Background: NF-κB regulation of COL1A1 in physiopathological situations is largely unknown.
Results: NF-κB down-regulates COL1A1 in normal and scleroderma fibroblasts, through its recruitment on COL1A1 by protein interactions with the Sp1/Sp3/c-Krox trans-activators, which are different in fibrotic fibroblasts.
Conclusion: Our findings highlight a new mechanism for COL1A1 regulation.
Significance: These data could allow the development of new antifibrotic strategies.
Keywords: Collagen; Fibroblast; Fibrosis; Gene Transcription; Skin; Sp1; Nuclear Factor-κB; Scleroderma; Transcription Factors, Sp3, c-Krox; Type I Collagen
Abstract
Transcriptional mechanisms regulating type I collagen genes expression in physiopathological situations are not completely known. In this study, we have investigated the role of nuclear factor-κB (NF-κB) transcription factor on type I collagen expression in adult normal human (ANF) and scleroderma (SF) fibroblasts. We demonstrated that NF-κB, a master transcription factor playing a major role in immune response/apoptosis, down-regulates COL1A1 expression by a transcriptional control involving the −112/−61 bp sequence. This 51-bp region mediates the action of two zinc fingers, Sp1 (specific protein-1) and Sp3, acting as trans-activators of type I collagen expression in ANF and SF. Knockdown of each one of these trans factors by siRNA confirmed the trans-activating effect of Sp1/Sp3 and the p65 subunit of NF-κB trans-inhibiting effect on COL1A1 expression. Despite no existing κB consensus sequence in the COL1A1 promoter, we found that Sp1/Sp3/c-Krox and NF-κB bind and/or are recruited on the proximal promoter in chromatin immunoprecipitation (ChIP) assays. Attempts to elucidate whether interactions between Sp1/Sp3/c-Krox and p65 are necessary to mediate the NF-κB inhibitory effect on COL1A1 in ANF and SF were carried out; in this regard, immunoprecipitation assays revealed that they interact, and this was validated by re-ChIP. Finally, the knockdown of Sp1/Sp3/c-Krox prevents the p65 inhibitory effect on COL1A1 transcription in ANF, whereas only the siRNAs targeting Sp3 and c-Krox provoked the same effect in SF, suggesting that particular interactions are characteristic of the scleroderma phenotype. In conclusion, our findings highlight a new mechanism for COL1A1 transcriptional regulation by NF-κB, and these data could allow the development of new antifibrotic strategies.
Introduction
The skin is the largest and one of the most important mammalian organs. It serves as a barrier that protects from a wide variety of chemical, microbial, and physical aggressions.
The extracellular matrix of the skin is composed mostly of collagen and elastin. Collagen represents 70–80% of the dry weight of the skin and gives to the dermis its mechanical and structural integrity. Type I collagen, the major isotype of skin dermis, is a heterotrimer consisting of two α1(I) chains and one α2(I) chain encoded by COL1A1 and COL1A2 genes, respectively, which are coordinately expressed (1, 2).
Because of its biomechanical properties, collagen plays a major role in tissue and organ development; cell migration, proliferation, and differentiation; wound healing; tissue remodeling; and homeostasis. In this last case, an imbalance between collagen aggregation and degradation leads to pathologies, such as localized scleroderma or systemic sclerosis, a connective disease characterized by excessive deposition of extracellular matrix proteins in the skin and various internal organs (3). Moreover, several studies have shown that this exaggerated tissue aggregation most likely results, at least in part, from a transcriptional activation of extracellular matrix genes, particularly collagen genes, in response to cytokines and other factors present in the prefibrotic/inflammatory lesions (4, 5). Among them, transforming growth factor-β (TGF-β) was the most investigated. Original works demonstrated that TGF-β-responsive activating sequences regarding the promoter of COL1A1 are located between 174 and 84 bp from the transcription start site. This region contains a binding site for the transcription factor Sp1 (specific protein-1) and an element with the canonical NF-1 (nuclear factor-1) binding motif (6). Similarly, TGF-β trans-activation of COL1A2 is governed via GC-rich sequences containing Sp1/Sp3 binding sites (7–9). Sp1 may therefore play a major role in fibrotic diseases, and in this regard, Sp1 binding activity is increased in human scleroderma fibroblasts compared with normal cells (6). We also studied another profibrogenic zinc finger protein, c-Krox, and found that it increases type I collagen synthesis through a transcriptional control in human foreskin fibroblasts (FF),6 ANF, and SF. Interestingly, the c-Krox binding activity on COL1A1 is also increased in SF as for Sp1 and CBF, and this is correlated with an increased production of type I collagen (10).
Sp1 and Sp3 have similar structural features with highly conserved DNA-binding domains and bind to DNA with similar specificity and affinity. Sp3 was shown as an activator or repressor of collagen genes. This “bifunctional” transcriptional activity could depend on the cellular/molecular context, such as the number of Sp1/3 DNA-binding sites (11), intranuclear organization of Sp3 (12), or protein-protein interaction between Sp3 and components of the general transcription complex (13) or other factors.
This last parameter is essential and determines the promoter context. As a matter of fact, like Sp3 repressing Sp1-mediated transcription (14), some studies examined the possibility that NF-κB inhibits collagen transcription by interfering with Sp1-mediated activation (15).
NF-κB was first described in 1986 as a nuclear factor essential for immunoglobulin κ light chain transcription in B cells (16). Since then, NF-κB has been found to play an active part in immune responses, apoptosis, and cellular growth and be involved in several diseases, such as arthritis and cancers (17). NF-κB transcription factors (p50, p52, p65, RelB, and c-Rel) share a highly conserved sequence of ∼300 amino acids within their N-terminal domain, termed the Rel homology domain, which contains a nuclear localization sequence and is involved in dimerization, sequence-specific DNA binding, and interaction with the inhibitory IκB proteins. Only RelA (p65), RelB, or c-Rel displays a C-terminal trans-activation domain and can modulate target gene expression.
In unstimulated cells, NF-κB family proteins exist as heterodimers or homodimers that are sequestered in the cytoplasm in an inactive form by virtue of their association with a member of the IκB family of inhibitory proteins (18, 19). About 200 extracellular signals can lead to activation through the dissociation of NF-κB from the IκB proteins. We can mention proinflammatory cytokines, including IL-1 and TNF-α (20–23), that lead to the activation of the IκB kinase, which in turn phosphorylates IκB proteins, preceding their ubiquitination and degradation by the proteasome. The destruction of IκB unmasks the nuclear localization sequence of NF-κB (18), allowing a rapid accumulation of free NF-κB dimers in the nucleus, where they bind to specific DNA elements present within the promoters of target genes.
Very little was discovered about the role of NF-κB in the regulation of type I collagen expression, particularly for the human COL1A1 gene; some authors showed that TNF-α increases NF-κB activity and produces a down-regulation in the expression of COL1A1 (24). Mori et al. (25) demonstrated that TNF-α suppressed COL1A1 promoter transcription activity through two elements located between −101 and −97 bp and between −46 and −38 bp of the COL1A1 promoter and that the suppression involved unidentified protein interactions. Recently, more and more evidence has suggested that the degradation of IκB and the subsequent liberation of NF-κB are not sufficient to activate NF-κB-dependent transcription. A second level of post-translational regulation of NF-κB (26–32) is essential for the regulation of NF-κB transcription activity, facilitating in some cases interaction between NF-κB and other factors. Rippe et al. (15) demonstrated that down-regulation of type I collagen expression in murine NIH-3T3 fibroblasts by TNF-α was mediated by a physical interaction between NF-κB and Sp1 and a probable recruitment in the −220/+116 bp minimal region of Col1a1.
In this study, we investigated the function of NF-κB in ANF and SF (from localized scleroderma). Our results revealed that NF-κB is a strong inhibitor of collagen synthesis and of COL1A1 transcription in healthy and scleroderma fibroblasts, the p65 subunit of NF-κB being responsible for the inhibitory action. We then demonstrated, for the first time, that this effect is mediated through a −112/−61 bp promoter sequence of COL1A1, including no consensus NF-κB binding site and thus requiring a cooperation by protein-protein interactions with three zinc fingers proteins, c-Krox, Sp1, and Sp3, all three activators of COL1A1 expression through their recruitment on a common promoter sequence (10). Moreover, the present study relates that some particular interactions in the recruitment of the p65 subunit of NF-κB on COL1A1 could account for the phenotypic differences between healthy and scleroderma fibroblasts.
EXPERIMENTAL PROCEDURES
Cell Culture
Foreskin fibroblasts from children were provided by Dr. P. Ravasse and Dr. B. Geffard (Caen University Hospital, Caen, France). Human ANF and SF were obtained from abdomen biopsies of healthy donors (Johnson & Johnson Laboratories, Val-de-Reuil, France) or patients suffering from localized scleroderma (Dr. Anne Dompmartin (Caen University Hospital) and Prof. François-Xavier Maquart (Laboratory of Medical Biochemistry and Molecular Biology, Reims, France)). Fibroblasts were obtained after explant cultures and seeded at 3 × 106 cells/m2 in 500-cm2 triple flasks (Dutscher, Brumath, France) in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS) (Invitrogen), gentamicin (4 μg/ml), ciprofloxacin (10 μg/ml), added extemporaneously, and fungizone (0.25 μg/ml) in a 5% CO2 environment. They were passaged with a trypsin (0.05%)/EDTA (0.25 mm) solution (Invitrogen) after reaching confluence. All of the experiments were performed on cells between 4 and 9 passages.
Collagen Labeling and Assay
To assay newly synthesized collagen, FF, ANF, and SF cultures, at 80% confluence, were preincubated in 10% FCS-containing DMEM in 9.6-cm2 dishes for 15 h in the presence of 50 μg/ml sodium ascorbate. Then the medium was changed to 2% FCS plus DMEM supplemented with β-APN (100 μg/ml), ascorbic acid (50 μg/ml), and [3H]proline (2 μCi/ml) (PerkinElmer Life Sciences), containing or not containing TNF-α (1 ng/ml), IL-1β (1 ng/ml), or parthenolide (5 μm). After 24 h, the culture medium was collected, and the amount of labeled collagen was assayed as explained below.
To assay neosynthesized collagen in the case of transfection experiments, fibroblasts were initially preincubated in 10% FCS-containing DMEM in 9.6-cm2 dishes for 15 h in the presence of 50 μg/ml sodium ascorbate. Then the cells were transiently transfected by the calcium phosphate co-precipitation method with p50, p65, Sp1, or Sp3 expression vectors (pSG5/p50-p65; pEVR2/Sp1; pN3/Sp3) and/or the corresponding insertless plasmids (pSG5; pEVR2; pN3). After 15 h, the medium was changed to 2% FCS plus DMEM supplemented with β-APN (100 μg/ml), ascorbic acid (50 μg/ml), and [3H]proline (2 μCi/ml). After 24 h, the culture medium was collected, and the labeled collagen was assayed, using the bacterial collagenase method as described previously (33). The cell layer was scraped and used to assay cell protein levels, using the Bradford colorimetric assay. Levels of collagen and noncollagenous proteins were corrected for the total protein amount.
An alternative method of transfection was also used to estimate collagen neosynthesis in the small interfering RNA (siRNA) knockdown experiments. Fibroblasts were passaged by trypsin at confluence and then distributed in Eppendorf tubes (1 × 106 cells/1.5-ml tube). Cells were centrifuged for 10 min at 200 × g and transfected using the AMAXA Nucleofector® apparatus (Lonza, Köln, Germany), according to the manufacturer's instructions. After p65, Sp1, Sp3, or c-Krox siRNA (1 μg) overnight transfection, the cells were plated and incubated in the same media, and the bacterial collagenase assay was performed as described above.
