Background: Human CYP3A4 metabolizes the majority of administered drugs including bromoergocryptine (BEC), a dopamine receptor agonist.
Results: Crystallographic and experimental data suggest the importance of Arg212 and Thr224 in BEC binding.
Conclusion: H-bonding interactions with Thr224 and conformational adjustments modulated by Arg212 are critical for the productive orientation of BEC.
Significance: Mechanistic insights on the CYP3A4-BEC interaction may help develop new and safer pharmaceuticals.
Keywords: Crystal Structure, Cytochrome P450, Drug Metabolism, Enzyme Mechanisms, Enzyme Mutation, Bromoergocryprine
Abstract
Cytochrome P4503A4 (CYP3A4), a major human drug-metabolizing enzyme, is responsible for the oxidation and clearance of the majority of administered drugs. One of the CYP3A4 substrates is bromoergocryptine (BEC), a dopamine receptor agonist prescribed for the inhibition of prolactin secretion and treatment of Parkinson disease, type 2 diabetes, and several other pathological conditions. Here we present a 2.15 Å crystal structure of the CYP3A4-BEC complex in which the drug, a type I heme ligand, is bound in a productive mode. The manner of BEC binding is consistent with the in vivo metabolite analysis and identifies the 8′ and 9′ carbons of the proline ring as the primary sites of oxidation. The crystal structure predicts the importance of Arg212 and Thr224 for binding of the tripeptide and lysergic moieties of BEC, respectively, which we confirmed experimentally. Our data support a three-step BEC binding model according to which the drug binds first at a peripheral site without perturbing the heme spectrum and then translocates into the active site cavity, where formation of a hydrogen bond between Thr224 and the N1 atom of the lysergic moiety is followed by a slower conformational readjustment of the tripeptide group modulated by Arg212.
Introduction
Cytochrome P450 enzymes are heme-thiolate proteins that catalyze a wide variety of monooxygenation reactions including hydroxylation, epoxidation, and heteroatom dealkylations (1). Among 57 human P450s, the 3A4 isoform (CYP3A4) is one of the most abundant and important because, in addition to oxidation of various endogenous molecules and xenobiotics, it contributes to the clearance of over 50% of administered pharmaceuticals (2, 3). The high capacity of CYP3A4 to oxidize molecules diverse in size and chemical structure is due to its large and malleable active site (4, 5). Another intriguing feature of CYP3A4 is the ability to accommodate more than one molecule in the substrate-binding pocket, where one molecule serves as a substrate while another acts as a modulator of substrate metabolism. Among the substrates that exhibit binding cooperativity with CYP3A4 are testosterone, progesterone, diazepam, α-naphthoflavone, and several others (6–8).
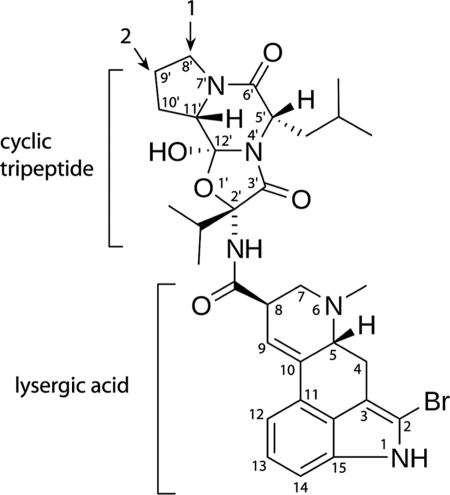
A CYP3A4 substrate that does not exhibit binding cooperativity is bromoergocryptine (BEC2; also known as bromocryptine) (9–11). BEC is an ergot alkaloid that acts as a dopamine receptor agonist. Currently, BEC is prescribed for inhibition of prolactin secretion, the treatment of Parkinson disease, type 2 diabetes, migraines, and pituitary tumors, and correction of abnormal secretion of growth hormone. BEC is one of the largest CYP3A4 substrates (molecular mass of 655) and consists of lysergic acid and a proline-containing cyclic tripeptide linked by an amide bond (Fig. 1). The tripeptide group is critical for the interaction with P450s of the 3A family (12). BEC has a high affinity for microsomal and recombinant CYP3A4 (Kd of 0.3–1 μm) and upon binding induces type I spectral changes (a low-to-high spin shift) accompanied by a 80-mV increase in the heme redox potential (11–17). In vivo metabolite analysis has revealed that BEC is oxidized primarily by CYP3A4 at the pyrrolidine moiety, with the 8′-mono- and 8′,9′-dihydroxy derivatives being the major products (Fig. 1) (18, 19).
FIGURE 1.
Structure of BEC. Two primary hydroxylation sites are indicated by arrows.
Although determination of the 2.7 Å crystal structure of BEC-bound CYP3A4 has been reported at a scientific meeting (20), the atomic coordinates are still unavailable. Moreover, to date, there is no structural information on any other type I ligand bound to CYP3A4 in a productive mode. To obtain insights into the mechanism of substrate binding to CYP3A4, we determined and report here the 2.15 Å crystal structure of the CYP3A4-BEC complex. The x-ray data suggest the importance of Arg212 and Thr224 for optimal BEC binding, which we confirmed experimentally using mutagenesis, spectroscopic, and kinetic techniques.
MATERIALS AND METHODS
Protein Expression, Purification, and Mutagenesis
R212A and T224A mutations were introduced to the CYP3A4Δ3–22 expression plasmid using a QuikChange mutagenesis kit (Stratagene). The wild type (WT) and mutants of CYP3A4 were expressed and purified as described previously (21).
