Background: Yeast hexokinase 2 shuttles in and out of nucleus in response to glucose availability.
Results: Hexokinase 2 enters the nucleus by binding Kap60 through a nuclear localization sequence in the presence of Kap95.
Conclusion: The karyopherins Kap60 and Kap95 are necessary for efficient nuclear import of hexokinase 2.
Significance: The current study has identified a new pathway for Hxk2 import into the nucleus.
Keywords: Glucose, Hexokinase, Nuclear Transport, Signaling, Yeast, Kap60, Kap95
Abstract
Hexokinase 2 (Hxk2) from Saccharomyces cerevisiae was one of the first metabolic enzymes described as a multifunctional protein. Hxk2 has a double subcellular localization and role, it functions as a glycolytic enzyme in the cytoplasm and as a regulator of gene transcription of several Mig1-regulated genes in the nucleus. However, the mechanism by which Hxk2 enters in the nucleus was unknown until now. Here, we report that the Hxk2 protein is an import substrate of the carriers α-importin (Kap60 in yeast) and β-importin (Kap95 in yeast). We also show that the Hxk2 nuclear import and the binding of Hxk2 with Kap60 are glucose-dependent and involve one lysine-rich nuclear localization sequence (NLS), located between lysine 6 and lysine 12. Moreover, Kap95 facilitates the recognition of the Hxk2 NLS1 motif by Kap60 and both importins are essential for Hxk2 nuclear import. It is also demonstrated that Hxk2 nuclear import and its binding to Kap95 and Kap60 depend on the Gsp1-GTP/GDP protein levels. Thus, our study uncovers Hxk2 as a new cargo for the α/β-importin pathway of S. cerevisiae.
Introduction
Transport between the nucleus and cytoplasm is mediated by soluble receptors that recognize specific cargoes and carry them through the nuclear pore complex, which are embedded between the inner and outer nuclear membranes. Whereas the protein components of the nuclear pore complex are largely stationary within the pore, soluble transport factors can be freely exchanged between the nuclear and cytoplasmic compartments. The majority of nucleocytoplasmic transport factors belong to the family of karyopherin proteins (1). In higher eukaryotes, as well as in the yeast Saccharomyces cerevisiae, karyopherins that mediate the nuclear import of proteins and RNAs are generally known as importins (2), whereas those mediating nuclear export are known as exportins (3). Importins associate with their macromolecular cargo in the cytoplasm, either directly or indirectly via adaptor proteins. They dock to components of the nuclear pore complex (known as nucleoporins), translocate to the opposite side of the nuclear envelope, and release their cargo there. Cargo release is achieved by association of the importins with the GTPase Ran (Gsp1 in yeast) in the GTP-bound form (Gsp1-GTP). Nuclear export is essentially the reciprocal process, with cargo recognition occurring in the nucleus in the presence of Gsp1-GTP and cargo complexes dissociating in the cytoplasm upon GTP hydrolysis. Gsp1 is present at high concentrations in the nucleus in the GTP-bound form, and mainly in the GDP-bound form in the cytoplasm (4). This compartmentalization depends on the localization of proteins that regulate the nucleotide state of Gsp1. GTP hydrolysis requires a GTPase-activating protein that is present in the cytoplasm. Conversely, the exchange of GDP to GTP is catalyzed by a guanine nucleotide exchange factor that is bound to chromatin in the nucleus (5–7). Importin β (Kap95 in yeast) was first identified as the transport factor for proteins carrying classical nuclear localization signals (NLS),5 such as those of nucleoplasmin or the SV40 T antigen (8). Importin β hardly ever binds these classical NLSs directly, but binds importin α (Kap60 in yeast), which in turn binds the NLS (9).