Transient Transfections with COL1A1 Constructs
Dermal fibroblasts were seeded in 500-cm2 triple flasks. At confluence, cells were trypsinized and resuspended in culture medium. They were divided in aliquots of 0.8–1.2 × 106 cells per the “nucleofection” sample. After centrifugation (200 × g, 10 min), they were suspended in 100 μl of human dermal fibroblast Nucleofector® solution at room temperature. Expression vectors (1 μg), reporter plasmids (2 μg), and pSV40/β-gal plasmid (2 μg) were added. The nucleofection was then performed with the AMAXA Nucleofector®, according to the manufacturer's recommendations. Then 500 μl of prewarmed culture medium was added, and cells were seeded in 9.6-cm2 wells. After 12 h at 37 °C, the culture medium was changed, and the cells were incubated for 6 h. In some experiments, fibroblasts were transiently transfected, at 80% confluence, by the calcium phosphate co-precipitation method as described above and described previously (10). The COL1A1-luciferase reporter vectors (−804/+42 bp, −311/+114 bp, −198/+28 bp, −112/+28 bp, −61/+28 bp) were described previously (10).
Then the samples were harvested, and protein content, luciferase, and β-galactosidase activities were determined. Luciferase activity was measured (Promega kit, Charbonnières, France) in a luminometer (Berthold Centro LB 960, Thoiry, France). β-Galactosidase activity was determined by a colorimetric assay as described previously (34), and protein amount was assayed by the Bradford procedure (Bio-Rad). Luciferase activities were normalized to transfection efficiency and protein amount, and means ± S.D. of three independent samples were expressed as relative luciferase units (RLU).
siRNA Experiments
To inhibit c-Krox, Sp1, Sp3, p50, and p65 expression, we used the technique of siRNA. The c-Krox siRNA has been designed by Qiagen, as described previously (10); control, SOX9, CBF (A subunit), Sp1/3, and p50/65 siRNAs were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) and directed against the human SOX9, CBF, Sp1/3, and p50/65 mRNAs, respectively.
ANF and SF were transfected with the AMAXA Nucleofector® as described earlier, with the following modifications. For each sample of 1 × 106 cells, 1 μg of siRNA or control siRNA or SOX9 siRNA was added to the transfection mixture. After overnight transfection, the medium was replaced with fresh medium, and 6 h later, cells were harvested to measure the transcriptional activity or to extract total RNAs.
Quantitative Real-time RT-PCR
Total RNA was extracted by the guanidium hydrochloride method as described previously (35), and the integrity of samples was checked by 1% agarose-formaldehyde gel electrophoresis. Then 2 μg of total RNA were reverse transcribed into cDNA in the presence of 50 pmol of random hexamers, 40 units/μl RNase inhibitor, 10 mm each dNTP, 5× first-strand buffer, and 60 units/μl Moloney murine leukemia virus in a final reaction volume of 25 μl. The mixture was incubated at 37 °C for 1 h, and the cDNA synthesis was stopped by heating the reaction mixture at 90 °C for 15 min. To test the efficiency of reverse transcription, GAPDH cDNA was submitted to 40 cycles of amplification (1 cycle of 94 °C for 1 min, 60 °C for 2 min, 72 °C for 2 min) in an Omni-E-Hybrid thermocycler and using a PCR kit (Invitrogen). Then the samples were separated on a 2% agarose gel and visualized by ethidium bromide staining. Real-time RT-PCR amplifications were performed, as described previously (10), using sequence-specific primers (Table 1) (Eurogentec, Angers, France), defined with the “Primer Express” software (Applied Biosystems, Courtaboeuf, France); analysis of relative gene expression was done by using the 2−ΔΔCT method (36).
TABLE 1.
Oligonucleotides used for this study
The CAT box, 5′ and 3′ GC-rich and c-Krox sequences are indicated in boldface type in the decoy oligonucleotides (10). The respective mutated sequences are underlined.
| Name | Sequences | Assay |
|---|---|---|
| RPL13 | Sense: 5′-GAGGTATGCTGCCCCACAAA-3′ | RT-PCR |
| Antisense: 5′-GTGGGATGCCGTCAAACAC-3′ | ||
| COL1A1 | Sense: 5′-CACCAATCACCTGCGGTACAGAA-3′ | RT-PCR |
| Antisense: 5′-CAGATCACGTCATCGCACAAC-3′ | ||
| COL1A2 | Sense: 5′-AAAACATCCCAGCCAAGAACTG-3′ | RT-PCR |
| Antisense: 5′-TCAAACTGGCTGCCAGCAT-3′ | ||
| COL3A1 | Sense: 5′-TCTTGGTCAGTCCTATGCGGATA-3′ | RT-PCR |
| Antisense: 5′-CATCGCAGAGAACGGATCCT-3′ | ||
| c-Krox | Sense: 5′-AGGGTTCTGGAAGATGAAATGAGT-3′ | RT-PCR |
| Antisense: 5′-TTCATAACCGATTTCAACCAGGAGAA-3′ | ||
| Sp1 | Sense: 5′-AGAATTGAGTCACCCAATGAGAACA-3′ | RT-PCR |
| Antisense: 5′-GTTGTGTGGCTGTGAGGTCAAG-3′ | ||
| Sp3 | Sense: 5′-ATCAAATGGTACAGTGTCCAGTGTTC-3′ | RT-PCR |
| Antisense: 5′-AAGAGCCTGTGAAACCAATTTGAA-3′ | ||
| p50 | Sense: 5′-TGAGTCCTGCTCCTTCCAA-3′ | RT-PCR |
| Antisense: 5′-CTTCGGTGTAGCCCATTTGT-3′ | ||
| p65 | Sense: 5′-TAGGAAAGGACTGCCGGGAT-3′ | RT-PCR |
| Antisense: 5′-CCGCTTCTTCACACACTGGA-3′ | ||
| NF-κB | 5′-AGTTGAGGGGACTTTCCCAGGC-3′ | GRA |
| Enh1 | 5′-AGCGCAGCCTGGCCCCGCCCCTGCGCCGGC-3′ | GRA |
| −112/−61 WT decoy | Sense: 5′-AGGCAGCTCTGATTGGCTGGGGCACGGGCGGCCGGCTCCCCCTCTCCGAGGG-3′ | Transfection |
| Antisense: 3′-TCCGTCGAGACTAACCGACCCCGTGCCCGCCGGCCGAGGGGGAGAGGCTCCC-5′ | ||
| −112/−61 mut decoy | Sense: 5′-AGGCAGCTCTGAAAAGCTGGTTTTCGGTTGGCCTGCAAACCTTCTCCGAGTT-3′ | Transfection |
| Antisense: 3′-TCCGTCGAGACTTTTCGACCAAAAGCCAACCGGACGTTTGGAAGAGGCTCAA-5′ | ||
| Sox9 decoy | Sense: 5′AGCCCCATTCATGAGAGACGAGGT-3′ | Transfection |
| Antisense: 3′-TCGGGGTAAGTACTCTCTGCTCCA-5′ |
Nuclear Extracts and Gel Retardation Assays
Nuclear extracts were prepared by a minipreparation procedure as described previously (37), and gel retardation assays were performed with the oligonucleotides presented in Table 1 (consensus NF-κB and Enh-1 probes).
The probes were end-labeled with [γ-32P]dATP (PerkinElmer Life Sciences) using T4 polynucleotide kinase (Promega, Charbonnières, France). ANF and SF nuclear extracts (10 μg) were incubated for 10 min at room temperature with the probe (2 fmol) in 20 μl of a specific binding buffer (with NF-κB probe: 20 mm HEPES (pH 7.5), 50 mm KCl, 0.5 mm DTT, 0.2 mm EDTA, 1 mg/ml BSA, 0.05% (v/v) Nonidet P-40, 20% (v/v) glycerol, 4 mm MgCl2; with Enh-1 probe (+2817/+2842 sequence of the COL2A1 gene), see Ref. 38) in the presence of 2 μg of poly(dI-dC)·poly(dI-dC) (Amersham Biosciences) used as a DNA nonspecific competitor. For antibody interference reactions, nuclear extracts were incubated with specific antibodies directed against p50 or p65 (Tebu-Bio SA, Le Perray en Yvelines, France) (2 μl) for 10 min at room temperature and then for 30 min at 4 °C. Finally, the probe was added, and a further 10-min incubation at room temperature was performed. Samples were fractionated by electrophoresis for 1.5 h at 150 V on a 5% polyacrylamide gel in 0.5× TBE (45 mm Tris borate, 1 mm Na2EDTA) and visualized by autoradiography.
Western Blot Analysis
Western blot analysis were performed on nuclear extracts which were submitted to SDS-PAGE under reducing conditions and electrotransferred to polyvinylidene difluoride transfer membrane (PVDF; PerkinElmer Life Sciences). Membranes were blocked for 1 h at room temperature in Tris-buffered saline (pH 7.6) with 0.1% Tween 20 (TBS-T) and 10% nonfat dry milk. Then they were rinsed twice in TBS-T and incubated overnight at 4 °C with the primary antibodies (rabbit anti-human p50, p65, Sp1, Sp3, and CBF, 1:500 dilutions (Tebu-Bio, Le Perray en Yvelines, France), rabbit anti-c-Krox, 1:300 dilution (antibody developed in collaboration between our laboratory and Novotec (Lyon, France)), or rabbit anti-human house-keeping β-tubulin, 1:300 dilution (Tebu-Bio) in TBST plus 2% nonfat dry milk. The membranes were then rinsed eight times for 5 min in TBST and incubated for 2 h with a secondary antibody (horseradish peroxidase (HRP)-conjugated goat anti-rabbit antibody) (1:7000 dilution). The blots were then washed eight times for 5 min in TBST. Proteins were then revealed, using a Western blot detection kit (West Pico® kit, Pierce). Finally, signals were captured with the Fluor-S Multimager® video system (Bio-Rad) and digitized into jpg files.
Immunoprecipitation (IP) Assays
100 μg of nuclear extracts from control ANF and SF were precleared with an appropriate amount of protein A-Sepharose at 4 °C for 2 h. Precleared nuclear extracts were then centrifuged for 2 min at 4,000 × g and 4 °C to pellet protein A-Sepharose. The supernatant was collected and immunoprecipitated overnight at 4 °C using either anti-Sp1, anti-Sp3, anti-p50, anti-p65, anti-c-Krox, or anti-CBF antibody. Then protein A-Sepharose was added to each tube for 2 h at 4 °C, and after IP, complexes were collected by centrifugation for 2 min at 4,000 × g. The unbound proteins were then removed by washing the solid phase four times with complete lysis buffer. Protein A was then resuspended in 50 μl of 2× reducing sample buffer and boiled for 10 min. Samples were centrifuged for 5 min at 14,000 × g, and supernatants were submitted to Western blot analysis by incubating PVDF membranes with anti-p50, anti-p65, anti-Sp1, anti-Sp3, anti-c-Krox, or anti-CBF primary antibodies.
Chromatin Immunoprecipitation (ChIP) and Re-ChIP Assays
ChIP assays were performed by using a commercially purchased chromatin immunoprecipitation kit (Active Motif). Briefly, fibroblasts (10 150-cm2 flasks) were cross-linked, scraped, and lysed according to the manufacturer's instructions. The DNA was then sheared using enzymatic digestion, and specific protein-DNA complexes were immunoprecipitated using either anti-c-Krox, anti-Sp1, anti-Sp3, anti-p50, or anti-p65 antibodies. Following overnight immunoprecipitation, cross-linking was reversed, the proteins were removed by treatment with proteinase K, and the DNA was purified and used as a template in a PCR assay. The primers used for the amplification of the COL1A1 promoter were 5′-CAGAGCTGCGAAGAGGGGA-3′ (forward) and 5′-AGACTCTTTGTGGCTGGGGAG-3′ (reverse). The amplicon corresponds to a 300-bp fragment that covers the core COL1A1 gene promoter (−200/+100 bp).
For re-ChIP experiments, the ChIP assay was performed with p65 antibody. Then complexes were dissociated with DTT at 10 mm for 30 min at 37 °C, and chromatin was diluted 25 times and again subjected to the ChIP procedure with anti-Sp1, anti-Sp3, or anti-c-Krox antibody.
Statistical Analysis
Each experiment was repeated at least three times with similar results. Results are expressed as means ± S.D. of triplicate determinations. Statistical significance was assessed using Student's t test.