Crystallization and Structure Determination
The CYP3A4-BEC complex was crystallized by a micro batch method under oil. Half a microliter of the BEC-bound protein (20–25 mg/ml) in 20 mm phosphate, pH 7.7, 20% glycerol, and 100 mm NaCl was mixed with 0.5 μl of 4% tacsimate, pH 5.0, and 12% polyethylene glycol 3350 (solution No. 11 from the Hampton Research PEG/Ion 2 kit) and then covered with 10 μl of paraffin oil. On the next day crystals were harvested and frozen in Paratone oil, used as a cryoprotectant. X-ray diffraction data were collected at the Stanford Synchrotron Radiation Laboratory beamline 7-1. The structure was solved by molecular replacement with PHASER (22) using ligand-free CYP3A4 (Protein Data Bank ID code 1TQN) as a search model. The initial model was rebuilt and refined with COOT (23) and REFMAC (22). The N and C termini as well as residues 266–268 and 280–286 are not present in the final structure because of conformational disorder. Data collection and refinement statistics are given in Table 1. The atomic coordinates have been deposited to the Protein Data Bank with the ID code 3UA1.
TABLE 1.
Data collection and refinement statistics
| Data statistics | |
| Space group | I222 |
| Unit cell parameters | a = 78 Å, b = 99 Å, c = 132 Å |
| α, β, γ = 90° | |
| Resolution range | 49.7–2.15 (2.27–2.15)a |
| Total reflections | 144,569 (15,811) |
| Unique reflections | 27,296 (3,721) |
| Redundancy | 5.3 (4.2) |
| Completeness | 97.2 (92.2) |
| Average I/σI | 12.1 (2.1) |
| Rmerge | 0.059 (0.53) |
| Refinement statistics | |
| Molecules per asymmetric unit | 1 |
| R/Rfreeb | 22.1/27.8 |
| Average B-factor (Å2) | 36.2 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.013 |
| Bond angles (°) | 1.64 |
a All values in parentheses are for the highest resolution shell.
b Rfree was calculated from a subset of 5% of the data that were excluded during refinement.
Spectral Binding Titrations
Binding of BEC (Sigma) to the WT and mutants of CYP3A4 was monitored in 50 mm phosphate buffer, pH 7.4, containing 20% glycerol and 1 mm dithiothreitol. Spectra were recorded after the addition of small amounts of BEC dissolved in dimethyl sulfoxide; the total volume of the solvent added was <2% (v/v). The difference in absorbance between the wavelength maximum and minimum was plotted against the concentration of BEC, and the spectral dissociation constant (Ks) was calculated using quadratic nonlinear regression analysis as described elsewhere (15).
Kinetics of Ligand Binding
The kinetics of BEC binding to CYP3A4 was monitored at ambient temperature in a SX.18MV stopped flow apparatus (Applied Photophysics) in the absence and presence of Emulgen 913 (Kao Chemicals, Japan), IGEPAL CA-630 (Sigma) and CHAPS (Sigma). CYP3A4 solutions (6 μm) in 50 mm phosphate buffer, pH 7.4, were mixed with 0.125–36 μm BEC to follow absorbance changes at 417 nm. Owing to low solubility of BEC, 36 μm was the maximal concentration that could be reached under our experimental conditions. In a separate experiment, 2 μm CYP3A4 was mixed with 2–36 μm BEC to confirm that the rate constant for BEC ligation starts to level off when the BEC:CYP3A4 ratio exceeds 2. Kinetic data were analyzed using the IgorPro program (WaveMetrics, Inc.).
RESULTS
Soluble CYP3A4 Has High Affinity for BEC
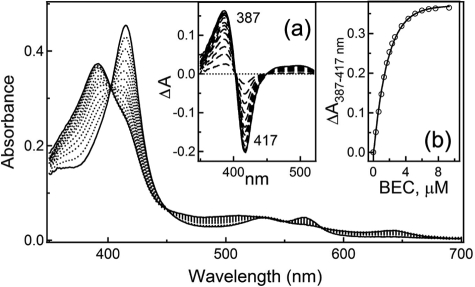
As interaction of truncated human CYP3A4Δ3–22 with BEC had not been investigated previously, we performed equilibrium titrations to determine a spectral dissociation constant (Ks) for the drug. As seen in Fig. 2, the addition of a 2-fold excess of BEC to soluble CYP3A4 leads to a nearly complete 417 to 387-nm shift in the absorbance maximum (type I spectral changes), indicative of displacement of the coordinated water molecule and conversion of the heme iron to a high spin form. The estimated Ks value (0.37 ± 0.02 μm) suggests that BEC binds to the truncated form of CYP3A4 as tightly as to the full-length hemoprotein, whose dissociation constant for BEC ranges from 0.3 to 1 μm depending on the determination method and protein source (11–13, 15, 16).
FIGURE 2.
Equilibrium titration of WT CYP3A4 with BEC. Inset a, spectral changes in 4.5 μm CYP3A4 with varying BEC concentration. Inset b, plot of ΔA387–417 nm (from Inset a) versus BEC concentration.
Crystal Structure of the CYP3A4-BEC Complex
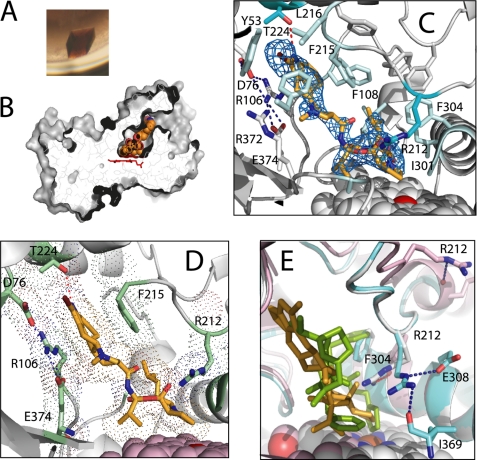
Owing to the high affinity of the CYP3A4-BEC complex, it was possible to maintain CYP3A4 in the BEC-bound form during crystallization. The deep brown color of the crystals was the first indication that crystalline CYP3A4 forms a high spin complex with BEC (Fig. 3A). The well defined electron density of the BEC molecule shows that the drug is bound in the active site in an extended conformation, with an angle between the tripeptide and lysergic groups of 130° (Fig. 3, B and C). As metabolic analyses have predicted (12, 18, 19), BEC approaches the heme via the tripeptide moiety (Fig. 3, C and D), with the primary sites of oxidation, the 8′ and 9′ carbons of the proline ring (Fig. 1), being 4.1 and 3.7 Å, respectively, away from the iron. Thus, in the crystal structure BEC is bound in a productive mode.
