program fails to extract any primer sequences from an island under all conditions, the complementary sequence prepared by a complementary program was subjected to the program.
Note that the presence of HpaII or HhaI sites in the primer sites may bias amplification. A problem would occur in the case of methylated CGIs with unmethylated primer sites, because digestion of the priming site leads to no amplification from HpaII- or HhaI-digested DNA as well as from McrBC-digested DNA and hence the CGI is judged as incomplete methylation. To avoid this, CGIs showing incomplete methylation by primers bearing HpaII or HhaI sites should be re-examined by bisulfite sequencing. In this study, only the CGI #88 has a possibility to be mis-classified, and we confirmed its incomplete methylation by bisulfite sequencing.
Preparation of Genomic DNA From Human Peripheral Blood Leukocytes and Placental Tissues
Normal human lymphocytes were prepared from peripheral blood using Lymphoprep (DAIICHI PURE CHEMICALS). Human placentas were obtained, with informed consent, from the Department of Obstetrics and Gynecology, Saga University Hospital, Saga, Japan. These tissues were derived from 7.4 to 39 wk after conception. To eliminate the contamination of maternal decidua, a sample of placental tissue, as thin as possible, was excised from fetal surface and washed in a series of chilled normal saline solutions, then frozen immediately. Genomic DNAs from human lymphocytes and placental tissues were extracted by standard methods.
HpaII-McrBC PCR Assay
Human genomic DNA (0.5 μg) was digested with 30 units of HpaII, HhaI, MspI (TaKaRa), or McrBC (New England Biolabs) overnight at 37°C in 50 μL of the buffers recommended by the suppliers. Following the addition of 50 μL of 5 M NH4OAc, digested DNAs were recovered by ethanol precipitation and dissolved in 10 μL of TE (10 mM Tris-HCl at pH 8.0 and 1 mM EDTA).
For PCR, 1.0 μL (50 ng) of genomic DNA digested with each enzyme was used in a 10-μL reaction mixture containing 2.5 U of Ex-Taq DNA polymerase (TaKaRa) and 2.5 pmoles of each primer in PCR buffer (10 mM Tris-HCl at pH 7.5, 50 mM KCl, 1.5 mM MgCl2, 1 mM DTT, 10 mM 2-mercaptoethanol, and 0.2 mM of each dNTP). For some amplicons, betaine (Nacalai Tesque), dimethyl sulfoxide (Sigma), or PCR enhancer (Invitrogen) was added to the reaction mixture to improve amplification (Baskaran et al. 1996). The thermal cycling parameter was optimized for each amplicon. See Supplemental Table S1 for detailed conditions for each PCR including primer sequences.
The amplified products were electrophoresed on a 1%-2% agarose gel, stained with ethidium bromide, and visualized by UV illumination.
Identification of SNPs in CGIs
Three CGIs (#112, #130, and #59) were PCR-amplified from Japanese individuals and directly sequenced using the sense or antisense primer used in PCR under the following thermal cycling: 1 min at 96°C + (10 sec at 96°C + 5 sec at 55°C + 90 sec at 60°C) times 25 cycles using the Big dye cycle sequencing Kit (Applied Biosystems). The following primers were used: CGI #112, 5′-AAGAGAAGCTCGCCTCGCTTCTA-3′, 5′-AAACATGCACCGGC AAAACCAAG-3′; CGI #130, 5′-GCGCCCGGCTTGAAATTTAGG AAA-3′, 5′-GGTTTGTGCATAGTGTGCATGGTT-3′; and CGI #59, 5′-GTCCGGCAGCAGCACCGATTG-3′, 5′-CCCTCTCTTAGGCC CGAAACCTGC-3′. Obtained sequence data were analyzed by an analysis software package, SEQUENCHER (Gene Codes).
Identification of Parental-Origin-Specific Methylation by Direct Sequencing of HpaII-McrBC PCR Products
Genomic DNA (500 ng) from human peripheral blood leukocytes or placental tissues was digested with 30 units of HpaII, HhaI, or McrBC overnight in 50 μL of the recommended buffer for each enzyme. For PCR, 50 ng of digested DNAs was used in a 10-μL reaction volume under the conditions described in Supplemental Table S1, and the PCR products obtained were subjected to direct cycle sequencing to reveal allelic identity.
Bisulfite Genomic Sequencing
Human genomic DNA (1-10 μg) from peripheral blood leukocytes were treated with sodium bisulfite according to the standard procedure (Clark et al. 1994). One-tenth of the bisulfite-treated DNA was used for PCR in a 10-μL reaction mixture (10 mM Tris-HCl at pH 7.5, 50 mM KCl, 1.5 mM MgCl2, 1 mM DTT, 10 mM 2-mercaptoethanol, 0.2 mM of each dNTP, 0.25 μM of each primer, and 2.5 U of Ex-Taq DNA polymerase [TaKaRa]). Other detailed conditions including primer sequences are described in Supplemental Table S1. The amplified products were subsequently cloned into pT7Blue vector (Novagen) and sequenced.