RESULTS
NF-κB Inhibits Type I Collagen Expression in FF, ANF, and SF
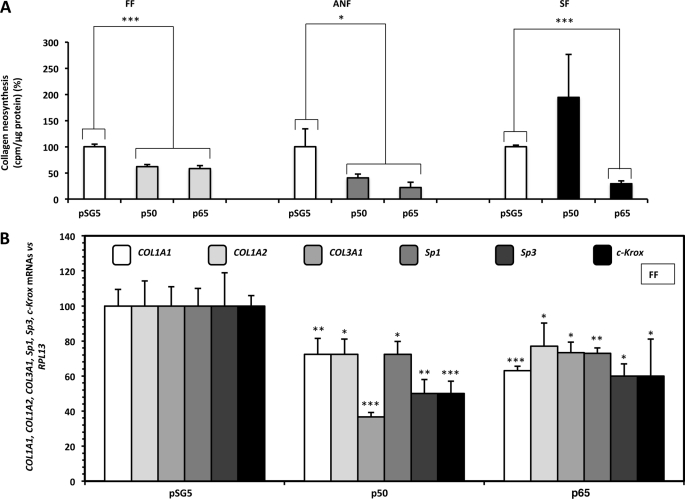
We first studied the effect of NF-κB p50 or p65 on total collagen neosynthesis in primary fibroblast cultures. Cells were transiently transfected with 1 μg of an expression vector containing the p50 or p65 subunit cDNA, and the collagen neosynthesis was determined after tritiated proline labeling. We found that p50 and p65 decreased the amount of newly synthesized collagen in FF, which includes type I isotype as the major form in the dermis. Similar data were observed in ANF and also in SF for the effect of the p65 subunit (Fig. 1A). By contrast, p50 forced expression was unable to modulate the production of newly synthesized collagens in SF (Fig. 1A).
FIGURE 1.
NF-κB inhibits type I collagen neosynthesis and decreases type I/III collagens, Sp1, Sp3, and c-Krox mRNA steady-state levels in cultured fibroblasts. A, FF, ANF, and SF were transiently transfected, at 80% confluence, by the calcium phosphate co-precipitation method with 1 μg of pSG5 expression vector containing or not (control) the NF-κB p50 or p65 subunit cDNA. After overnight transfection, cells were incubated in DMEM plus 2% FCS containing sodium ascorbate (50 μg/ml), β-APN (100 μg/ml), and 2 μCi/ml [3H]proline. 24 h later, the culture medium was collected, and the amount of radiolabeled collagen was assayed using the bacterial collagenase method. The values, normalized to total protein amount, are expressed as counts/min/μg of protein and represent the means ± S.D. (error bars) of triplicate dishes. The data are expressed as percentage versus control transfected with the insertless expression vector pSG5. B, FF were transfected as in A. 18 h after transfection, total RNAs were extracted, and 1 μg was reverse-transcribed into cDNA. The resulting products were diluted (1:100) and analyzed by real-time PCR using specific primers for COL1A1, COL1A2, COL3A1, Sp1, Sp3, c-Krox, and RPL13 cDNAs. Results were normalized to RPL13 cDNA and represented graphically as relative levels of COL1A1, COL1A2, COL3A1, Sp1, Sp3, and c-Krox mRNAs, expressed as percentage versus control transfected with the insertless expression vector pSG5. Statistical significance was evaluated using Student's t test (*, p < 0.05; **, p < 0.01; ***, p < 0.001); n ≥ 5.
To determine whether the effect induced by NF-κB on the synthesis of the two major isotypes produced by fibroblasts (i.e. type I and III collagens) was accompanied by a similar effect at the transcriptional level, the steady-state levels of COL1A1, COL1A2, and COL3A1 were estimated by real-time RT-PCR performed on total RNA extracts of primary FF. Both p50 and p65 decreased COL1A1, COL1A2, and COL3A1 mRNA steady-state levels (Fig. 1B).
The effects of NF-κB on the steady-state amounts of transcription factors, such as c-Krox, known to be a strong activator of collagen expression, and Sp1/3 (also activators; see below), with which c-Krox interacts (10), have been studied, and were all found to be decreased under p50 or p65 forced expressions (Fig. 1B).
IL-1β and TNF-α Enhance NF-κB Expression in Fibroblasts
In order to validate the preceding results, we used another approach, which consisted in increasing physiologically the NF-κB cellular levels. To this end, we used the cytokines IL-1β and TNF-α, two well known activators of NF-κB (23).
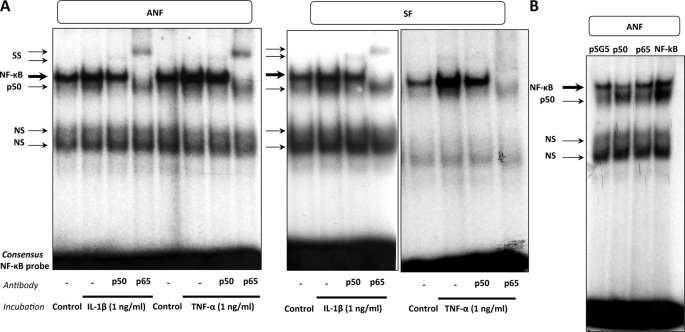
Initially, NF-κB activation by both cytokines was carried out by evaluating the binding activity, and EMSA analysis was therefore performed using as a probe a consensus NF-κB binding site and nuclear extracts from ANF and SF. As shown in Fig. 2A, one specific complex was formed upon incubation with the NF-κB probe when the binding reaction was performed with control nuclear extracts. DNA binding activity of the transcription factor involved in this complex (i.e. NF-κB) was increased when nuclear extracts from ANF and SF treated with cytokines were used. The implication of p50 and p65 subunits in the formation of the specific complex was demonstrated by the use of antibodies directed against these two subunits. The addition of a p50 or a p65 antibody was found to prevent the NF-κB complex formation. More precisely, the higher complex area concerns p50-p65 heterodimers and p65 homodimers, because the addition in the binding reaction of a p50 antibody is not capable of abolishing the complex formation, indicating that p65 homodimers co-migrate in the complex. By contrast, when the p65 antibody was used, the upper area of the complex completely disappeared, indicating that both p50-p65 and p65-p65 co-migrate in this upper area complex. The lower area corresponds to p50 homodimers because the use of a p50 antibody is able to prevent completely the formation of this complex.
FIGURE 2.
IL-1β and TNF-α enhance NF-κB expression in ANF and SF. A, DNA binding activity was analyzed by EMSA. EMSA reactions were performed using the 5′-end-labeled consensus NF-κB probe, incubated with 10 μg of nuclear extracts from ANF or SF treated or not treated (control) with IL-1β (1 ng/ml) or TNF-α (1 ng/ml), in the presence or absence of anti-p50 or anti-p65 antibody (2 μl). The arrows indicate the complexes formed between DNA and nuclear proteins. The NF-κB complex includes p50 homodimers, p50-p65 heterodimers, and p65 homodimers. B, fibroblasts (ANF) were transiently transfected by the AMAXA Nucleofector® method, with 1 μg of pSG5 expression vector containing or not containing (control) NF-κB subunit cDNA (p50, p65, or both p50 and p65 (labeled NF-κB)). 12 h after transfection, the medium was changed. 6 h later, the samples were harvested, and nuclear extracts were prepared. EMSA reactions were carried out using the 5′-end-labeled consensus NF-κB probe, incubated with 10 μg of nuclear extracts. The arrows indicate the complexes formed between DNA and nuclear proteins. The NF-κB complex includes p50 homodimers, p50-p65 heterodimers, and p65 homodimers. SS, supershift; NS, nonspecific complex.
After characterization of the different NF-κB complexes, the effectiveness of the forced expression experiments of p50 and/or p65 subunits of NF-κB was verified in EMSA. As shown in Fig. 2B, the overexpressions of the p50, p65, and both subunits were correlated with an increase in the corresponding DNA binding activities to the probe of the p50 homodimers, p65 homodimers, and p50-p65 heterodimers.
NF-κB activation by IL-1β/TNF-α was also confirmed in Western blotting and quantitative RT-PCR experiments. The cytokines exerted a transcriptional control and increase the steady-state levels of p50-p65 mRNA in ANF and SF (supplemental Fig. S1, A and B).
To further corroborate the specific activating effect of TNF-α on NF-κB, parthenolide (PT), a post-translational inhibitor of NF-κB activation (39, 40), was used. NF-κB binding activity was found to be abolished by PT in a dose-dependent manner in basal or TNF-α-stimulated conditions (supplemental Fig. S1C, i).
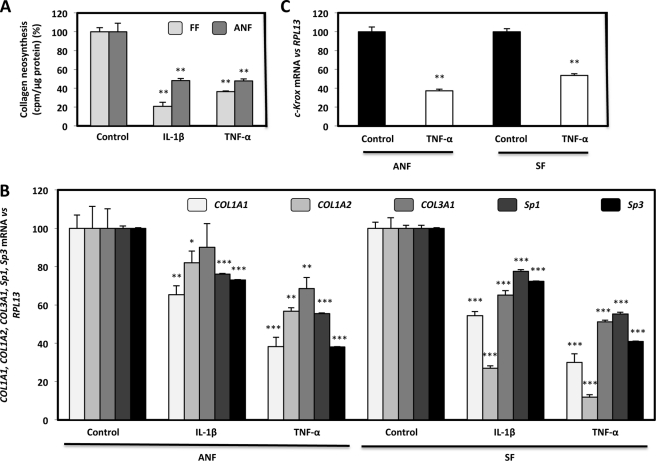
NF-κB Activation by IL-1β and TNF-α Inhibits Type I Collagen Expression in FF, ANF, and SF
As shown in Fig. 1, an inhibitory effect of NF-κB on type I collagen expression was demonstrated in p50 and p65 forced expression experiments. Through an alternative approach, NF-κB induction by IL-1β and TNF-α was found to decrease collagen neosynthesis in FF and ANF (Fig. 3A). These effects resulted, as for forced expression experiments, from a decrease in the steady-state levels of COL1A1, COL1A2, and COL3A1 under IL-1β and TNF-α incubation and also from a reduced expression in the mRNA amounts of the transcription activators of COL1A1 (i.e. Sp1, Sp3, and c-Krox) (Fig. 3, B and C).
FIGURE 3.
IL-1β and TNF-α decrease type I collagen neosynthesis and COL1A1, COL1A2, COL3A1, Sp1, Sp3, and c-Krox mRNA steady-state levels in normal and scleroderma fibroblasts. A, FF and ANF (80% confluence) were cultured in DMEM + 10% FCS containing sodium ascorbate (50 μg/ml). 24 h later, cells were incubated for 24 h in DMEM + 2% FCS containing sodium ascorbate (50 μg/ml), β-APN (100 μg/ml), 2 μCi/ml [3H]proline in the presence or absence (control) of IL-1β or TNF-α (1 ng/ml). At the end of the experiment, the amount of radiolabeled collagen was assayed in culture medium as collagenase-digestible material. The values, normalized to the amount of total protein assayed by the Bradford colorimetric method, were expressed as cpm/μg protein. The data are expressed as percentage versus control incubated without cytokines and represent the means ± S.D. (error bars) of triplicate dishes. B and C, total RNAs from normal and scleroderma fibroblasts treated or not treated (control) with IL-1β or TNF-α (1 ng/ml), were reverse-transcribed into cDNA. The resulting products were diluted (1:100) and analyzed by real-time PCR using specific primers for COL1A1, COL1A2, COL3A1, Sp1, Sp3, c-Krox, and RPL13 cDNAs. Results were normalized to RPL13 cDNA and represented graphically as relative levels of COL1A1, COL1A2, COL3A1, Sp1, and Sp3 (B) and c-Krox (C) mRNAs, expressed as percentage versus control incubated without cytokine. Statistical significance was evaluated using Student's t test (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
The same experiments were performed in order to evaluate the resulting collagen neosynthesis in ANF in the presence of NF-κB blockade by PT. IL-1β and TNF-α decreased the production of newly synthesized collagens, and the concomitant addition of PT with both cytokines indicated that NF-κB, upon cytokine activation, inhibits collagen neosynthesis because PT was able to prevent the IL-1β- and TNF-α-induced inhibition of collagen production (Fig. S1C, ii). In conclusion, these experiments indicate that NF-κB activation by IL-1β and TNF-α is associated with an inhibition of collagen synthesis, which corroborates the inhibitory effect of NF-κB obtained in gain of function experiments.