FIGURE 3.
Crystal structure of the CYP3A4-BEC complex. A, brown-colored crystals of BEC-bound CYP3A4 indicate that the heme iron is in a high spin form. B, BEC binds to CYP3A4 in an extended conformation. The active site cavity is connected to the solvent and has a volume of 563 Å3 (calculated with LIGSITE (32)). C, the active site residues interacting with BEC. Residues making van der Waals contacts with the drug are in pale cyan. The critical residues, Arg212 and Thr224, are highlighted in darker cyan. Arg212 controls the BEC approach to the heme iron, whereas Thr224 establishes an H-bond with the BEC N1 atom. The 2Fo − Fc electron density map around BEC is contoured at 1 σ. D, another view at the active site showing the lysergic moiety of BEC sandwiched between the parallel Arg106 and Phe215 side chains. E, comparison of the Arg212 conformations in substrate-free (1TQN (gray)), BEC-bound (cyan), and ritonavir-bound CYP3A4 (3NXU (pink)). BEC is in yellow and ritonavir in green. To accommodate BEC, the Arg212 side chain moves aside and establishes hydrogen bonds with the side and main chain atoms of Glu308 and Ile369.
The tripeptide group does not establish any specific polar or electrostatic interactions but makes extensive van der Waals contacts with Ile301, Phe304, and Ala305 from the I-helix and with the side and main chains of Arg105, Arg212, Ala370, and Arg372. To allow the tripeptide moiety to approach the heme iron, Arg212 adopts a new rotamer relative to the ligand-free structure (29). This conformation is stabilized by two hydrogen bonds formed between the Arg212 guanidinium group, the carboxyl of Glu308, and the carbonyl oxygen of Ile369 (Fig. 3E). The lysergic moiety of BEC is sandwiched between the parallel Arg106 and Phe215 side chains and H-bonded to Thr224 from the G′-helix via the N1 atom (Fig. 3, C and D). To accommodate this bulky group, the Pro107–Gly109 peptide shifts aside by 2.4 Å. The fact that the drug establishes only one hydrogen bond and there are no well defined waters or water-mediated contacts in the active site allows us to conclude that the CYP3A4-BEC interactions are predominantly non-polar.
Rationale for Mutagenesis
Despite the large size and complex chemical structure of BEC, only small changes in the CYP3A4 conformation are needed to position the substrate optimally for oxidation. As mentioned, one notable rearrangement is in the Arg212 side chain but not the associated peptide backbone. This is in contrast to other large ligands such as ritonavir, ketoconazole, and erythromycin, where a major conformational change in the Arg212-containing FF′-loop occurs upon binding (Fig. 3E) (4, 21). Because the tripeptide moiety of BEC binds next to Arg212, we anticipated that elimination of the Arg212 side chain would increase the volume of the active site cavity near the heme iron and, hence, should affect the affinity and/or binding kinetics of the drug. Thr224, on the other hand, assists BEC binding by H-bonding with the lysergic group (Fig. 3, C and D). Elimination of this hydrogen bond could increase motional freedom and prevent proper positioning of the drug, which could be manifested through changes in the extent and kinetics of a spin shift in CYP3A4. To test these predictions, we replaced Arg212 and Thr224 with alanine to determine how single and double mutations affect the CYP3A4-BEC interaction.
Arg212 and Thr224 Define Affinity and Facilitate BEC Binding to CYP3A4
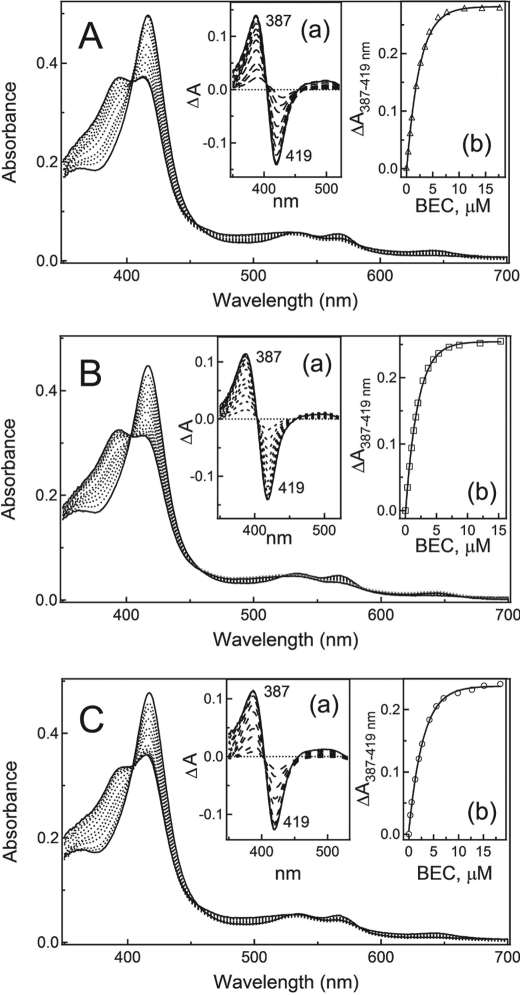
Equilibrium titrations show (Fig. 4) that, at saturating BEC levels, there is only a partial conversion of the mutants to a high spin form: 56, 60, and 52% for CYP3A4 R212A, T224A, and R212A/T224A, respectively, as opposed to ∼90% for the WT. The Ks value calculated for the T224A variant was close to that of the WT, whereas the corresponding constants for the R212A and double mutants were 6-fold higher (Table 2). This implies that during equilibrium binding, Arg212 is more critical for the BEC binding than Thr224.