RT-PCR
For expression analysis of PPP1R2P2, HSF2BP, H2B-like, and DKFZp434A171-like, total RNA (2.5 μg) from various human tissues (Clontech) was reverse-transcribed and used as templates for PCR using the following thermal cycling parameters: 3 min at 95°C + (30 sec at 95°C + 30 sec at 65°C + 30 sec at 72°C) times 30 cycles. The primers used were as follows: PPP1R2P2, 5′-AT CAAGGAGAACCTCAAGAACAACTT-3′, and 5′-CGAATTTCTTA GCTAAGATATCTCGTT-3′; HSF2BP, 5′-CTGGCTGGAATTGT CACGAATGTTG-3′, and 5′-GGCCGACTTGGAGAAGACTTCAG-3′; H2B-like, 5′-GAGCTACTCCGTATACGTGTACAAG-3′, and 5′-GTGATGGTCGAGCGCTTGTTGTA-3′. DKFZp434A171-like, 5′-GCCTTGTGGATCTTCTGCAGTTC-3′, and 5′-GGCTGCGAGTGT CGTTGCTGAAG-3′. The PCR products were electrophoresed on a 7%-9% polyacrylamide gel, stained with SYBR Green (TaKaRa), and visualized by UV illumination. Note that the products for H2B-like were digested by HhaI prior to electrophoresis so that they can be distinguished from that for H2B.
Allelic Expression Analysis
The RT-PCR products from PBL RNAs were analyzed either by direct sequencing (DSCR3) or HhaI-RFLP (SLC19A1). The primer sequences used for PCR are as follows: DSCR3, 5′-AACCT CCCTGGCTCAAGCGATC-3′ and 5′-AGAGGCAGACCAAATT CATCAAGTC-3′; SLC19A1, 5′-GCGCAAGAGGCGCTGGAGCA TTTC-3′ and 5′-GAGGTAGGGGGTGATGAAGCTC-3′.
Acknowledgments
This work was in part supported by research grants from the Ministry of Education, Culture, Sports, Science and Technology, Japan. Both Y.Y. and F.M. are supported by the Japan Society for Promotion of Science.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1351604.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Antequera, F. and Bird, A. 1993. Number of CpG islands and genes in human and mouse. Proc. Natl. Acad. Sci. 90: 11995-11999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskaran, N., Kandpal, R.P., Bhargava, A.K., Glynn, M.W., Bale, A., and Weissman, S.M. 1996. Uniform amplification of a mixture of deoxyribonucleic acid with varying GC content. Genome Res. 6: 633-638. [DOI] [PubMed] [Google Scholar]
- Chen, C.X., Cho, D.S., Wang, Q., Lai, F., Carter, K.C., and Nishikura, K. 2000. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA 6: 755-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, S.J., Harrison, J., Paul, C.L., and Frommer, M. 1994. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 22: 2990-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eads, C.A., Danenberg, K.D., Kawakami, K., Saltz, L.B., Blake, C., Shibata, D., Danenberg, P.V., and Laird, P.W. 2000. MethyLight: A high-throughput assay to measure DNA methylation. Nucleic Acids Res. 28: e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden, M. and Frommer, M. 1987. CpG islands in vertebrate genomes. J. Mol. Biol. 196: 261-282. [DOI] [PubMed] [Google Scholar]
- Grunau, C., Hindermann, W., and Rosenthal, A. 2000. Large-scale methylation analysis of human genomic DNA reveals tissue-specific differences between the methylation profiles of genes and pseudogenes. Hum. Mol. Genet. 9: 2651-2663. [DOI] [PubMed] [Google Scholar]
- Hagiwara, Y., Hirai, M., Nishiyama, K., Kanazawa, I., Ueda, T., Sakaki, Y., and Ito, T. 1997. Screening for imprinted genes by allelic message display: Identification of a paternally expressed gene Impact on mouse chromosome 18. Proc. Natl. Acad. Sci. 94: 9249-9254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori, M., Fujiyama, A., Taylor, T.D., Watanabe, H., Yada, T., Park, H.S., Toyoda, A., Ishii, K., Totoki, Y., Choi, D.K., et al. 2000. The DNA sequence of human chromosome 21. Nature 405: 311-319. [DOI] [PubMed] [Google Scholar]
- Ioshikhes, I.P. and Zhang, M.Q. 2000. Large-scale human promoter mapping using CpG islands. Nat. Genet. 26: 61-63. [DOI] [PubMed] [Google Scholar]
- Kafri, T., Ariel, M., Brandeis, M., Shemer, R., Urven, L., McCarrey, J., Cedar, H., and Razin, A. 1992. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes & Dev. 6: 705-714. [DOI] [PubMed] [Google Scholar]