A −112/−61 bp Region of COL1A1 Gene Promoter Mediates Inhibitory Effect of NF-κB
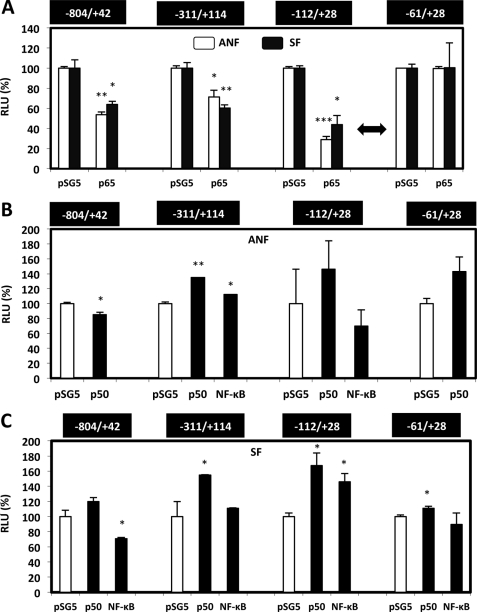
Because previous results suggest a putative transcriptional control induced by NF-κB (p65 subunit or NF-κB induced by IL-1β and TNF-α) and to better understand the molecular mechanisms that mediate the down-regulation of COL1A1 gene transcription, primary fibroblasts (ANF and SF) were transfected by the AMAXA Nucleofector® method, with several constructs, including the 5′-flanking region of the COL1A1 gene surrounding the transcription start site. The expression of the reporter gene was always found to be reduced after p65 forced expression, except for the shortest construct, including only 61 bp of the proximal promoter, indicating that the transcription control is mediated by the −112/−61 bp region (Fig. 4A). These results were observed in both ANF and SF.
FIGURE 4.
NF-κB p65 subunit down-regulates COL1A1 gene transcription activity through a region localized between −112 and −61 bp. A–C, fibroblasts were transiently co-transfected by the AMAXA Nucleofector® method, with 2 μg of different COL1A1 reporter plasmids together with 1 μg of pSV40/β-gal expression vector and 1 μg of pSG5 expression vector containing or not containing (control) NF-κB subunit(s) cDNA. These experiments were performed in ANF and SF by co-transfecting p65 subunit (A) or p50 subunit (B and C). In some cases, these two NF-κB subunits were cotransfected (labeled NF-κB). 12 h after transfection, the medium was changed. 6 h later, the samples were harvested, and protein content, luciferase, and β-galactosidase activities were assayed. Each set of transfections was performed in triplicate. Transcriptional activity of each construct was expressed as RLU after correction for transfection efficiency and protein amounts, and expressed as a percentage versus the respective controls transfected with the insertless expression vector pSG5. Values represent the means ± S.D. (error bars) of three independent samples of a representative experiment. Statistical significance was evaluated using Student's t test (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
In parallel experiments, the overexpression of the NF-κB p50 subunit in ANF and SF seems to have no clear significant effect on COL1A1 transcriptional activity by itself; however, it appeared that p50 expression diminished the p65 inhibitory effect when the two subunits were co-expressed (Fig. 4, B and C).
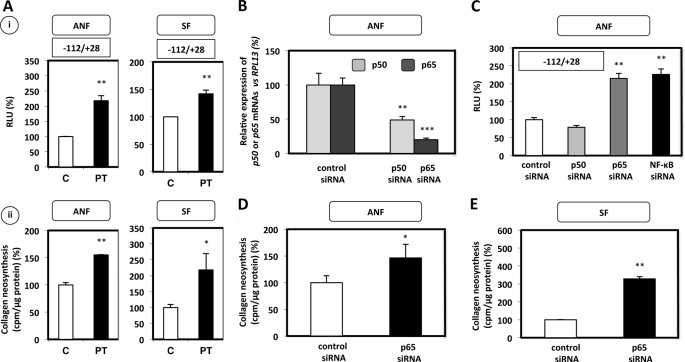
Parthenolide and p65 siRNA Prevent NF-κB-induced Inhibition of COL1A1 Gene Transcription and Type I Collagen Neosynthesis in Normal and Scleroderma Fibroblasts
To finally demonstrate that NF-κB decreased specifically type I collagen expression, we used two experimental approaches. In the first one, we blocked NF-κB activation through the use of PT. As shown in Fig. 5A, inhibition of NF-κB by PT led to an increased transcription activity of COL1A1 (especially through the region by which NF-κB inhibits classically the transcription) and type I collagen neosynthesis in both ANF and SF, suggesting, one more time, that NF-κB is a strong inhibitor of type I collagen production in human primary fibroblasts.
FIGURE 5.
Parthenolide and a p65 siRNA inhibit the p65-induced inhibition of type I collagen expression in ANF and SF. A, i, ANF and SF were transiently co-transfected by the AMAXA Nucleofector® method, both with 2 μg of pGL2–112-bp COL1A1 construct and 1 μg of pSV40/β-gal expression vector, and were incubated for 12 h with or without (control), PT at 5 μm. After overnight transfection, cells were treated for 6 h in DMEM + 10% FCS containing or not containing PT (5 μm). Treatment of the samples and expression of the results are the same as in Fig. 4. ii, incubations with PT (5 μm) were carried out in a bacterial collagenase assay, as described under “Experimental Procedures.” The values, normalized to total protein amount, are expressed as cpm/μg protein and represent the means ± S.D. of triplicate dishes. The data are expressed as percentage versus control incubated without PT. B, to verify the efficiency of p50 and p65 silencing, fibroblasts were transiently transfected using the AMAXA Nucleofector® with a control siRNA, p50 siRNA, or p65 siRNA (1 μg). 12 h after transfection, the cell culture medium was changed, and 6 h later, the samples were harvested. 1 μg of total RNAs extracted was reverse-transcribed into cDNA. 2 μl of cDNA were diluted (1:100) and used in real-time PCR to amplify p50 or p65 and RPL13 cDNAs. The relative expression of p50 and p65 mRNAs was normalized to RPL13 mRNA according to the 2−ΔΔCT method. C, ANF were transiently cotransfected by the AMAXA Nucleofector® with 1 μg of a control siRNA or p50 siRNA, or p65 siRNA, or with both p50 and p65 siRNA (NF-κB siRNA), together with 2 μg of pGL2–112-bp construct and 1 μg of the expression vector pSV40/β-gal. 12 h after transfection, the medium was replaced. 6 h later, the samples were harvested, and transcriptional activities were analyzed as described in the legend to Fig. 4. D and E, in parallel, ANF (D) and SF (E) were transiently transfected by the AMAXA Nucleofector® with 1 μg of control or p65 siRNA. 12 h after transfection, the culture medium was changed, and a bacterial collagenase assay was performed, as described under “Experimental Procedures.” The values, normalized to total protein amount, are expressed as cpm/μg protein and represent the means ± S.D. of triplicate dishes. The data are expressed as percentage versus siRNA control. Statistical significance was evaluated using Student's t test (*, p < 0.05; **, p < 0.01; ***, p < 0.001). PT, Partherolide.
In the second approach and in order to better characterize the NF-κB subunit(s) implicated in type I collagen regulation, experiments with siRNAs directed against p50 and p65 were carried out. We first determined the efficiency of these siRNAs to effectively block p50 and p65 expression by real-time RT-PCR assays. As shown in Fig. 5B, the significant decrease of p50-p65 mRNA steady-state levels (∼50–80%) validates the siRNA efficiency. Next, ANF were transfected with the pGL2–112-bp construct, which includes the COL1A1 gene region mediating the NF-κB effect in the presence of control or NF-κB subunit siRNAs. Fig. 5C demonstrates that the p65 siRNA (but not the p50 siRNA), in the presence or absence of the p50 siRNA, increases the COL1A1 transcription activity in normal fibroblasts, suggesting that NF-κB, and p65 in particular, interacts probably with trans factors binding to the −112/−61 bp sequence, through which it induces decreased transcriptional activity of the human COL1A1 gene. These data have been obtained in fibroblasts cultured in basal conditions (not treated by IL-1β and/or TNF-α), indicating therefore that the endogenous NF-κB produced by the cells already inhibits type I collagen production.
Additionally, we determined if the observed effects at the transcriptional level in the p65 knockdown experiments were also detected at the protein level. For that purpose, ANF and SF cultured in basal conditions were transiently transfected with control or p65 siRNAs, and the collagen neosynthesis was evaluated. As shown in Fig. 5, D and E, the production of newly synthesized collagens was increased by ∼40–300% when a p65 siRNA was transfected in ANF and SF, respectively, confirming that the endogenous p65 is an inhibitor of type I collagen synthesis. In conclusion, these data suggest that the −112/−61 bp sequence of the proximal promoter mediates p65-induced inhibition of COL1A1 gene transcription in both ANF and SF.
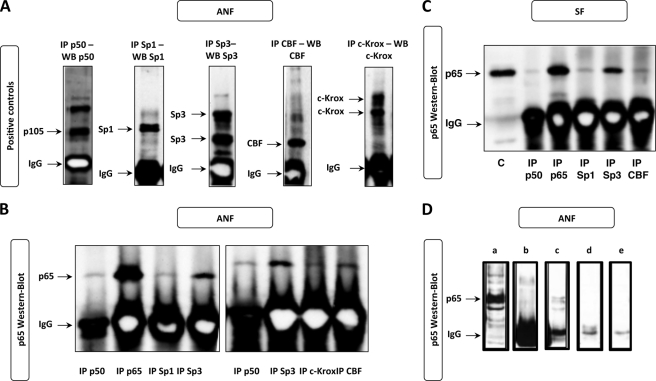
p65 Physically Interacts with p50, c-Krox, CBF, Sp1, and Sp3 in ANF and SF
The nucleotide sequence analysis of the COL1A1 proximal promoter does not reveal any binding site for NF-κB, especially in the −112/−61 bp region. The effect of this trans factor on type I collagen expression through the −112/−61 bp region is thus certainly mediated by interaction with other transcription factors; for instance, we have shown in a previous study that c-Krox, CBF, Sp1, and Sp3 bind to the −112/−61 bp sequence of the COL1A1 promoter in fibroblasts (10). Therefore, to address this issue, IP experiments were performed with nuclear extracts prepared from ANF and SF, using antibodies directed against p50, Sp1, Sp3, CBF, and c-Krox. Then immunoprecipitated proteins were subjected to Western blotting, and the membranes were incubated with the p65 antibody. As shown in Fig. 6A, controls were used to verify the efficiency of the IPs. The Western blots presented in Fig. 6, B and C, indicate that all of the trans factors tested interact with p65 in ANF and SF. However, the affinity of the interaction between p65 and Sp3 seems to be the highest. These interactions are specific because a control precipitation using only protein A-Sepharose did not reveal any p65 band (Fig. 6D).
FIGURE 6.
p65 interacts physically with p50, c-Krox, CBF, Sp1, and Sp3 in ANF and SF. After immunoprecipitations using either anti-p50, anti-p65, anti-c-Krox, anti-CBF, anti-Sp1, or anti-Sp3 antibody, nuclear extracts from ANF (A, B, and D) or SF (C) were subjected to Western blotting (WB). The membranes were immunoblotted with antibodies directed against p65 (1:500 dilution). Then the membranes were processed as described under “Experimental Procedures.” As positive control (C), 15 μg of nuclear extracts that were not subjected to immunoprecipitation were also loaded onto SDS-polyacrylamide gels. In parallel, to verify the immunoprecipitation efficiency, each immunoprecipitate was subjected to Western blotting by incubating the membranes with the respective antibodies used during immunoprecipitation (A). Some controls used during the IP procedure are presented in D. a, total nuclear extracts without IP; b, IP with IgG antibody; c, protein A-Sepharose + Sp1 IP; d, protein A-Sepharose after preclearing; e, total nuclear extracts + protein A-Sepharose.