FIGURE 4.
Equilibrium titration of CYP3A4 R212A (A), T224A (B), and R212A/T224A (C) with BEC. Absorbance changes in 4.3–4.7 μm CYP3A4 (a insets) were plotted against the BEC concentration (b insets). The derived Ks values are given in Table 2.
TABLE 2.
Equilibrium and kinetic parameters for the reaction of BEC binding to WT and mutants of CYP3A4
| WT | R212A | T224A | R212A/T224A | |
|---|---|---|---|---|
| Ksa (μm) | 0.37 ± 0.02 | 2.3 ± 0.3 | 0.44 ± 0.03 | 2.5 ± 0.2 |
| kfastb (s−1) | 0.66 ± 0.02 | 0.87 ± 0.03 | 0.46 ± 0.03 | 0.56 ± 0.02 |
| kslowc (s−1) | 0.035 ± 0.002 | 0.034 ± 0.002 | 0.039 ± 0.003 | 0.074 ± 0.002 |
a Spectroscopic dissociation constant.
b Rate constant for ligand binding in the fast phase, determined at BEC:CYP3A4 = 5.
c Rate constant for ligand binding in the slow phase, determined at BEC:CYP3A4 = 5.
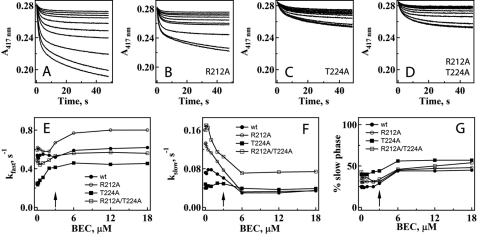
To determine the mutational effects on the kinetics of BEC association, we monitored the disappearance of the low spin form as an absorbance decrease at 417 nm after mixing CYP3A4 and BEC in a stopped flow spectrophotometer. Consistent with the incomplete shift from low to high spin when BEC was added to the mutants, the change in amplitude at 417 nm was less for both mutants: 33% for CYP3A4 R212A, and 63% for the T224A and double mutants compared with the WT (Fig. 5, A–D). In addition, the kinetics of BEC binding to the WT and variants was biphasic under all studied conditions. The rate constants for the fast phase of BEC binding (kfast) were independent of the BEC concentration at subequimolar BEC:CYP3A4 ratios (<0.5) and then gradually increased and remained unchanged after the BEC:CYP3A4 ratio exceeded 2 (Fig. 5E). A comparison of the kfast values calculated at saturating levels of BEC (Table 2) indicates that BEC binds faster to the R212A mutant (∼32% increase in kfast), whereas the T224A mutation slows down the spin conversion by 30%.
FIGURE 5.
Effect of R212A, T224A, and R212A/T224A mutations in CYP3A4 on the kinetics of BEC binding. A–D, kinetics of BEC binding to the WT and R212A, T224A, and R212A/T224A mutants of CYP3A4, respectively. Solutions of 0.12, 0.25, 0.5, 1, 2, 4, 6, 12, 24, and 36 μm BEC (top to bottom traces, respectively) were mixed with 6 μm CYP3A4, and the low-to-high spin conversion was monitored at 417 nm. The kinetics were biphasic for the whole range of BEC concentrations studied. E and F, observed rate constants for the fast (kfast) and slow phases (kslow), respectively, are plotted versus BEC concentration. The rate constants calculated at saturating BEC are given in Table 2. G, effect of the BEC concentration on the relative percentage of the slow phase. The arrows indicate where the BEC and CYP3A4 concentrations are equal.
For CYP3A4 T224A, the rate constant for the slow phase (kslow) was independent of BEC concentration (Fig. 5F). For other proteins, kslow decreased to a different extent until the BEC:CYP3A4 ratio reached 2; this remained unchanged at higher drug concentrations. The most notable changes in the slow phase were observed for the R212A and double mutants. Stopped flow measurements also revealed that regardless of whether or not mutations were present, the percentage of the slow phase changed sharply from ∼30–35% to 50–55% when the BEC:protein ratio exceeded unity (Fig. 5G). Because the latter effect could be due to conformational heterogeneity of CYP3A4 (14), we checked whether the biphasicity of the BEC binding reaction could be eliminated in the presence of detergents.