In S. cerevisiae, hexokinase 2 (Hxk2) is the predominant glucose kinase in cells growing in high-glucose conditions (10), and it shuttles in and out of the nucleus (11, 12). In the cytoplasm, Hxk2 catalyzes glucose phosphorylation at C6, whereas the nuclear Hxk2 plays a nonclassical role involved in the glucose repression signaling of several Mig1-regulated genes. The relative abundance of Hxk2 in the nucleus is Mig1 and glucose-dependent (12). Recent data revealed that the mechanism of Hxk2 sequestration in the nucleus is based on direct Hxk2 interaction with Mig1. This interaction is mediated by a 10-amino acid motif located between Lys-6 and Met-15 of Hxk2 and serine 311 of Mig1 (12–14). These findings suggest that the main regulatory role of Hxk2 is caused by its interaction with Mig1 generating a repressor complex located in the nucleus of S. cerevisiae. In high glucose, nuclear Hxk2 stabilizes the repressor complex by blocking Mig1 phosphorylation in serine 311 by the Snf1 protein kinase. In low glucose, Hxk2 leaves the complex and does not block serine 311 of Mig1 so Snf1 can phosphorylate it. Under these conditions the repressor complex is disorganized and Hxk2 and phosphorylated Mig1 leave the nucleus (12, 13).
The mechanism by which Hxk2 leaves the nucleus is Xpo1-dependent and was recently described (15), but its nuclear import mechanism is largely unknown. Because Hxk2 is too large to translocate through the nuclear pore complex by diffusion from the cytoplasm to the nucleus, its transport across the nuclear envelope must be a mediated process, which may depend on carrier proteins. In this study, we show that the Hxk2 protein is an import cargo of the α/β-importin pathway in S. cerevisiae. We also show that Hxk2 interacts with Kap95 and Kap60 directly without the participation of other auxiliary proteins. Moreover, the Hxk2-Kap60 interaction is glucose-dependent and requires a NLS in the Hxk2 protein located between Lys6 and Lys12. We also demonstrate that Hxk2 recognition occurs in the cytoplasm in the presence of Gsp1-GDP and cargo complexes dissociate in the nucleus upon the exchange of GDP/GTP in the Gsp1 protein.
EXPERIMENTAL PROCEDURES
Strains and Growth Conditions
The S. cerevisiae strains used throughout this study were derived from W303-1A (16) and DBY1315 (17) haploid wild type strains and are listed in Table 1. We created two mutant NLS alleles of HXK2 by mutating the HXK2 gene in the YEp352-HXK2 plasmid to generate HXK2(K6A,K7A,K12A) (encodes Hxk2nls1(Ala) protein) and HXK2(K406A,K407A,K410A) (encodes Hxk2nls2(Ala) protein) genes using a PCR-based mutagenesis protocol. The KanR gene was inserted into the XbaI site, located in the yeast genome 371 bp from the HXK2 stop codon, of plasmids YEp352-HXK2nls1(Ala) and YEp352-HXK2nls2(Ala). Then, by using these constructs, recombination cassettes were obtained by PCR; these cassettes contained the mutated HXK2 locus and the KanR marker together with 515 bp from the 5′-genomic region downstream from the HXK2 gene. These linear DNAs were integrated into the HXK2 locus of strains DBY1315 and JCY1410. To confirm the correct insertion, we PCR amplified the HXK2 locus from strains FMY304, which contains the HXK2 gene replaced by the HXK2nls1(Ala) gene, FMY305, which contains the HXK2 gene replaced by the HXK2nls2(Ala) gene to detect, respectively, the presence of the HXK2(K6A,K7A,K12A) and HXK2(K406A,K407A, K410A) mutant alleles by sequencing analysis. FMY306 is a conditional kap60tsHXK2nls1(Ala) mutant derivative from the JCY1410 strain (18). FMY307 is a conditional xpo1-1Δmig1 mutant derivate from the xpo1-1 strain (3). Escherichia coli DH5α (FØ80dlacZ ΔM15 recA1 endA1 gyrA96 thi-1 hsdR17(rk−rk−) supE44 relA1 deoRΔ 99U169) was the host bacterial strain for the recombinant plasmid constructions.
TABLE 1.