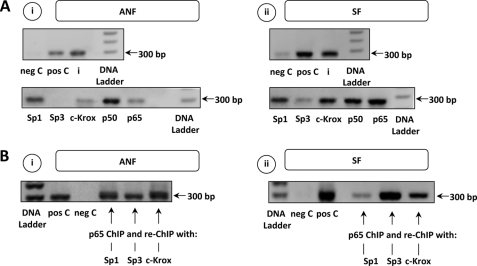
p50, p65, c-Krox, Sp1, and Sp3 Bind to COL1A1 Promoter in Vivo
In order to determine whether p50, p65, c-Krox, Sp1, and Sp3, behind their protein/protein interactions, are effectively recruited on the COL1A1 gene and involved in its regulation and to validate our in vitro experimental data, ChIP assays were performed. As shown in Fig. 7A, the presence of the 300-bp amplicon reveals that all transcription factors bind to the −200/+100 bp promoter region of COL1A1 in ANF and SF. It also seems that higher amounts of trans factors interact with this sequence in SF. Conversely, interaction between Sp3 and the COL1A1 promoter stays very weak in ANF. Moreover, re-ChIP experiments were performed on ANF and SF and validate that protein-protein interactions do exist in vivo between Sp1/3, c-Krox, and p65 (Fig. 7B). Moreover, re-ChIP experiments performed on ANF demonstrated that CBF interacts also with p65 (data not shown).
FIGURE 7.
p50, p65, c-Krox, Sp1, and Sp3 bind to the COL1A1 promoter in ChIP/re-ChIP assay. A, to evaluate in vivo interactions between DNA and transcription factors, ANF (i) or SF (ii) cultures were fixed, chromatin was digested, and specific protein: DNA complexes were immunoprecipitated with Sp1, Sp3, c-Krox, p50, or p65 antibodies, followed by PCR amplification with primers flanking the −200/+100 bp COL1A1 promoter region. As positive control (input (i)), the same primers were used on the genomic DNA. Samples were also immunoprecipitated with anti-RNA polymerase II used as another positive control (pos C) and with nonspecific immunoglobulin antibody used as a negative control (neg C). B, re-ChIP experiments were also carried out (i, ANF; ii, SF). In this respect, the ChIP assay was performed first with a p65 antibody. Then complexes were dissociated with DTT at 10 mm for 30 min at 37 °C, and chromatin was diluted 25-fold and again subjected to the ChIP procedure with anti-Sp1, anti-Sp3, and anti-c-Krox antibodies. DNA Ladder, molecular marker.
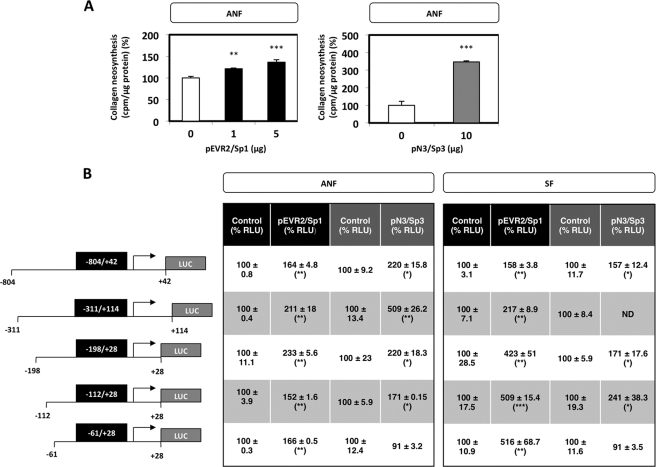
Sp1 and Sp3 Increase Type I Collagen Expression in ANF and SF
The c-Krox role on COL1A1 transcription in ANF and SF was already well documented by our laboratory (10). We demonstrated in the present study that this factor, Sp1, and Sp3 interact in vivo with the COL1A1 minimal promoter. But if Sp1 data exist and seem contradictory, data for Sp3 on the regulation of type I collagen expression in fibroblasts remained to be determined. For these reasons, we examined the effect of Sp1 and Sp3 on total collagen neosynthesis. In ANF, Sp1 and Sp3 forced expression were found to increase total collagen neosynthesis (Fig. 8A), which is known to be mainly formed of type I collagen in dermal fibroblasts.
FIGURE 8.
Effect of Sp1 and Sp3 on the transcriptional activity of the human COL1A1 gene and on collagen neosynthesis. A, ANF were transiently transfected by the calcium phosphate co-precipitation method with 1–10 μg of pEVR2/Sp1 and pN3/Sp3 expression vectors. For each amount of Sp1/Sp3, the insertless expression vectors pEVR2 and pN3, respectively, were used as complement to the maximum quantity of expression vector used, to transfect each sample with the same amount of DNA. After overnight transfection, the cell culture medium was changed, and a bacterial collagenase assay was carried out as described under “Experimental Procedures.” The values, normalized to total protein amount, are expressed as cpm/μg protein and represent the means ± S.D. (error bars) of triplicate dishes. The data are expressed as percentage versus the respective control transfected with the insertless expression vectors pEVR2 or pN3. B, ANF and SF were transiently co-transfected by the AMAXA Nucleofector® method with 2 μg of different COL1A1 reporter plasmids together with 1 μg of the pSV40-β-gal expression vector and 1 μg of pEVR2 or pN3 expression vector containing or not containing (control) the Sp1 or Sp3 cDNA, respectively. After overnight transfection, the medium was changed, and 6 h later, the samples were harvested, and protein content, luciferase, and β-galactosidase activities were assayed. Each series of transfections was performed in triplicate. Transcriptional activity of each construct was expressed as RLU, after correction for both protein amounts and transfection efficiency, and expressed as percentage versus the respective control transfected with the insertless expression vectors pEVR2 or pN3. Values represent the means ± S.D. of three independent samples of a representative experiment. Statistical significance was evaluated using Student's t test (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
To investigate the potential transcription function of Sp1 and Sp3 on the human COL1A1 gene, ANF and SF were transiently transfected with plasmid constructs covering different regions of the human COL1A1 promoter in the presence of an expression vector containing or not containing the Sp1 or Sp3 cDNA. Our data showed that Sp1 and Sp3 up-regulate the human COL1A1 gene in a similar fashion in both ANF and SF, except, concerning Sp3, for the shortest construct containing only 61 bp of the COL1A1 promoter (Fig. 8B). Therefore, Sp1 activates the transcription, and this effect is mediated by a short promoter containing a 61-bp region upstream from the start site of COL1A1, whereas the activating Sp3 effect is governed by a sequence located between −112 and −61 bp.
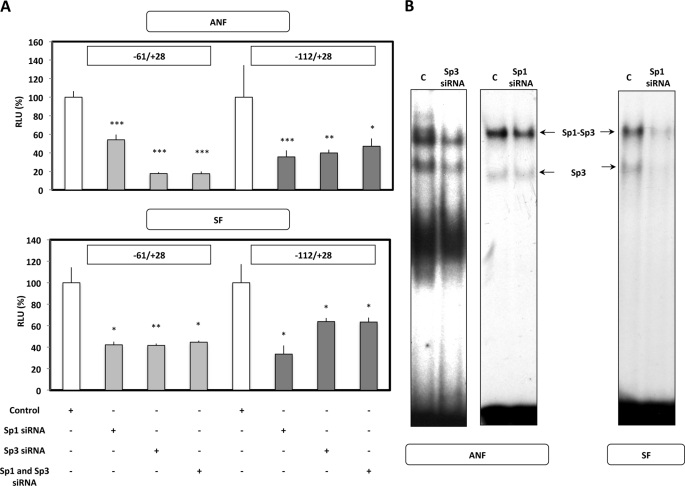
An siRNA strategy was used to determine the trans-activating potential of Sp1 and Sp3 in fibroblastic cells. ANF and SF were transfected with a short plasmid construct that includes the minimal COL1A1 gene domain mediating trans factor effect (i.e. −61/+28 bp for Sp1 and −112/+28 bp for Sp3) in the presence of a control or Sp1/3 siRNA. In these conditions, Sp1 and Sp3 knockdown led to an inhibition of COL1A1 transcriptional activity in ANF and SF (Fig. 9A). Furthermore, because Sp3 forms a complex with Sp1 and works with it cooperatively in some instances, we tested the simultaneous addition of the siRNAs on transcription activity. The promoter activity of the −61/+28 bp construct with Sp1 siRNA was not further decreased after Sp3 siRNA addition. In the same sense, the inclusion of the Sp1 siRNA did not further reduce the −112/+28 bp promoter activity when the Sp3 siRNA was added concomitantly, indicating that there is no synergistic effect of both factors in our conditions in ANF and SF.
FIGURE 9.
Sp1 and Sp3 siRNA inhibit the Sp1/3-induced activation of human COL1A1 gene expression in ANF and SF. A, ANF and SF were transiently co-transfected by the AMAXA Nucleofector® method with 1 μg of the pSV40-β-gal expression vector; 1 μg of control, Sp1, Sp3, or both Sp1 and Sp3 siRNA; and 2 μg of the −61/+28 bp or −112/+28 bp reporter constructs. After overnight transfection, the medium was replaced, and 6 h later, the samples were harvested, and transcriptional activities were analyzed as described in the legend to Fig. 8. RLU are expressed as percentage versus control siRNA and represent the means ± S.D. (error bars) of three independent samples. B, in parallel, to verify the efficiency of Sp1/3 silencing, 10 μg of nuclear extracts (NE) from ANF and SF, in which Sp1/Sp3 expression was silenced by the use of Sp1/3 siRNA, were incubated with radiolabeled Enh1 probe, which represents the +2817/+2842 bp sequence of the human COL2A1 gene we previously demonstrated to be highly specific for Sp1/Sp3 (37). The arrows indicate the complexes formed between DNA and nuclear proteins. C, NE without extinction of Sp1/3.
Silencing of Sp1 or Sp3 has been validated in EMSA experiments. For this purpose, we have used a double-stranded Sp1/Sp3 probe called Enh1 (a GC-rich sequence found in the intronic specific enhancer of COL2A1 (37)) that allows us to discriminate, at least in part, Sp1 and Sp3 formed upon incubation with the probe. As shown in Fig. 9B, transfection of an Sp3 siRNA in ANF provoked a significant decrease in the DNA binding activity in both complexes in which Sp3 is included. By contrast, the Sp1 siRNA induces a decrease in the Sp1 DNA binding activity in the complex migrating with the lower electrophoresis mobility in ANF, indicating that the effect of the siRNA is specific. However, in SF cells and by contrast to ANF, the Sp1 siRNA decreases the binding of Sp3 (higher and lower migrating complexes) to the probe, suggesting an indirect effect of Sp1, which is a much more potent trans-activator of the SP3 gene in SF than normal fibroblasts (supplemental Fig. S2). In conclusion, these data suggest that Sp1 and Sp3 interact with cis elements (i.e. Sp1) or with other transcription factors (i.e. Sp3) that bind to DNA (10) in the minimal promoter, through which they induce increased transcriptional activity of the human COL1A1 gene (Fig. 9A).
Effect of siRNAs or Decoy Oligonucleotides Targeting Some Transcription Activators of Type I Collagen Gene Expression on p65-induced Inhibition of COL1A1 Transcription in Dermal Fibroblasts
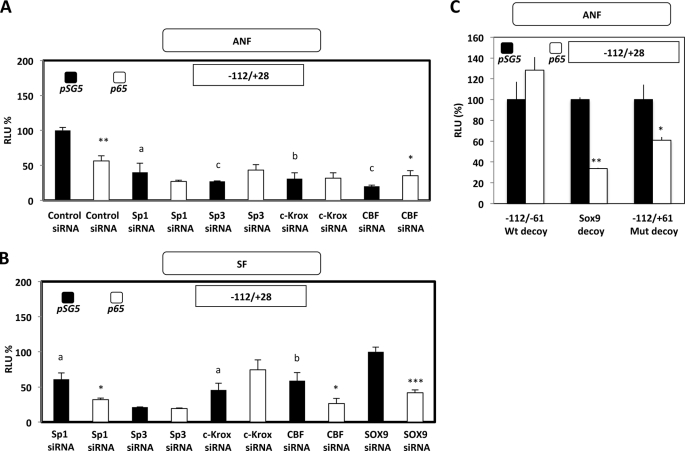
To gain insight into the mechanisms of transcription inhibition induced by the p65 subunit of NF-κB on COL1A1, experiments using siRNA were performed. These experiments were based on the fact that the NF-κB transcription function is dependent on the recruitment of this factor on the COL1A1 promoter through protein interactions with trans-activators (Sp1/3, c-Krox, and CBF), which was evidenced by IP and ChIP/re-ChIP assays. For that purpose, ANF and SF were co-transfected with the pGL2–112-bp construct by which NF-κB mediates its transcription effect on COL1A1, together with the expression vector encoding or not encoding (pSG5) the p65 subunit (p65), and siRNAs directed against Sp1, Sp3, c-Krox, and CBF. As shown in Fig. 10A, Sp1, Sp3, c-Krox, and CBF are transcription activators of COL1A1 in ANF because transfection of siRNAs against these regulators in the presence of the empty expression vector pSG5 was found to decrease the expression of the reporter gene. As expected, forced expression of the p65 subunit of NF-κB together with the transfection of a control siRNA provoked an inhibition of the transcription of the short COL1A1 promoter (∼50%). However, the p65-induced inhibitory effect was prevented when siRNAs against Sp1, Sp3, c-Krox, and CBF were co-transfected, suggesting that all of these trans factors form a multimolecular complex necessary for the NF-κB transcription function, and each one of these transcription factors is required in ANF.