BEC Binding Kinetics Remains Biphasic in the Presence of Detergents
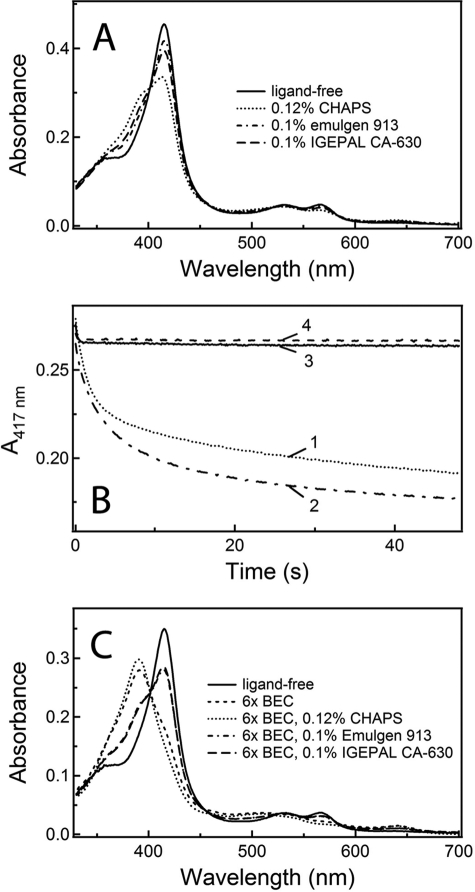
The kinetics of BEC binding to WT CYP3A4 was measured in the presence of two non-ionic detergents, Emulgen 913 and IGEPAL CA-630, as well as zwitterionic CHAPS. All detergents were used at a final concentration of 0.1–0.12%, which is lower than the critical micelle concentration and sufficient to dissociate aggregates of full-length CYP1A2 and 2B4 to catalytically active dimers (24).
Spectral measurements showed that the addition of CHAPS, Emulgen 913, and IGEPAL CA-630 to CYP3A4 caused partial low-to-high spin shifts (53, 14, and 27%, respectively (Fig. 6A)). This means that each compound binds near the heme and displaces the distal water ligand or partially stabilizes a conformer that favors water ligand displacement. When CHAPS-bound CYP3A4 was mixed with BEC, a large decrease in 417-nm absorbance was detected (Fig. 6B). Further, as observed in a detergent-free buffer, the kinetics of CYP3A4-BEC complex formation in the presence of CHAPS was biphasic, with kfast and kslow values of 0.66 and 0.06 s−1, respectively, and the percentage of the slow phase was ∼50%. According to an absorbance spectrum recorded at the end of the stopped flow experiment (Fig. 6C), the conversion of CYP3A4 to a high spin form was near completion (∼100%) at saturating BEC levels.
FIGURE 6.
Effect of detergents on the CYP3A4-BEC interaction. A, spectral changes in CYP3A4 caused by 0.1–0.12% CHAPS, Emulgen 913, and IGEPAL CA-630. B, absorbance changes at 417 nm observed upon mixing 6 μm WT CYP3A4 with 36 μm BEC in the absence and presence of 0.12% CHAPS, 0.1% Emulgen 913, and 0.1% IGEPAL CA-630 (traces 1–4, respectively). C, absorbance spectra of CYP3A4 recorded at the end of the stopped flow experiments shown in B.
When Emulgen 913 or IGEPAL CA-630 was present in the medium, the BEC-induced absorbance changes were very small (Fig. 6B). Nevertheless, the reaction remained biphasic at BEC:CYP3A4 ratios of >1 (kfast and kslow of 5–8 and 0.02–0.04 s−1, respectively), with the fast phase accounting for 90% of the total absorbance change. At subequimolar ratios, the ligation of BEC was slow (0.01–0.05 s−1) and monophasic. A BEC-dependent increase in the high spin content reached only 21 and 7% for Emulgen- and IGEPAL-bound CYP3A4, respectively (Fig. 6C). Thus, non-ionic detergents significantly interfere with the BEC binding and, similar to CHAPS, do not eliminate the biphasicity of the reaction.
DISCUSSION
Co-crystallization of CYP3A4 with substrates/type I ligands is very challenging because most of these have low affinity (Kd of 5–150 μm) and low solubility in aqueous solutions. In the two currently available substrate-bound structures of CYP3A4, erythromycin is bound in a nonproductive mode (4), whereas progesterone is docked outside of the active site pocket (25). The present CYP3A4-BEC complex is the first in which the substrate is bound in a mode suitable for oxidation.
Surprisingly, no major conformational changes in CYP3A4 were required to accommodate a large BEC molecule (the root mean square deviation between the ligand-free and BEC-bound structures is only 0.32 Å). The crystal structure shows that two residues, Arg212 and Thr224, may be important for association and optimal orientation of the drug. Arg212, part of the FF′-loop (residues 210–214), is thought to be actively involved in substrate binding and the mediation of cooperativity in steroid hydroxylation reactions (26, 27). Although Arg212 plays no significant role in the hydroxylation of relatively small steroids (26, 27), it is predicted to influence the action of the effector molecule and control the substrate orientation via interactions with Phe304, supposedly located at the interface between the active and the effector binding sites (28).
In our structure, Arg212 is positioned strategically near the tripeptide group of BEC and establishes interactions with Glu308 and Ile369. This led us to hypothesize that Arg212, as well as the H-bond-forming Thr224, could be important for the affinity and kinetics of BEC binding. Indeed, our experimental data show that substitution of Arg212 or/and Thr224 with alanine alters both the extent of the low-to-high spin shift and the rate of BEC binding. The reaction of BEC ligation to the WT or mutants of CYP3A4 was biphasic, with non-hyperbolic dependence of kfast on BEC concentration. As the drug is known to bind to CYP3A4 stoichiometrically and non-cooperatively (11, 15), such kinetics cannot be explained by the allosteric properties of the enzyme.
One possible reason follows from a previous study in which three kinetic phases were distinguished (20, ∼1, and 0.009 s−1) based on fluorescence and absorbance changes observed during association of BEC to the full-length CYP3A4 (15). The fastest step, detected only by fluorescence spectroscopy, was proposed to correspond to the peripheral binding of BEC, proceeding without perturbations in the heme spectrum. The two subsequent steps, in turn, are thought to reflect the interaction of the second BEC molecule with the active site and the heme. Our data are consistent with this model and provide further insights into the mechanism of BEC association.