S. cerevisiae strains used in this study
| Name | Relevant genotype | Source/Ref. |
|---|---|---|
| W303-1A | Matα ura3–52 trp1–289 leu2–3,112 his3-Δ1 ade2–1 can1–100 | 16 |
| DBY1315 | Matα ura3–52 leu2–3,2–112 lys2–801 gal2 | 17 |
| DBY2052 | Mat α hxk1::LEU2 hxk2–202 ura3–52 leu2–3,2–112 lys2–801 gal2 | 17 |
| THG1 | MATα leu2–1 ura3–1 lys1–1 hxk1::LEU2 hxk2::LEU2 glk1::LEU2 | 31 |
| Y03694 | Matα his3-Δ1 leu2Δ0 met15 Δ0 ura3Δ0 msn5::kanMX4 | Euroscarf |
| B0119B | Matα ura3–52 his3D1 leu2–3_112 trp1–289 nmd5::HIS3 | Euroscarf |
| JCY1410 | Matα ade2–1 ura3–1 his3–11 15 trp16̂3 leu2–3 112 can1–100 srp11–31 | 18 |
| JCY1407 | Matα, ade2–1 ura3–1, his3–11, 15 trp1–1, leu2–3, 112 can1–100 kap95Δ::HIS3, pSW509 [CEN LEU],pkap95-L63A | 18 |
| FMY304 | Matα ura3–52 leu2–3,2–112 lys2–801 gal2 hxk2::HXK2nls1(Ala) | This work |
| FMY305 | Matα ura3–52 leu2–3,2–112 lys2–801 gal2 hxk2::HXK2nls2(Ala) | This work |
| FMY306 | Mat α ade2–1 ura3–1 his3–11 15 trp1–63 leu2–3 112 can1–100 srp11–31 hxk2::HXK2nls1(Ala) | This work |
| FMY307 | xpo1::LEU2[pRS313(CEN HIS3) xpo1–1]mig1::kanMX4 | This work |
Yeast cells were grown in the following media: YEPD, high-glucose (2% glucose, 2% peptone, and 1% yeast extract), YEPGly, low-glucose (0.05% glucose, 3% glycerol, 2% peptone, and 1% yeast extract), and synthetic media containing the appropriate carbon source and lacking appropriate supplements to maintain selection for plasmids (2% glucose (SD) or 3% glycerol and 0.05% glucose (SGly); and 0.67% yeast nitrogen base without amino acids). Amino acids and other growth requirements were added at a final concentration of 20–150 μg/ml. The solid media contained 2% agar in addition to the components described above.
Plasmids
The yeast expression plasmids YEp352-HXK2, YEp352-HXK2/GFP were constructed as indicated previously (12). To create Hxk2 mutant alleles Hxk2nls1(Ala) (K6A, K7A, and K12A) and Hxk2nls2(Ala) (K406A, K407A, and K410A), plasmids YEp352-HXK2, YEp352-HXK2nes2(Ala), and YEp352-HXK2/GFP were used as templates in conjunction with oligonucleotides OL1-d, TCATTTAGGTCCAGCAGCACCACAAGCCGCAGCGGGTTCCATGGCCGATG; OL1-r, CATCGCCATGGAACCCGCTGCGTTGTGGTGCTGCTGGACCTAAATGA; OL2-d, CTGCTATCTGTCAAGCGAGCGGTTCAGCGACCGGTCACATCGCTGC; OL2-r, GCAGCGATGTGACCGGTCGCGTAACCGCTCGCTTGACAGATSGCAGC, in the PCR-based site-directed mutagenesis method (19). All nucleotide changes were verified by DNA sequencing.
GST fusion vector pGEX-HXK2 was constructed as indicated in Ref. 12. Plasmid pGEX-GSP1 for expression of GST-Gsp1 in E. coli was a gift from E. Hurt (20) and plasmids pGEX-Kap95 and pGEX-Kap60 for expression of GST-Kap95 and GST-Kap60 in E. coli were gifts from M. P. Rout (21).
Fluorescence Microscopy
Yeast strains expressing Hxk2-GFP, Hxk2nes2(Ala)-GFP, Hxk2nes2(Ala)nls1(Ala)-GFP, or Hxk2nes2(Ala)nls2(Ala)-GFP were grown to early-log phase (A600 of less than 0.8) in synthetic high-glucose medium (SD-ura). Half of the culture was shifted to synthetic low-glucose medium (SGly-ura) for 1 h. The media contained the appropriate carbon source and lacked the appropriate supplements to maintain selection of plasmids. Cells (25 μl) were loaded onto poly-l-lysine-coated slides, and the remaining suspension was immediately withdrawn by aspiration. One microliter of DAPI (2.5 μg/ml in 80% glycerol) was added, and a covert slide was placed over the microscope slide. GFP and DAPI localization in live cultures was monitored by direct fluorescence using a Leica DM5000B microscope. To avoid the nonlinear range of fluorescent signals, cells highly overexpressing GFP-tagged fusion protein were excluded from further analyses. The localization of proteins was monitored by visual inspection of the images. At least 100 cells were scored in each of at least three independent experiments. The distribution of fluorescence was scored in the following way: N, denotes a nuclear fluorescence signal; C, cytoplasmic fluorescence signal without nuclear fluorescence signal. Images representative of the results obtained were shown. Images were processed in Adobe Photoshop CS.