FIGURE 10.
siRNAs targeting some transcription activators of COL1A1 are able to differentially prevent, in ANF and SF, the p65-induced inhibition of COL1A1 transcription. A, ANF were transiently co-transfected by the AMAXA Nucleofector® method with 1 μg of the pSV40-β-gal expression vector, 1 μg of control or Sp1/Sp3/c-Krox/CBF siRNA, 1 μg of pSG5 expression vector containing or not containing (control, pSG5) p65 subunit cDNA, and 2 μg of the −112/+28 bp reporter constructs. After overnight transfection, the medium was replaced, and 6 h later, the samples were harvested, and transcriptional activities were analyzed as described in the legend to Fig. 8. RLU are expressed as percentage versus control siRNA and pSG5 co-transfected cells and represent the means ± S.D. (error bars) of three independent samples. B, SF were transiently co-transfected by the AMAXA Nucleofector® method with 1 μg of the pSV40-β-gal expression vector, 1 μg of SOX9 (control siRNA) or Sp1/Sp3/c-Krox/CBF siRNA, 1 μg of pSG5 expression vector containing or not (control, pSG5) p65 subunit cDNA, and 2 μg of the −112/+28 bp reporter constructs. After overnight transfection, the medium was replaced, and 6 h later, the samples were harvested. Processing of the samples as well as expression of the results were the same as described in A. Statistical significance was evaluated using Student's t test (comparison of p65 forced expression versus respective control transfected with the same siRNA and the pSG5 empty expression vector (*, p < 0.05; **, p < 0.01; ***, p < 0.001); comparison of the effect of Sp1, Sp3, CBF, and c-Krox siRNA versus control siRNA (A) or SOX9 siRNA (B) (a, p < 0.05; b, p < 0.01; c, p < 0.001)). C, ANF were transiently co-transfected by the AMAXA Nucleofector® method with 1 μg of the pSV40-β-gal expression vector, 30 μg of −112/−61 wild-type (Wt) or −112/−61 mutant (Mut) or SOX9 oligonucleotide sequences used as a decoy, 1 μg of pSG5 expression vector containing or not containing (control, pSG5) p65 subunit cDNA, and 2 μg of the −112/+28 bp reporter constructs. After 6 h of transfection, the medium was replaced by the same one including 30 μg of decoy sequences, and 12 h later, the samples were harvested, and transcriptional activities were analyzed as in A. RLU are expressed as percentage versus pSG5 co-transfected cells and represent the means ± S.D. of three independent samples. Statistical significance was evaluated using Student's t test (*, p < 0.05; **, p < 0.01).
In SF, the SOX9 siRNA was used as another control because this factor has not been shown to modulate the transcription of COL1A1 in dermal fibroblasts (Fig. 10B and supplemental Fig. S3). In such conditions and when the cells were co-transfected with the empty expression plasmid pSG5, Sp1, Sp3, c-Krox, and CBF were also trans-activators of COL1A1, as was the case in ANF. As in ANF, p65 overexpression in SF decreased by ∼60% the expression of the reporter gene, but by contrast, only the siRNAs targeting Sp3 and c-Krox were able to prevent the p65-induced inhibition of transcription (Fig. 10B).
The data presented in Fig. 10 correspond to a representative experiment. Because there is a differential effect between ANF and SF concerning the Sp1 siRNA effect on p65-induced inhibition of COL1A1 transcription, we have made the average of the three experiments performed. In ANF, the data are as follows: Sp1 siRNA + pSG5 = 100 ± 28.5% and Sp1 siRNA + p65 = 72.4 ± 5.4%. For the SF, the values are as follows: Sp1 siRNA + pSG5 = 100 ± 13.2% and Sp1 siRNA + p65 = 27.1 ± 0.7 (p < 0.05). In conclusion, these data indicate that although protein-protein interactions between p65, Sp1, Sp3, c-Krox, and CBF have been evidenced in IP/ChIP/re-ChIP experiments in SF, apparently only Sp3 and c-Krox are absolutely required for the transcription function of p65. In parallel experiments, forced expression of p50 in SF confirmed the absence of effect of this subunit on the transcription of the −112 bp promoter of COL1A1, and incubation with siRNAs against Sp1, Sp3, c-Krox, and CBF did not modify the observed absence of effect (data not shown).
As an alternative approach to finally demonstrate the involvement of Sp1, Sp3, c-Krox, and CBF in the p65 recruitment and inhibition of COL1A1 transcription through protein interactions in ANF, decoy experiments were performed. As shown in Fig. 10C, the −112/+28 promoter activity is decreased by p65 forced expression when a SOX9 or a mutated −112/−61 bp decoy oligonucleotide (preventing the recruitment of the trans-activators as a result of mutations in the c-Krox, and CBF 3′- and 5′GC-rich sequences) was transfected in the cells. By contrast, the p65-induced inhibition of COL1A1 transcription was prevented when a wild-type −112/−61 decoy oligonucleotide was transfected in the ANF cultures. Taken together, these data demonstrate that, surprisingly, transcriptional activators, such as Sp1, Sp3, and c-Krox, are essential to the recruitment and the activity of NF-κB p65 subunit in the down-regulation of type I collagen expression.
DISCUSSION
In this study, we have tried to delineate the NF-κB regulation of COL1A1 gene expression, one of the two genes encoding type I collagen, a phenotypic marker of the skin. We found that the NF-κB p65 subunit inhibits collagen synthesis at protein and mRNA steady-state levels. We also demonstrated that proinflammatory cytokines acting via NF-κB activation, such as IL-1β and TNF-α, down-regulate collagen neosynthesis in human healthy and scleroderma fibroblasts, this protein decrease being correlated with the steady-state levels of mRNAs encoding the different α chains of type I and III collagens and the cytokine inhibitory effect on Sp1, Sp3, and c-Krox mRNA levels, all being trans-activators of type I collagen. Indeed, it has been demonstrated that TNF-α is implicated in the regulation of both of the type I collagen genes (i.e. COL1A1 (15, 25) and COL1A2 (41, 42), in fibroblasts or in other related cell types (24, 43), via NF-κB or other transcription factors (44–46). For example, in rat hepatic stellate cells, a TNF-α-inhibitory responsive element (TαRE) between −378 and −345 bp has been characterized in Col1a1; it binds a complex including p20C/EBPβ, p35C/EBPβ, and C/EBPδ, following p38 mediation, and surprisingly, this DNA-binding element colocalizes with the TGF-β-responsive element (43–45). Another mechanism of down-regulation of collagen synthesis by TNF-α, associated with p65, has been shown to be necessary for the inhibition of TGF-β-induced phosphorylation, nuclear translocation, and DNA binding of Smad signaling complexes. This mechanism implicates a TNF-α activation of the inhibitory Smad 7 by p65 (46) (for a review, see Ref. 4) and TβRII expression down-regulation in human dermal fibroblasts (48).
Our data showed that p65-induced collagen down-regulation takes place at the transcriptional level via a 51-bp region localized between −112 and −61 bp in the COL1A1 gene, whereas the p50 subunit has no effect by itself. These results, validated by complementary approaches, employing forced expression, a NF-κB inhibitor (parthenolide), and p50-p65 siRNAs, highlighted that the NF-κB inhibitory effect on type I collagen expression is essentially due to the p65 subunit.
Although p50-p65 NF-κB heterodimers generally activate target gene transcription (23), they are involved in the decreased collagen synthesis as well (49, 50). Rippe et al. (15) and Kouba et al. (42) were among the first to demonstrate the NF-κB inhibitory role on collagen regulation in fibroblasts. The inhibitory action of this trans factor was evidenced in other cell types, and the need for interactions with HDAC1 and HDAC2 or two types of corepressor complexes, SMRT-HDAC1 and NCoR-HDAC3, was required (51–53). Additionally, some studies mention a p50 activating effect, particularly in the presence of Bcl-3 (54–56), although others consider it as a repressor, p50 sometimes binding κB sites usually activated by the heterodimer p50-p65 (57, 58). Eventually, the importance of the p50 trans-regulatory effect is relative because p65 is much more implicated in Col1a1 inhibition compared with p50 or c-Rel (15). Although p50 has no trans-activation domain, unlike p65, it is nevertheless a fact that p50 has a crucial role because it allows p50-p65 “anchoring” to the DNA in a half-site κB (59), the other half-site, more degenerated, binding p65 with less affinity (60).
The data of Franzoso et al. (57) and Udalova et al. (58) could explain why in our results COL1A1 transcription activity is not down-regulated after p50 forced expression. However, this result is associated with an inhibitory effect of p50 on the steady-state amounts of COL1A1 and COL1A2 mRNA and collagen neosynthesis as well in FF and ANF. Moreover, p50 forced expression affects neither collagen neosynthesis nor COL1A1 transcription in SF, and this could explain at least in part the scleroderma phenotype characterized by an increased production of extracellular matrix proteins. The fact that the p50 forced expression has no effect on SF, given that the p50 allows anchoring of the p50/65 heterodimer on both types of fibroblasts to the collagen promoter via protein interactions, could be explained by the relative endogenous abundance of p50, whose binding activity would not be affected, defining as a consequence p65 as the limiting factor.
Because of the absence in the −112/−61 bp sequence of a consensus NF-κB binding site governing NF-κB-mediated COL1A1 down-regulation, co-immunoprecipitation assays were performed and revealed that p65 interacts with Sp1, Sp3, CBF, and c-Krox, both in ANF and SF, with an apparently increased affinity for Sp3. There is nothing surprising about NF-κB interacting with other proteins because it has been shown that p65 interacts with at least two components of the transcription basal machinery via the transcription activation domain, TBP and TFIIB, allowing in that way an activation of NF-κB target genes (61, 62).
In the context of TNF-α-induced transcription inhibition of COL1A1, Mori et al. (25) characterized through functional analyses the responsive elements that mapped the sequences −101/−97 bp and −46/−38 bp binding unidentified protein interactions. These data, further corroborated by Rippe et al. (15), revealed that TNF-α effects on type I collagen expression in NIH-3T3 fibroblasts do not act by a consensus NF-κB binding site but via a −220 bp promoter, with a physical and functional interaction between Sp1 and p65.
In the present study, we validated the functional Sp1/p65 interaction by ChIP/re-ChIP but showed additionally that p50, Sp3, and c-Krox interact not only in vitro with p65 but also interact in vivo prior to their binding on the COL1A1 proximal promoter. Although we do not know whether they interact with p65 directly or not (through another common partner/cofactor, for example), these results were correlated with co-immunoprecipitation assays.
In parallel, we demonstrated that Sp1 and Sp3 interact in vivo with the −200/+100 bp region of COL1A1 in ANF and SF. This region was characterized by Artlett et al. (63) as a promoter region sufficient to confer maximal COL1A1 transcription activity. Our results also showed that Sp1 and Sp3 are able to trans-activate COL1A1 in both ANF and SF, which is mediated by a short proximal promoter located 61 bp upstream from the transcription start site or a region located between −112 and −61 bp, respectively, for Sp1 and Sp3. Many studies demonstrated the Sp1 trans-activating function on target genes (64, 65), including genes encoding dermis matrix proteins, such as type I collagen (15, 66, 67). Moreover, Azakie et al. (68) showed that Sp1 up-regulates the expression of troponin, and Sp3 represses this activation. On the contrary, many other studies reported an activating effect of Sp3 (69, 70). This bifunctional activity of Sp3 was already described and could depend on the cellular context, including the nature of the present coactivators and corepressors (14).