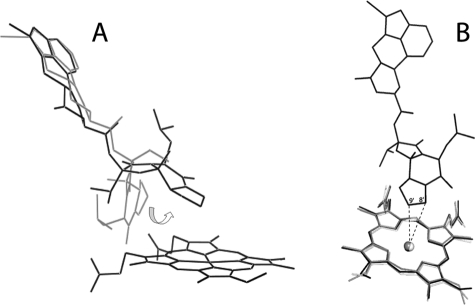
Because BEC has low solubility in aqueous solutions, it was not possible to study the binding kinetics under pseudo-first order conditions over a wide range of BEC concentrations (Fig. 5, E and F). Nonetheless, the range studied was sufficient to estimate the limiting kfast/slow values, because BEC is a high affinity ligand whose binding rate becomes concentration-independent when the BEC:CYP3A4 ratio exceeds 2. Small changes in binding rates at low BEC concentrations mean that an event taking place remotely from the heme (e.g. binding of BEC to a peripheral site or structural changes within the protein) precedes the spin state change and is rate-limiting. Once this remote site is saturated, the reaction is first order and presumably is limited by movement of BEC from the remote site to the active site, which leads to the low-to-high spin shift. Another factor that can affect binding is conformational reorganization in the BEC molecule itself. The BEC conformation currently deposited in the DrugBank database (Fig. 1), which we used as a starting model, differs from that in the x-ray model. To fit into the CYP3A4 active site, the tripeptide group of BEC rotates by ∼180° around the amide bond (Fig. 7A). The existence and interconversion of different BEC conformers may complicate the binding process and limit the overall reaction rate.
FIGURE 7.
A, comparison of BEC conformations in the initial model (gray) and the x-ray structure (black). To fit into the CYP3A4 active site, the tripeptide moiety of BEC rotates by ∼180° around the amide bond as indicated by an arrow. B, conformational changes in the heme of CYP3A4 induced by BEC binding. Compared with the ligand-free structures (Protein Data Bank ID 1WOE (white) and 1TQN (gray)), the heme cofactor in BEC-bound CYP3A4 (black) is distorted slightly, and the vinyl groups are in different conformations. The primary sites of BEC oxidation, 8′ and 9′ carbons (indicated), are 4.1 and 3.7 Å, respectively, away from the heme iron.
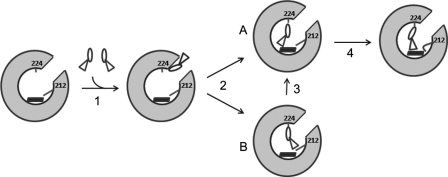
Our kinetic data did not allow us to differentiate whether binding of BEC to CYP3A4 occurs before or after a conformational change in CYP3A4 and/or BEC takes place (“induced fit” versus “conformational selection” mechanism). However, the structure-based mutagenesis identified two possible events through which the entering BEC molecules may cause a spin shift. The T224A substitution slows down and the R212A speeds up the fast phase of the BEC binding kinetics. The T224A mutation has little effect on the slow phase, whereas the R212A replacement increases the slow phase. Therefore, these residues play a role in the translocation of BEC from the remote site to the heme active site, with Thr224 primarily affecting the early stage and Arg212 the later stage of binding near the heme. Based on this scenario and data reported previously (15), we propose that upon translocation from the peripheral binding site into the active site cavity, BEC first establishes a hydrogen bond with Thr224 via the lysergic head group (Fig. 8). This interaction directs the tripeptide moiety toward the heme and, if the orientation is favorable, leads to partial displacement of the water ligand and low-to-high spin shift (fast kinetic phase). When a different BEC conformer enters the active site, the tripeptide group must rotate prior to heme ligation. The final step could be positional adjustment of the tripeptide moiety assisted by Arg212. A conformational switch in the Arg212 side chain, stabilized through H-bonding interactions with Glu308 and Ile369, would allow the tripeptide group to come closer to the heme iron, thereby leading to further changes in spin equilibrium.
FIGURE 8.
Proposed mechanism for the binding reaction between CYP3A4 and BEC. BEC exists in different conformations and, as previously suggested (15), may first associate with a peripheral site remote from the heme (step 1). Upon moving into the active site cavity, BEC establishes a hydrogen bond with Thr224 via the lysergic moiety (step 2), which directs the tripeptide group toward the heme cofactor. If the BEC conformation is favorable (A), partial displacement of the distal water ligand takes place. If another conformer binds to P450 (B), the tripeptide moiety must rotate prior to heme ligation (step 3). The position of BEC may be further optimized upon rearrangement of the Arg212 side chain, which would lead to additional changes in the spin equilibrium (step 4).
In addition to the aforementioned factors, the BEC binding reaction may be affected by intrinsic properties of CYP3A4 such as conformational heterogeneity and aggregation. High pressure spectroscopy studies, for instance, have suggested that there are two conformers of full-length CYP3A4 that have distinct spin equilibrium, barotropic properties and reactivity toward BEC (14). The relative content of these conformers, 70 and 30%, was unaffected by BEC concentration but modulated by Emulgen 913. This prompted us to check whether conformational heterogeneity or/and hydrophobic interactions between the CYP3A4 molecules contribute to the multiphasicity of the BEC binding reaction. To do so, we monitored BEC-dependent spin shift in CYP3A4 in the presence of Emulgen 913 and two other detergents frequently used for CYP3A4 purification, zwitterionic CHAPS and non-ionic IGEPAL CA-630 (25, 29).
One complication in these experiments was that all three detergents were able to enter the CYP3A4 active site and cause type I spectral changes, which supports the notion that detergents can serve as substrates for CYP3A4 (30). Interestingly, despite the ability to cause the largest spin perturbations (Fig. 6A), CHAPS had virtually no effect on the BEC binding kinetics. Moreover, a low-to-high spin shift was complete when both CHAPS and BEC were present (Fig. 6C) as opposed to 90% conversion induced by BEC in a detergent-free buffer. We attribute this phenomenon to a cumulative effect of the two compounds, because it is unclear at the moment whether CHAPS can be displaced by BEC or the detergent stabilizes a conformer disfavoring coordination of the water ligand.