Statistical Analysis
Data result from at least 3 independent experiments. Results are shown as the mean ± S.E. Statistical analysis was performed using Statistical Package for the Social Sciences 15.0 (SPSS). The Kolmogorov-Smirnov test with the correction of Lilliefors was used to evaluate the fit of the data to a normal distribution and the test of Levene to evaluate the homogeneity of variance. Significance was tested by the one-way analysis of variance test followed by a Turkey's multiple range test or Student's t test to compare sample means. Significant differences were considered when p < 0.01.
Preparation of Crude Protein Extracts
Yeast protein extracts were prepared as follows: yeast were grown in 10 to 20 ml of YEPD or synthetic high-glucose medium (SD-ura) at 28 °C to an optical density at 600 nm of 0.8–1.0. Half of the culture was shifted to YEPGly or synthetic low-glucose medium (SGly-ura) for 1 h. Cells were collected, washed twice with 1 ml of 1 m sorbitol, and suspended in 500 μl of PBS buffer (150 mm NaCl, 100 mm Na2HPO4, 18 mm NaH2PO4, pH 7.3). The cells were broken in the presence of glass beads by one pulse of 20 s at 6.0 m/s using a FastPrep homogenizer (Thermo Electron Co.). After centrifugation at 17,000 × g for 15 min at 4 °C, the supernatant was used as the crude protein extract. After centrifugation at 6,100 × g for 15 min at 4 °C, the supernatant was used as the crude protein extract in immunoprecipitation and GST pulldown experiments.
Enzyme Assay
Invertase activity was assayed in whole cells as previously described (22) and expressed as micromoles of glucose released per minute per 100 mg of cells (dry weight).
Immunoblot Analysis
Mutant or wild-type yeast cells were grown to an optical density at 600 nm of 0.8–1.0 in selective medium containing high-glucose (2%) and shifted to low-glucose conditions for 1 h. The cells were collected by centrifugation (3,000 × g, 4 °C, 2 min), and crude extracts were prepared as described above. For Western blotting, 20 to 40 μg of proteins were separated by SDS-12% polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a enhanced chemiluminescence PVDF transfer membrane (Amersham Biosciences, HybondTM-P) by electroblotting, which was then incubated with anti-Hxk2, anti-Kap60 (Santa Cruz Biotechnology), or anti-Kap95 (Santa Cruz Biotechnology) as primary antibodies and then the appropriate secondary antibody. Horseradish peroxidase-conjugated protein-A was used as the secondary reactant. The West Pico Chemiluminescent system (Pierce) was used for detection.
Co-immunoprecipitation Assay
Immunoprecipitation experiments were performed using whole cell extracts from different strains. The extracts were incubated with anti-Kap60, anti-Kap95, or anti-Pho4 polyclonal antibodies for 3 h at 4 °C. Protein A-Sepharose beads (GE Healthcare) were then added and incubated for 3 h at 4 °C in a spinning wheel. After extensive washing with PBS buffer, immunoprecipitated samples were boiled in SDS-loading buffer (50 mm Tris-HCl, pH 6.8, 100 mm DTT, 2% SDS, 0.1% bromphenol blue, 10% glycerol). The supernatant was subjected to 12% SDS-PAGE. The proteins were transferred to an enhanced chemiluminescence PVDF membrane and immunoblotted as described above using anti-Hxk2 polyclonal antibody. Values shown are representative results from individual experiments.