Our ChIP experiments, although these assays could not be considered as quantitative, showed that the three zinc fingers up-regulating type I collagen expression (i.e. Sp1/3 and c-Krox), as well as p50 and p65, seem to be more recruited in vivo on the COL1A1 promoter in SF compared with ANF. These zinc fingers, more expressed in SF, could contribute to the drastic increase of collagen synthesis during scleroderma, as demonstrated previously (10, 71). The fact that Sp1 and Sp3 siRNA-induced extinction of the COL1A1 transcription activity appears more pronounced in SF compared with ANF corroborates this hypothesis. Concerning the interaction of p50 and p65 on the COL1A1 promoter that seemed to be also increased in SF, this is much more surprising because the scleroderma phenotype is generally associated with an enhancement of collagen synthesis, whereas NF-κB has an inhibitory effect on collagen production. However, this NF-κB increase, certainly due to the inflammatory status observed in scleroderma, could contribute to the induction of collagen synthesis if the trans-inhibition domain of p65 is masked and/or becomes inactive through particular interactions with specific cofactors of the scleroderma phenotype.
Concerning the particularity of the scleroderma phenotype in fibroblasts, we also demonstrated that, by knocking down Sp1, Sp3, and c-Krox expression in ANF, the p65 inhibitory effect on the type I collagen regulation was no longer observed, whereas only if Sp3 or c-Krox was inhibited in SF was p65 function blocked. These latter data suggest therefore that Sp1, together with Sp3 and c-Krox, act not only as constitutive transcription factors but also as cofactors for the transcription function of p65 in ANF, whereas Sp1 is not necessary for the function of p65 in SF.
Despite these data, the regulatory mechanisms by which p65 regulates COL1A1 activity cannot be defined precisely, and unquestionably, other cofactors or particular interactions, in addition to those mentioned, seem to be essential to the p65 inhibitory effect, which is slightly different between ANF and SF.
In fact, the NF-κB-mediated transcriptional response implicates the participation of numerous coactivators and corepressors, involving direct effects on this transcription factor and changes in chromatin structure (72). More than 120 proteins are able to interact with NF-κB, directly or not, and at least 150 genes can be regulated by NF-κB (47, 72, 73). Moreover, the situation is rendered complex by the fact that the NF-κB corepressors and coactivators can be recruited together to the same promoter, and mutually interfere, suggesting that the NF-κB trans-regulation of a target gene depends on the dynamic balance of these complexes, as demonstrated for the IκBα gene promoter (53). Interestingly, in our model in which p65 inhibits collagen synthesis, the silencing of this subunit (3.5-fold decrease in p65 expression) is associated with an increase in type I collagen synthesis and of the steady-state mRNA amounts of p300 by ∼2-fold (data not shown), indicating that p65 is an inhibitor of this coactivator in ANF.
Overall, our findings reveal that the regulation of human COL1A1 transcription in human dermal fibroblasts involves a multimolecular complex implying at least three trans-activators, Sp1, Sp3, and c-Krox, which could act in concert to up-regulate COL1A1 transcription activity or down-regulate promoter activity when an interaction with NF-κB is taking place in a particular context with corepressors.
Our study also highlighted a new transcription regulatory mechanism for NF-κB, where interaction with other factors becomes as necessary as interaction with basal transcriptional machinery and where recruitment with a consensus κB sequence is not absolutely required because NF-κB can interact with other trans factors, via their own consensus binding sites. Moreover, this study offers new perspectives that may aid in the development of therapeutic strategies that could be applied to localized fibrotic diseases and aging-related skin disorders.
Supplementary Material
Acknowledgments
We thank P. Jalinot (Laboratoire de Biologie Moléculaire de la Cellule, UMR CNRS 5239, Ecole Normale Supérieure de Lyon, IFR 128, Lyon, France) for the generous gift of the p50 and p65 expression vectors. G. Suske (Institut für Molekularbiologie und Tumorforschung, Philipps-Universität, Marburg, Germany) is gratefully acknowledged for providing the Sp1 and Sp3 expression vectors.
This work was supported by the Regional Council of Lower Normandy, the French Ministry of Regional Development, and Johnson & Johnson Consumer France.

This article contains supplemental Figs. S1–S3.
- FF
- foreskin fibroblast(s)
- ANF
- adult normal fibroblast(s)
- SF
- scleroderma fibroblast(s)
- RLU
- relative luciferase units
- IP
- immunoprecipitation
- CBF
- CCAAT-binding factor.
REFERENCES
- 1. Vuorio E., de Crombrugghe B. (1990) The family of collagen genes. Annu. Rev. Biochem. 59, 837–872 [DOI] [PubMed] [Google Scholar]
- 2. Ramirez F., Di Liberto M. (1990) Complex and diversified regulatory programs control the expression of vertebrate collagen genes. FASEB J. 4, 1616–1623 [DOI] [PubMed] [Google Scholar]
- 3. Jimenez S. A., Hitraya E., Varga J. (1996) Pathogenesis of scleroderma. Collagen. Rheum. Dis. Clin. North Am. 22, 647–674 [DOI] [PubMed] [Google Scholar]
- 4. Verrecchia F., Mauviel A. (2004) TGF-β and TNF-α. Antagonistic cytokines controlling type I collagen gene expression. Cell. Signal. 16, 873–880 [DOI] [PubMed] [Google Scholar]
- 5. Trojanowska M., LeRoy E. C., Eckes B., Krieg T. (1998) Pathogenesis of fibrosis. Type 1 collagen and the skin. J. Mol. Med. 76, 266–274 [DOI] [PubMed] [Google Scholar]
- 6. Jimenez S. A., Varga J., Olsen A., Li L., Diaz A., Herhal J., Koch J. (1994) Functional analysis of human α 1(I) procollagen gene promoter. Differential activity in collagen-producing and -nonproducing cells and response to transforming growth factor β1. J. Biol. Chem. 269, 12684–12691 [PubMed] [Google Scholar]
- 7. Tamaki T., Ohnishi K., Hartl C., LeRoy E. C., Trojanowska M. (1995) Characterization of a GC-rich region containing Sp1 binding site(s) as a constitutive responsive element of the α 2(I) collagen gene in human fibroblasts. J. Biol. Chem. 270, 4299–42304 [DOI] [PubMed] [Google Scholar]
- 8. Ihn H., Trojanowska M. (1997) Sp3 is a transcriptional activator of the human α2(I) collagen gene. Nucleic Acids Res. 25, 3712–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ihn H., LeRoy E. C., Trojanowska M. (1997) Oncostatin M stimulates transcription of the human α2(I) collagen gene via the Sp1/Sp3-binding site. J. Biol. Chem. 272, 24666–24672 [DOI] [PubMed] [Google Scholar]
- 10. Kypriotou M., Beauchef G., Chadjichristos C., Widom R., Renard E., Jimenez S. A., Korn J., Maquart F. X., Oddos T., Von Stetten O., Pujol J. P., Galéra P. (2007) Human collagen Krox up-regulates type I collagen expression in normal and scleroderma fibroblasts through interaction with Sp1 and Sp3 transcription factors. J. Biol. Chem. 282, 32000–32014 [DOI] [PubMed] [Google Scholar]
- 11. Majello B., De Luca P., Lania L. (1997) Sp3 is a bifunctional transcription regulator with modular independent activation and repression domains. J. Biol. Chem. 272, 4021–4026 [DOI] [PubMed] [Google Scholar]
- 12. He S., Sun J. M., Li L., Davie J. R. (2005) Differential intranuclear organization of transcription factors Sp1 and Sp3. Mol. Biol. Cell 16, 4073–4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. De Luca P., Majello B., Lania L. (1996) Sp3 represses transcription when tethered to promoter DNA or targeted to promoter proximal RNA. J. Biol. Chem. 271, 8533–8536 [DOI] [PubMed] [Google Scholar]
- 14. Majello B., De Luca P., Suske G., Lania L. (1995) Differential transcriptional regulation of c-myc promoter through the same DNA binding sites targeted by Sp1-like proteins. Oncogene 10, 1841–1848 [PubMed] [Google Scholar]
- 15. Rippe R. A., Schrum L. W., Stefanovic B., Solís-Herruzo J. A., Brenner D. A. (1999) NF-κB inhibits expression of the α1(I) collagen gene. DNA Cell Biol. 18, 751–761 [DOI] [PubMed] [Google Scholar]
- 16. Ghosh S., May M. J., Kopp E. B. (1998) NF-κB and Rel proteins. Evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 16, 225–260 [DOI] [PubMed] [Google Scholar]
- 17. Karin M. (2006) Nuclear factor-κB in cancer development and progression. Nature 441, 431–436 [DOI] [PubMed] [Google Scholar]
- 18. Karin M., Ben-Neriah Y. (2000) Phosphorylation meets ubiquitination. The control of NF-κB activity. Annu. Rev. Immunol. 18, 621–663 [DOI] [PubMed] [Google Scholar]
- 19. Baeuerle P. A., Baltimore D. (1996) NF-κB. Ten years after. Cell 87, 13–20 [DOI] [PubMed] [Google Scholar]
- 20. Lu Y., Fukuda K., Li Q., Kumagai N., Nishida T. (2006) Role of nuclear factor-κB in interleukin-1-induced collagen degradation by corneal fibroblasts. Exp. Eye Res. 83, 560–568 [DOI] [PubMed] [Google Scholar]
- 21. Baeuerle P. A., Baichwal V. R. (1997) NF-κB as a frequent target for immunosuppressive and anti-inflammatory molecules. Adv. Immunol. 65, 111–137 [PubMed] [Google Scholar]
- 22. Okamoto T., Sakurada S., Yang J. P., Merin J. P. (1997) Regulation of NFκB and disease control. Identification of a novel serine kinase and thioredoxin as effectors for signal transduction pathway for NF-κB activation. Curr. Top. Cell Regul. 35, 149–161 [DOI] [PubMed] [Google Scholar]
- 23. Pahl H. L. (1999) Activators and target genes of Rel/NF-κB transcription factors. Oncogene 18, 6853–6866 [DOI] [PubMed] [Google Scholar]
- 24. Solis-Herruzo J. A., Brenner D. A., Chojkier M. (1988) Tumor necrosis factor α inhibits collagen gene transcription and collagen synthesis in cultured human fibroblasts. J. Biol. Chem. 263, 5841–5845 [PubMed] [Google Scholar]
- 25. Mori K., Hatamochi A., Ueki H., Olsen A., Jimenez S. A. (1996) The transcription of human α 1(I) procollagen gene (COL1A1) is suppressed by tumour necrosis factor-α through proximal short promoter elements. Evidence for suppression mechanisms mediated by two nuclear factor binding sites. Biochem. J. 319, 811–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Buss H., Dörrie A., Schmitz M. L., Frank R., Livingstone M., Resch K., Kracht M. (2004) Phosphorylation of serine 468 by GSK-3β negatively regulates basal p65 NF-κB activity. J. Biol. Chem. 279, 49571–49574 [DOI] [PubMed] [Google Scholar]
- 27. Zhong H., SuYang H., Erdjument-Bromage H., Tempst P., Ghosh S. (1997) The transcriptional activity of NF-κB is regulated by the IκB-associated PKAc subunit through a cyclic AMP-independent mechanism. Cell 89, 413–424 [DOI] [PubMed] [Google Scholar]
- 28. Greene W. C., Chen L. F. (2004) Regulation of NF-κB action by reversible acetylation. Novartis Found. Symp. 259, 208–217; discussion 218–225 [PubMed] [Google Scholar]