Unlike CHAPS, Emulgen 913 and IGEPAL CA-630 strongly inhibited the CYP3A4-BEC interaction. This agrees with a previous investigation showing that non-ionic detergents inhibit CYP3A4 catalysis by interfering with substrate binding (30). The fact that neither studied detergent could completely eliminate the biphasicity of BEC association undermines the possibility that protein heterogeneity and/or hydrophobic interactions between the truncated CYP3A4 molecules are major factors complicating the BEC binding reaction. On the other hand, a 30:70% distribution between the slow and fast kinetic phases observed at subequimolar concentrations of BEC and its sharp change to ∼50:50% when the BEC:CYP3A4 ratio exceeded unity (Fig. 5G) favor the hypothesis on CYP3A4 conformers with different reactivity toward BEC (14). Our data suggest that the relative content of such conformers might be affected by both substrate and detergent binding. One cause for protein heterogeneity could be variations in the Arg212 conformation. In the two crystal structures of substrate-free CYP3A4 available to date, the Arg212 side chain faces either the solvent or the active site (25, 29). As follows from our study, even this minor structural deviation could significantly affect the rate of BEC binding.
Finally, resonance Raman spectroscopy studies on nanodisc-incorporated CYP3A4 detected small changes in the modes associated with disposition of the heme peripheral groups and out-of-plane macrocycle distortion caused by BEC binding (31). A comparison of the ligand-free and BEC-bound structures showed that, indeed, the BEC-dependent change in the heme coordination state distorts the heme plane and vinyl group conformation (Fig. 7B). If the energy barrier between the 6- and 5-coordinated states is high, it could modulate the dynamics of BEC binding and, hence, contribute to the complexity of the reaction.
In conclusion, the crystallographic complex between CYP3A4 and BEC provides the first insights into the productive binding mode of a type I ligand. It also suggests the mechanism of BEC association, helps better understand previously accumulated data, and most importantly, may be useful for developing new and safer drugs.
Acknowledgments
Research was carried out at the Stanford Synchrotron Radiation Laboratory (SSRL), a national user facility operated by Stanford University on behalf of the U. S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research and by the National Institutes of Health, National Center for Research Resources (NCRR) Biomedical Technology Program and the National Institute of General Medical Sciences (NIGMS).
This work was supported, in whole or in part, by National Institutes of Health Grant GM33688. This study was also supported by Gilead Sciences, Inc. and the California Center for Antiviral Drug Discovery.
The atomic coordinates and structure factors (code 3UA1) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- BEC
- bromoergocryptine
- CYP3A4
- 3A4 isoform of cytochrome P450.
REFERENCES
- 1. Sono M., Roach M. P., Coulter E. D., Dawson J. H. (1996) Heme-containing oxygenases. Chem. Rev. 96, 2841–2887 [DOI] [PubMed] [Google Scholar]
- 2. Guengerich F. P., Hosea N. A., Parikh A., Bell-Parikh L. C., Johnson W. W., Gillam E. M., Shimada T. (1998) Twenty years of biochemistry of human P450s: purification, expression, mechanism, and relevance to drugs. Drug Metab. Dispos. 26, 1175–1178 [PubMed] [Google Scholar]
- 3. Wrighton S. A., Schuetz E. G., Thummel K. E., Shen D. D., Korzekwa K. R., Watkins P. B. (2000) The human CYP3A subfamily: practical considerations. Drug Metab. Rev. 32, 339–361 [DOI] [PubMed] [Google Scholar]
- 4. Ekroos M., Sjögren T. (2006) Structural basis for ligand promiscuity in cytochrome P450 3A4. Proc. Natl. Acad. Sci. U.S.A. 103, 13682–13687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Scott E. E., Halpert J. R. (2005) Structures of cytochrome P450 3A4. Trends Biochem. Sci. 30, 5–7 [DOI] [PubMed] [Google Scholar]
- 6. Niwa T., Murayama N., Yamazaki H. (2008) Heterotropic cooperativity in oxidation mediated by cytochrome p450. Curr. Drug Metab. 9, 453–462 [DOI] [PubMed] [Google Scholar]
- 7. Szklarz G. D., Halpert J. R. (1998) Molecular basis of P450 inhibition and activation: implications for drug development and drug therapy. Drug Metab. Dispos. 26, 1179–1184 [PubMed] [Google Scholar]
- 8. Ueng Y. F., Kuwabara T., Chun Y. J., Guengerich F. P. (1997) Cooperativity in oxidations catalyzed by cytochrome P450 3A4. Biochemistry 36, 370–381 [DOI] [PubMed] [Google Scholar]
- 9. Denisov I. G., Baas B. J., Grinkova Y. V., Sligar S. G. (2007) Cooperativity in cytochrome P450 3A4. Linkages in substrate binding, spin state, uncoupling, and product formation. J. Biol. Chem. 282, 7066–7076 [DOI] [PubMed] [Google Scholar]