GST Pulldown Experiments
E. coli cells from the BL21(DE3) pLysS strain were transformed with fusion protein expression vectors pGEX-KAP95 or pGEX-KAP60. Cells were grown to A600 0.5–0.8, induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside at 37 °C for 3 h, and collected by centrifugation. Cell pellets were resuspended in PBS buffer (150 mm NaCl, 100 mm Na2HPO4, 18 mm NaH2PO4, pH 7.3) and sonicated. Insoluble material was removed by centrifugation (17,000 × g for 20 min at 4 °C). Soluble extract was incubated with glutathione-Sepharose 4B beads (GE Healthcare) for 1 h at 4 °C, washed extensively with PBS buffer, and resuspended in the same buffer. The Kap95-GST and Kap60-GST fusion proteins coupled to glutathione-Sepharose were incubated with yeast whole cell extracts from the wild-type and mutant yeast strains expressing, respectively, Hxk2, HXK2nls1(Ala) or Hxk2nls2(Ala) proteins, for 1 h at 4 °C in PBS buffer. The cell extracts were obtained from yeast cells grown in YEPD medium containing high glucose (2%) and shifted to 0.05% glucose plus 3% glycerol (low glucose) for 1 h. Beads were gently washed five times with 2.5 ml of PBS buffer, boiled in 25 μl of sample loading buffer, and analyzed by SDS-PAGE followed by Western blot using anti-Hxk2 antibodies and horseradish peroxidase-conjugated protein-A. Bound antibodies were detected using the West Pico chemiluminescent system (Pierce).
GST fusion protein expression vectors pGEX-HXK2 and pGEX-GSP1 were transformed into E. coli strain BL21(DE3) pLysS. Cells were grown to A600 0.5–0.8, induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside at 37 °C for 3 h, and collected by centrifugation. Cell pellets were resuspended in PBS buffer and sonicated. The GST-Hxk2 and GST-Gsp1 fusion proteins coupled to glutathione-Sepharose beads were incubated with 2.5 units of thrombin (2 h at 4 °C) for site-specific separation of the GST affinity tag from Hxk2 and Gsp1 proteins. Affinity purified Gsp1 was immediately loaded with GTP or GDP by adding 30 mm K2PO4 (pH 7.5), 1 mm GTP or 1 mm GDP, and 10 mm EDTA (pH 8.0), incubating at room temperature for 1 h, adding 20 mm magnesium acetate, incubating on ice for 30 min, and freezing at −80 °C (20). Gsp1-mediated Kap95-Kap60 interactions with Hxk2 were analyzed by incubating purified Hxk2 with GST-Kap95 or GST-Kap60 bound to glutathione-Sepharose beads in the presence of Gsp1(GTP) or Gsp1(GDP) for 1 h at 4 °C in PBS buffer. Beads were washed and analyzed as described above.
RESULTS
The Nuclear Import Proteins Msn5 (Kap142) and Nmd5 (Kap119) Are Not Essential for Hxk2 Nuclear Import
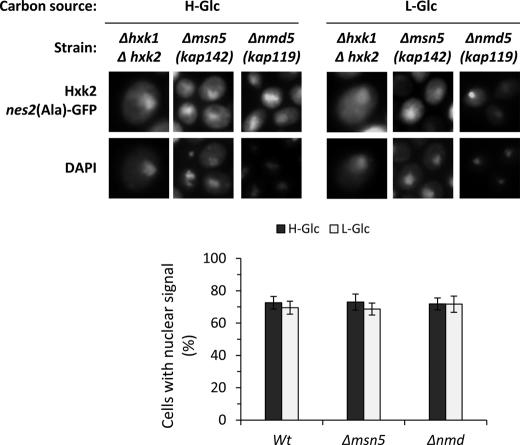
In contrast to the fairly well defined Hxk2 export system (15), the mechanism underlying Hxk2 translocation from the cytoplasm to the nucleus in high-glucose conditions is still unknown. We have tested the possible role of Msn5 and Nmd5 in Hxk2 nuclear import. Msn5, an import-export protein, has been shown to be involved in the export of Mig1 from the nucleus to the cytoplasm in response to glucose removal (23). Nmd5 is a classical import factor implicated in the nuclear import of some stress response proteins like Hog1 and Crz1 (24, 25). Because the Hxk2nes2(Ala)-GFP mutant protein shows nuclear accumulation in both high- and low-glucose conditions because it is unable to interact with the export factor Xpo1 (15), we have determined the Hxk2nes2(Ala)-GFP nuclear accumulation in Δmsn5 and Δnmd5 mutants cells. Wild-type cells and Δmsn5 and Δnmd5 mutants cells show identical nuclear distribution to Hxk2nes2(Ala)-GFP growing at high- or low-glucose conditions. The Hxk2nes2(Ala)-GFP mutant protein accumulates in the nucleus in high-glucose conditions and it keeps its nuclear location after glucose exhaustion (Fig. 1). These results demonstrate that Hxk2nes2(Ala)-GFP can enter into the nucleus in the wild-type cells and both mutants, so Msn5 and Nmd5 are not implicated in Hxk2 nuclear import.