- 29. Quivy V., Van Lint C. (2004) Regulation at multiple levels of NF-κB-mediated transactivation by protein acetylation. Biochem. Pharmacol. 68, 1221–1229 [DOI] [PubMed] [Google Scholar]
- 30. Ryo A., Suizu F., Yoshida Y., Perrem K., Liou Y. C., Wulf G., Rottapel R., Yamaoka S., Lu K. P. (2003) Regulation of NF-κB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol. Cell 12, 1413–1426 [DOI] [PubMed] [Google Scholar]
- 31. Desterro J. M., Rodriguez M. S., Hay R. T. (1998) SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell 2, 233–239 [DOI] [PubMed] [Google Scholar]
- 32. Marshall H. E., Merchant K., Stamler J. S. (2000) Nitrosation and oxidation in the regulation of gene expression. FASEB J. 14, 1889–1900 [DOI] [PubMed] [Google Scholar]
- 33. Peterkofsky B., Diegelmann R. (1971) Use of a mixture of proteinase-free collagenases for the specific assay of radioactive collagen in the presence of other proteins. Biochemistry 10, 988–994 [DOI] [PubMed] [Google Scholar]
- 34. Goldberg H., Helaakoski T., Garrett L. A., Karsenty G., Pellegrino A., Lozano G., Maity S., de Crombrugghe B. (1992) Tissue-specific expression of the mouse α 2(I) collagen promoter. Studies in transgenic mice and in tissue culture cells. J. Biol. Chem. 267, 19622–19630 [PubMed] [Google Scholar]
- 35. Chomczynski P., Sacchi N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159 [DOI] [PubMed] [Google Scholar]
- 36. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔ C(T)) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 37. Andrews N. C., Faller D. V. (1991) A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19, 2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ghayor C., Chadjichristos C., Herrouin J. F., Ala-Kokko L., Suske G., Pujol J. P., Galera P. (2001)) Sp3 represses the Sp1-mediated transactivation of the human COL2A1 gene in primary and dedifferentiated chondrocytes. J. Biol. Chem. 276, 36881–36895 [DOI] [PubMed] [Google Scholar]
- 39. Reuter U., Chiarugi A., Bolay H., Moskowitz M. A. (2002) Nuclear factor-κB as a molecular target for migraine therapy. Ann. Neurol. 51, 507–516 [DOI] [PubMed] [Google Scholar]
- 40. Tassorelli C., Greco R., Morazzoni P., Riva A., Sandrini G., Nappi G. (2005) Parthenolide is the component of tanacetum parthenium that inhibits nitroglycerin-induced Fos activation. Studies in an animal model of migraine. Cephalalgia 25, 612–621 [DOI] [PubMed] [Google Scholar]
- 41. Inagaki Y., Truter S., Tanaka S., Di Liberto M., Ramirez F. (1995) Overlapping pathways mediate the opposing actions of tumor necrosis factor-α and transforming growth factor-β on α 2(I) collagen gene transcription. J. Biol. Chem. 270, 3353–3358 [DOI] [PubMed] [Google Scholar]
- 42. Kouba D. J., Chung K. Y., Nishiyama T., Vindevoghel L., Kon A., Klement J. F., Uitto J., Mauviel A. (1999) Nuclear factor-κB mediates TNF-alpha inhibitory effect on α 2(I) collagen (COL1A2) gene transcription in human dermal fibroblasts. J. Immunol. 162, 4226–4234 [PubMed] [Google Scholar]
- 43. Iraburu M. J., Domínguez-Rosales J. A., Fontana L., Auster A., García-Trevijano E. R., Covarrubias-Pinedo A., Rivas-Estilla A. M., Greenwel P., Rojkind M. (2000) Tumor necrosis factor α down-regulates expression of the α1(I) collagen gene in rat hepatic stellate cells through a p20C/EBPβ- and C/EBPδ-dependent mechanism. Hepatology 31, 1086–1093 [DOI] [PubMed] [Google Scholar]
- 44. García-Trevijano E. R., Iraburu M. J., Fontana L., Domínguez-Rosales J. A., Auster A., Covarrubias-Pinedo A., Rojkind M. (1999) Transforming growth factor β1 induces the expression of α1(I) procollagen mRNA by a hydrogen peroxide-C/EBPβ-dependent mechanism in rat hepatic stellate cells. Hepatology 29, 960–970 [DOI] [PubMed] [Google Scholar]
- 45. Varela-Rey M., Montiel-Duarte C., Osés-Prieto J. A., López-Zabalza M. J., Jaffrèzou J. P., Rojkind M., Iraburu M. J. (2002) p38 MAPK mediates the regulation of α1(I) procollagen mRNA levels by TNF-α and TGF-β in a cell line of rat hepatic stellate cells. FEBS Lett. 528, 133–138 [DOI] [PubMed] [Google Scholar]
- 46. Bitzer M., von Gersdorff G., Liang D., Dominguez-Rosales A., Beg A. A., Rojkind M., Böttinger E. P. (2000) A mechanism of suppression of TGF-β/SMAD signaling by NF-κB/RelA. Genes Dev. 14, 187–197 [PMC free article] [PubMed] [Google Scholar]
- 47. Zhou A., Scoggin S., Gaynor R. B., Williams N. S. (2003) Identification of NF-κB-regulated genes induced by TNFα utilizing expression profiling and RNA interference. Oncogene 22, 2054–2064 [DOI] [PubMed] [Google Scholar]
- 48. Yamane K., Ihn H., Asano Y., Jinnin M., Tamaki K. (2003) Antagonistic effects of TNF-α on TGF-β signaling through down-regulation of TGF-beta receptor type II in human dermal fibroblasts. J. Immunol. 171, 3855–3862 [DOI] [PubMed] [Google Scholar]
- 49. Vermes C., Roebuck K. A., Chandrasekaran R., Dobai J. G., Jacobs J. J., Glant T. T. (2000) Particulate wear debris activates protein tyrosine kinases and nuclear factor κB, which down-regulates type I collagen synthesis in human osteoblasts. J. Bone Miner. Res. 15, 1756–1765 [DOI] [PubMed] [Google Scholar]
- 50. Aoki T., Kataoka H., Ishibashi R., Nozaki K., Morishita R., Hashimoto N. (2009) Reduced collagen biosynthesis is the hallmark of cerebral aneurysm. Contribution of interleukin-1β and nuclear factor-κB. Arterioscler. Thromb. Vasc. Biol. 29, 1080–1086 [DOI] [PubMed] [Google Scholar]
- 51. Schreiber J., Efron P. A., Park J. E., Moldawer L. L., Barbul A. (2005) Adenoviral gene transfer of an NF-κB super-repressor increases collagen deposition in rodent cutaneous wound healing. Surgery 138, 940–946 [DOI] [PubMed] [Google Scholar]
- 52. Ashburner B. P., Westerheide S. D., Baldwin A. S. (2001) The p65 (RelA) subunit of NF-κB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol. Cell. Biol. 21, 7065–7077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gao Z., Chiao P., Zhang X., Zhang X., Lazar M. A., Seto E., Young H. A., Ye J. (2005) Coactivators and corepressors of NF-κB in IκB α gene promoter. J. Biol. Chem. 280, 21091–21098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dechend R., Hirano F., Lehmann K., Heissmeyer V., Ansieau S., Wulczyn F. G., Scheidereit C., Leutz A. (1999) The Bcl-3 oncoprotein acts as a bridging factor between NF-κB/Rel and nuclear co-regulators. Oncogene 18, 3316–3323 [DOI] [PubMed] [Google Scholar]
- 55. Kim J. M., Oh Y. K., Kim Y. J., Youn J., Ahn M. J. (2004) Escherichia coli up-regulates proinflammatory cytokine expression in granulocyte/macrophage lineages of CD34 stem cells via p50 homodimeric NF-κB. Clin. Exp. Immunol. 137, 341–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kurland J. F., Kodym R., Story M. D., Spurgers K. B., McDonnell T. J., Meyn R. E. (2001) NF-κB1 (p50) homodimers contribute to transcription of the bcl-2 oncogene. J. Biol. Chem. 276, 45380–45386 [DOI] [PubMed] [Google Scholar]
- 57. Franzoso G., Bours V., Park S., Tomita-Yamaguchi M., Kelly K., Siebenlist U. (1992) The candidate oncoprotein Bcl-3 is an antagonist of p50/NF-κB-mediated inhibition. Nature 359, 339–342 [DOI] [PubMed] [Google Scholar]
- 58. Udalova I. A., Richardson A., Denys A., Smith C., Ackerman H., Foxwell B., Kwiatkowski D. (2000) Functional consequences of a polymorphism affecting NF-κB p50-p50 binding to the TNF promoter region. Mol. Cell. Biol. 20, 9113–9119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Urban M. B., Baeuerle P. A. (1991) The role of the p50 and p65 subunits of NF-κB in the recognition of cognate sequences. New Biol. 3, 279–288 [PubMed] [Google Scholar]
- 60. Baeuerle P. A. (1991) The inducible transcription activator NF-κB. Regulation by distinct protein subunits. Biochim. Biophys. Acta 1072, 63–80 [DOI] [PubMed] [Google Scholar]
- 61. Schmitz M. L., Stelzer G., Altmann H., Meisterernst M., Baeuerle P. A. (1995) Interaction of the COOH-terminal transactivation domain of p65 NF-κB with TATA-binding protein, transcription factor IIB, and coactivators. J. Biol. Chem. 270, 7219–7226 [DOI] [PubMed] [Google Scholar]
- 62. Zhu N. L., Li C., Huang H. H., Sebald M., Londhe V. A., Heisterkamp N., Warburton D., Bellusci S., Minoo P. (2007) TNF-α represses transcription of human bone morphogenetic protein-4 in lung epithelial cells. Gene 393, 70–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Artlett C. M., Chen S. J., Varga J., Jimenez S. A. (1998) Modulation of basal expression of the human α1(I) procollagen gene (COL1A1) by tandem NF-1/Sp1 promoter elements in normal human dermal fibroblasts. Matrix Biol. 17, 425–434 [DOI] [PubMed] [Google Scholar]
- 64. Chun J. Y., Hu Y., Pinder E., Wu J., Li F., Gao A. C. (2007) Selenium inhibition of survivin expression by preventing Sp1 binding to its promoter. Mol. Cancer Ther. 6, 2572–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kum Y. S., Kim K. H., Park T. I., Suh I. S., Oh H. K., Cho C. H., Park J. B., Chang Y. C., Park J. H., Lee K. G., Park K. K. (2007) Antifibrotic effect via the regulation of transcription factor Sp1 in lung fibrosis. Biochem. Biophys. Res. Commun. 363, 368–374 [DOI] [PubMed] [Google Scholar]
- 66. Ihn H., Tamaki K. (2000) Increased phosphorylation of transcription factor Sp1 in scleroderma fibroblasts. Association with increased expression of the type I collagen gene. Arthritis Rheum. 43, 2240–2247 [DOI] [PubMed] [Google Scholar]
- 67. Verrecchia F., Rossert J., Mauviel A. (2001) Blocking sp1 transcription factor broadly inhibits extracellular matrix gene expression in vitro and in vivo. Implications for the treatment of tissue fibrosis. J. Invest. Dermatol. 116, 755–763 [DOI] [PubMed] [Google Scholar]
- 68. Azakie A., Fineman J. R., He Y. (2006) Sp3 inhibits Sp1-mediated activation of the cardiac troponin T promoter and is down-regulated during pathological cardiac hypertrophy in vivo. Am. J. Physiol. Heart Circ. Physiol. 291, H600–H611 [DOI] [PubMed] [Google Scholar]
- 69. Teunissen B. E., van Amersfoorth S. C., Opthof T., Jongsma H. J., Bierhuizen M. F. (2002) Sp1 and Sp3 activate the rat connexin40 proximal promoter. Biochem. Biophys. Res. Commun. 292, 71–78 [DOI] [PubMed] [Google Scholar]
- 70. Wang J., Bannon M. J. (2005) Sp1 and Sp3 activate transcription of the human dopamine transporter gene. J. Neurochem. 93, 474–482 [DOI] [PubMed] [Google Scholar]
- 71. Hitraya E. G., Varga J., Artlett C. M., Jiménez S. A. (1998) Identification of elements in the promoter region of the α1(I) procollagen gene involved in its up-regulated expression in systemic sclerosis. Arthritis Rheum. 41, 2048–2058 [DOI] [PubMed] [Google Scholar]
- 72. Chen L. F., Greene W. C. (2003) Regulation of distinct biological activities of the NF-κB transcription factor complex by acetylation. J. Mol. Med. 81, 549–557 [DOI] [PubMed] [Google Scholar]
- 73. Karin M., Cao Y., Greten F. R., Li Z. W. (2002) NF-κB in cancer. From innocent bystander to major culprit. Nat. Rev. Cancer 2, 301–310 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.