- 10. Davydov D. R., Baas B. J., Sligar S. G., Halpert J. R. (2007) Allosteric mechanisms in cytochrome P450 3A4 studied by high-pressure spectroscopy: pivotal role of substrate-induced changes in the accessibility and degree of hydration of the heme pocket. Biochemistry 46, 7852–7864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fernando H., Halpert J. R., Davydov D. R. (2006) Resolution of multiple substrate binding sites in cytochrome P450 3A4: the stoichiometry of the enzyme-substrate complexes probed by FRET and Job's titration. Biochemistry 45, 4199–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peyronneau M. A., Delaforge M., Riviere R., Renaud J. P., Mansuy D. (1994) High affinity of ergopeptides for cytochromes P450 3A. Importance of their peptide moiety for P450 recognition and hydroxylation of bromocriptine. Eur. J. Biochem. 223, 947–956 [DOI] [PubMed] [Google Scholar]
- 13. Renaud J. P., Davydov D. R., Heirwegh K. P., Mansuy D., Hui Bon Hoa G. H. (1996) Thermodynamic studies of substrate binding and spin transitions in human cytochrome P450 3A4 expressed in yeast microsomes. Biochem. J. 319, 675–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Davydov D. R., Halpert J. R., Renaud J. P., Hui Bon Hoa G. (2003) Conformational heterogeneity of cytochrome P450 3A4 revealed by high pressure spectroscopy. Biochem. Biophys. Res. Commun. 312, 121–130 [DOI] [PubMed] [Google Scholar]
- 15. Isin E. M., Guengerich F. P. (2006) Kinetics and thermodynamics of ligand binding by cytochrome P450 3A4. J. Biol. Chem. 281, 9127–9136 [DOI] [PubMed] [Google Scholar]
- 16. Nath A., Grinkova Y. V., Sligar S. G., Atkins W. M. (2007) Ligand binding to cytochrome P450 3A4 in phospholipid bilayer nanodiscs. The effect of model membranes. J. Biol. Chem. 282, 28309–28320 [DOI] [PubMed] [Google Scholar]
- 17. Das A., Grinkova Y. V., Sligar S. G. (2007) Redox potential control by drug binding to cytochrome P450 3A4. J. Am. Chem. Soc. 129, 13778–13779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maurer G., Schreier E., Delaborde S., Loosli H. R., Nufer R., Shukla A. P. (1982) Fate and disposition of bromocriptine in animals and man. I: Structure elucidation of the metabolites. Eur. J. Drug Metab. Pharmacokinet. 7, 281–292 [DOI] [PubMed] [Google Scholar]
- 19. Maurer G., Schreier E., Delaborde S., Nufer R., Shukla A. P. (1983) Fate and disposition of bromocriptine in animals and man. II: Absorption, elimination, and metabolism. Eur. J. Drug Metab. Pharmacokinet. 8, 51–62 [DOI] [PubMed] [Google Scholar]
- 20. He Y. A., Zientek M., Parge H. E., Burke B. J., Lee C. A., Wester M. R. (2009) Crystal structures of CYP3A4 in complex with bromocriptine and clotrimazole:evidence of structural plasticity in the active site. In Book of Abstracts for the 16th International Conference on Cytochrome P450, p. 91, Okinawa, Japan [Google Scholar]
- 21. Sevrioukova I. F., Poulos T. L. (2010) Structure and mechanism of the complex between cytochrome P450 3A4 and ritonavir. Proc. Natl. Acad. Sci. U.S.A. 107, 18422–18427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 23. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 24. Viner R. I., Novikov K. N., Ritov V. B., Kagan V. E., Alterman M. A. (1995) Effect of different solubilizing agents on the aggregation state and catalytic activity of two purified rabbit cytochrome P450 isozymes, CYP1A2 (LM4) and CYP2B4 (LM2). Biochem. Biophys. Res. Commun. 217, 886–891 [DOI] [PubMed] [Google Scholar]
- 25. Williams P. A., Cosme J., Vinkovic D. M., Ward A., Angove H. C., Day P. J., Vonrhein C., Tickle I. J., Jhoti H. (2004) Crystal structures of human cytochrome P450 3A4 bound to metyrapone and progesterone. Science 305, 683–686 [DOI] [PubMed] [Google Scholar]
- 26. Harlow G. R., Halpert J. R. (1997) Alanine-scanning mutagenesis of a putative substrate recognition site in human cytochrome P450 3A4. Role of residues 210 and 211 in flavonoid activation and substrate specificity. J. Biol. Chem. 272, 5396–5402 [DOI] [PubMed] [Google Scholar]
- 27. Harlow G. R., Halpert J. R. (1998) Analysis of human cytochrome P450 3A4 cooperativity: construction and characterization of a site-directed mutant that displays hyperbolic steroid hydroxylation kinetics. Proc. Natl. Acad. Sci. U.S.A. 95, 6636–6641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fishelovitch D., Hazan C., Shaik S., Wolfson H. J., Nussinov R. (2007) Structural dynamics of the cooperative binding of organic molecules in the human cytochrome P450 3A4. J. Am. Chem. Soc. 129, 1602–1611 [DOI] [PubMed] [Google Scholar]
- 29. Yano J. K., Wester M. R., Schoch G. A., Griffin K. J., Stout C. D., Johnson E. F. (2004) The structure of human microsomal cytochrome P450 3A4 determined by X-ray crystallography to 2.05 Å resolution. J. Biol. Chem. 279, 38091–38094 [DOI] [PubMed] [Google Scholar]
- 30. Hosea N. A., Guengerich F. P. (1998) Oxidation of nonionic detergents by cytochrome P450 enzymes. Arch. Biochem. Biophys. 353, 365–373 [DOI] [PubMed] [Google Scholar]
- 31. Mak P. J., Denisov I. G., Grinkova Y. V., Sligar S. G., Kincaid J. R. (2011) Defining CYP3A4 structural responses to substrate binding. Raman spectroscopic studies of a nanodisc-incorporated mammalian cytochrome P450. J. Am. Chem. Soc. 133, 1357–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hendlich M., Rippmann F., Barnickel G. (1997) LIGSITE: automatic and efficient detection of potential small molecule-binding sites in proteins. J. Mol. Graph. Model. 15, 359–363 [DOI] [PubMed] [Google Scholar]