FIGURE 1.
Localization of Hxk2nes2(Ala) in Δhxk1Δhxk2, Δmsn5, and Δnmd5 yeast cells. The DBY2052, Y03694, and B0119B strains expressing Hxk2nes2(Ala)-GFP from plasmid YEp352-HXK2nes2(Ala)-GFP were grown in high-glucose synthetic medium (H-Glc) until an A600 nm of 1.0 was reached and then transferred to low-glucose synthetic medium (L-Glc) for 60 min. Cells were stained with DAPI and imaged for GFP and DAPI fluorescence. The nuclear localization of the Hxk2nes2(Ala)-GFP protein was determined in at least 100 cells per growth condition. No statistically significant differences were detected between mutants.
Mig1 Does Not Participate in Hxk2 Nuclear Import
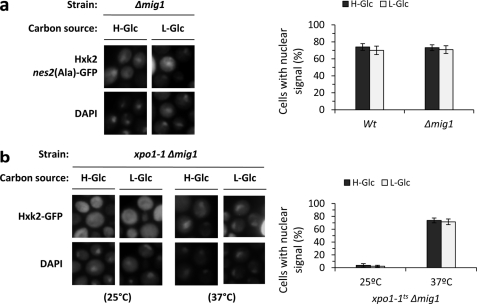
In high-glucose conditions Hxk2 interacts with Mig1 to generate a glucose repressor complex in the nucleus. After glucose exhaustion, both Mig1 and Hxk2 exit the nucleus (12, 13). Because the nucleocytoplasmic traffic of both Hxk2 and Mig1 is glucose-dependent we have studied whether Mig1 is required for Hxk2 nuclear import. As can be seen in Fig. 2a, in Δmig1 cells, the fluorescent Hxk2nes2(Ala)-GFP protein does not shuttle between the nucleus and cytoplasm, it presents a constitutive nuclear location not affected by glucose levels. To confirm this result, we have also generated a novel mutant xpo1–1 Δmig1 to study the subcellular location of Hxk2-GFP in the presence of a defective Xpo1-dependent nuclear export pathway. The xpo1–1 Δmig1 double mutant cells were grown at the permissive temperature (25 °C) and then shifted to 37 °C (nonpermissive temperature) for 1 h. No accumulation of Hxk2 was detected in the nuclei of cells grown at the permissive temperature (Fig. 2b), and Hxk2-GFP showed identical subcellular distribution in a wild-type strain (15) in both high- and low-glucose conditions. However, in cells shifted to the nonpermissive temperature, an accumulation of Hxk2 in the nuclei of cells was already observed after 1 h of culture in high- (74%) and low (72%)-glucose conditions (Fig. 2b). This nuclear location of the protein demonstrates that Hxk2-GFP is able to enter the nucleus in the absence of Mig1. Thus, these results suggest that Mig1 was not required for Hxk2 nuclear import and support the idea that both proteins enter the nucleus independently to form the SUC2 repressor complex in the nucleus when there is a high-glucose level in the medium.
FIGURE 2.