- Kubota, T., Das, S., Christian, S.L., Baylin, S.B., Herman, J.G., and Ledbetter, D.H. 1997. Methylation-specific PCR simplifies imprinting analysis. Nat. Genet. 16: 16-17. [DOI] [PubMed] [Google Scholar]
- Kuromitsu, J., Yamashita, H., Kataoka, H., Takahara, T., Muramatsu, M., Sekine, T., Okamoto, N., Furuichi, Y., and Hayashizaki, Y. 1997. A unique downregulation of h2-calponin gene expression in Down syndrome: A possible attenuation mechanism for fetal survival by methylation at the CpG island in the trisomic chromosome 21. Mol. Cell. Biol. 17: 707-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macleod, D., Ali, R.R., and Bird, A. 1998. An alternative promoter in the mouse major histocompatibility complex class II I-Aβ gene: Implications for the origin of CpG islands. Mol. Cell. Biol. 18: 4433-4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morison, I.M. and Reeve, A.E. 1998. A catalogue of imprinted genes and parent-of-origin effects in humans and animals. Hum. Mol. Genet. 7: 1599-1609. [DOI] [PubMed] [Google Scholar]
- Neumann, B., Kubicka, P., and Barlow, D.P. 1995. Characteristics of imprinted genes. Nat. Genet. 9: 12-13. [DOI] [PubMed] [Google Scholar]
- Norris, D.P., Brockdorff, N., and Rastan, S. 1991. Methylation status of CpG-rich islands on active and inactive mouse X chromosomes. Mamm. Genome 1: 78-83. [DOI] [PubMed] [Google Scholar]
- Okamura, K., Hagiwara-Takeuchi, Y., Li, T., Vu, T.H., Hirai, M., Hattori, M., Sakaki, Y., Hoffman, A.R., and Ito, T. 2000. Comparative genome analysis of the mouse imprinted gene Impact and its nonimprinted human homolog IMPACT: Toward the structural basis for species-specific imprinting. Genome Res. 10: 1878-1889. [DOI] [PubMed] [Google Scholar]
- Plass, C., Shibata, H., Kalcheva, I., Mullins, L., Kotelevtseva, N., Mullins, J., Kato, R., Sasaki, H., Hirotsune, S., Okazaki, Y., et al. 1996. Identification of Grf1 on mouse chromosome 9 as an imprinted gene by RLGS-M. Nat. Genet. 14: 106-109. [DOI] [PubMed] [Google Scholar]
- Ponger, L., Duret, L., and Mouchiroud, D. 2001. Determinants of CpG islands: Expression in early embryo and isochore structure. Genome Res. 11: 1854-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer-Sam, J., LeBon, J.M., Tanguay, R.L., and Riggs, A.D. 1990. A quantitative HpaII-PCR assay to measure methylation of DNA from a small number of cells. Nucleic Acids Res. 18: 687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen, B.B., Manel, E., Michael, R.R., Kurtis, E.B., Kornel, S., and James, G.H. 2001. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum. Mol. Genet. 10: 687-692. [DOI] [PubMed] [Google Scholar]
- Stewart, F.J. and Raleigh, E.A. 1998. Dependence of McrBC cleavage on distance between recognition elements. Biol. Chem. 379: 611-616. [PubMed] [Google Scholar]
- Strichman-Almashanu, L.Z., Lee, R.S., Onyango, P.O., Perlman, E., Flam, F., Frieman, M.B., and Feinberg, A.P. 2002. A genome-wide screen for normally methylated human CpG islands that can identify novel imprinted genes. Genome Res. 12: 543-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland, E., Coe, L., and Raleigh, E.A. 1992. McrBC: A multi-subunit GTP-dependent restriction endonuclease. J. Mol. Biol. 225: 327-348. [DOI] [PubMed] [Google Scholar]
- Wutz, A., Smrzka, O.W., Schweifer, N., Schellander, K., Wagner, E.F., and Barlow, D.P. 1997. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature 389: 745-749. [DOI] [PubMed] [Google Scholar]
- Xiong, Z. and Laird, P.W. 1997. COBRA: A sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 25: 2532-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, B.J., Herman, H., Sikora, A., Smith, L.T., Plass, C., and Soloway, P.D. 2002. Regulation of DNA methylation of Rasgrf1. Nat. Genet. 30: 92-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://hgp.gsc.riken.go.jp/CGI/; RIKEN.
- http://repeatmasker.genome.washington.edu; RepeatMasker.
- http://www.uk.embnet.org/Software/EMBOSS/; prima software.
- http://genome.ucsc.edu/; UCSC genome browser.