Localization of Hxk2nes2(Ala) in Δmig1 and xpo1–1Δmig1 yeast cells. a, the Δmig1 strain expressing Hxk2nes2(Ala)-GFP from plasmid YEp352-HXK2nes2(Ala)-GFP was grown in high-glucose synthetic medium (H-Glc) until an A600 nm of 1.0 was reached and then transferred to low-glucose synthetic medium (L-Glc) for 60 min. Cells were stained with DAPI and imaged for GFP and DAPI fluorescence. Nuclear localization of the Hxk2nes2(Ala)-GFP protein was determined in at least 100 cells per growth condition. Error bars represent mean ± S.D. for three independent experiments. b, the xpo1–1Δmig1 double mutant strain expressing Hxk2-GFP, from plasmid YEp352-HXK2/GFP, were grown in high-glucose synthetic medium (H-Glc) until an A600 nm of 1.0 was reached and then transferred to low-glucose synthetic medium (L-Glc). The cells were grown at 25 °C (permissive temperature) and then shifted to 37 °C (nonpermissive temperature) for 1 h. The localization of Hxk2-GFP was analyzed by fluorescence microscopy (GFP). Nuclear DNA was stained with DAPI. Nuclear localization of the Hxk2-GFP protein was determined in at least 100 cells per growth condition. No statistically significant differences were detected between mutants.
Nuclear Import of Hxk2 Is Altered in kap60ts and kap95ts Mutants
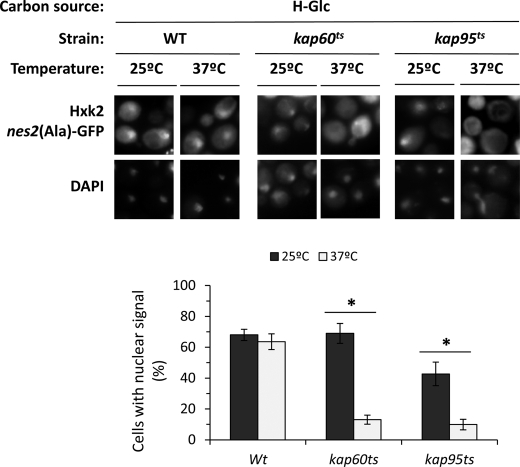
Kap60 normally binds proteins through a highly basic stretch within their N terminus, resulting in the recruitment of Kap95 and formation of a heterotrimer complex that facilitates nuclear import. The carrier Kap60 recognizes two classes of NLSs matching the consensus K(K/R)X1–3(K/R)1–2 and (K/R)(K/R)X1-3(K/R)X10–12(K/R)3–5. The first class, known as monopartite NLS, have a single cluster of basic amino acid residues and the second class, known as bipartite NLS, have two clusters of basic amino acids separated by a 10–12-amino acid linker (26). Because Hxk2 presents two such monopartite NLS motives, 6KKPQARK12 (NLS1) and 406KRGYK410 (NLS2), we hypothesized that the Kap60-Kap95 complex may mediate the nuclear import of Hxk2. To test this, we analyzed the subcellular localization of Hxk2nes2(Ala)-GFP in temperature-sensitive kap60 and kap95 mutant cells. The kap60 and kap95 mutants were grown at the permissive temperature (25 °C) in high-glucose conditions and then shifted to 37 °C (nonpermissive temperature) for 1 and 5 h. A similar accumulation of Hxk2 was detected in the nuclei of wild-type, kap60 and kap95 mutant cells grown at the permissive temperature (Fig. 3). However, in kap60 and kap95 mutant cells incubated at the nonpermissive temperature, no nuclear accumulation of Hxk2 was observed after 1 and 5 h of culture (Fig. 3).
FIGURE 3.
Nuclear import of Hxk2-GFP is inhibited in xpo1–1 cells at the nonpermissive temperature. The DBY2052, kap60ts, and kap95ts strains expressing Hxk2nes2(Ala)-GFP, from plasmid YEp352-HXK2nes2(Ala)/GFP, were grown in high-glucose synthetic medium (H-Glc) until an A600 nm of 1.0 was reached and then transferred to low-glucose synthetic medium (L-Glc). The cells were grown at 25 °C (permissive temperature) and then shifted to 37 °C (nonpermissive temperature) for 1 h. Cells were stained with DAPI and imaged for GFP and DAPI fluorescence. Nuclear localization of the Hxk2nes2(Ala)-GFP protein was determined in at least 100 cells per growth condition from 3 different experiments. *, statistically significant differences 25 versus 37 °C in Kaps mutants, p < 0.01.
Mapping Residues Necessary for NLS Function
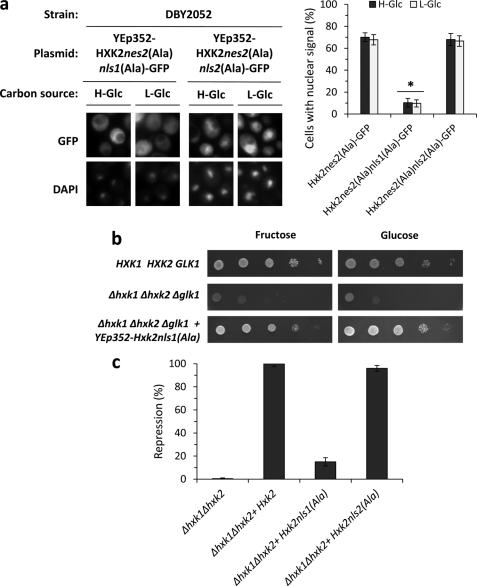
Next we determined whether the two putative NLS sequences of Hxk2 are necessary for the entry of Hxk2 to the nucleus. For this aim, we performed a mutational analysis inside the putative NLSs sequences to confirm its functionality. The presence of basic amino acids such as lysine and arginine appears to be an important feature of the NLSs (26). Thus, we created two HXK2nes2(Ala)-GFP mutants in which the basic residues of NLS1 and NLS2 were replaced with alanine residues to generate HXK2nes2(Ala)K6AK7AK12A-GFP (coding for Hxk2nes2(Ala)nls1(Ala)-GFP protein) and HXK2nes2(Ala)K406AR407AK410A-GFP (coding Hxk2nes2(Ala)nls2(Ala)-GFP protein) genes. The analysis of intracellular localization of the different Hxk2 variants by fluorescent microscopy showed that the Hxk2nes2(Ala)nls2(Ala)-GFP fusion protein accumulated in the nuclei of Δhxk2 mutant cells expressing the HXK2nes2(Ala)K406AR407AK410A-GFP mutant allele both at high- and low-glucose conditions. However, the Hxk2nes2(Ala)nls1(Ala)-GFP protein was excluded from the nuclei of Δhxk2 mutant cells carrying the HXK2nes2(Ala)K6AK7AK12A-GFP mutant allele (Fig. 4a). Our data show that cells expressing the Hxk2nes2(Ala)nls2(Ala)-GFP mutant allele showed nuclear accumulation of Hxk2-GFP similar to that found in a Hxk2nes2(Ala) strain (68 and 67%, respectively). Because cells expressing the Hxk2nes2(Ala)nls1(Ala)-GFP lost the Hxk2 nuclear import function, the nls1(Ala) mutation is strongly defective causing an import defect in 90% of the cells (Fig. 4a).
FIGURE 4.
Identification of Hxk2 NLSs. a, the DBY2052 (Δhxk1Δhxk2) mutant strain was transformed with plasmids YEp352-HXK2nes2(Ala)nls1(Ala)/GFP and YEp352-HXK2nes2(Ala)nls2(Ala)/GFP. Transformed cells were grown in high-glucose synthetic medium (H-Glc) until an A600 nm of 1.0 was reached and then transferred to low-glucose synthetic medium (L-Glc) for 60 min. The cells were visualized by fluorescence microscopy, DAPI staining revealed nuclear DNA. Nuclear localization of fluorescent reporter proteins was determined in at least 100 cells in three independent experiments. Mean ± S.D. are shown for at least three independent experiments. *, statistically significant differences between groups, p < 0.001. b, the THG1 (Δhxk1Δhxk2Δglk1) triple mutant strain was transformed with the YEp352-HXK2nls1(Ala) plasmid and grown overnight in synthetic media with galactose (2%) as carbon source (SGal ura−). 5-Fold serial dilutions of a 1.0 A600 overnight culture were plated on SFru ura− (fructose, 2%) medium and SD ura− medium and photographed after 48 h. c, the Δhxk1Δhxk2 double mutant strain was transformed with YEp352, YEp352-HXK2, YEp352-HXK2nls1(Ala), and YEp352-HXK2nls2(Ala) plasmids. Transformed cells were grown in high-glucose synthetic medium until an A600 nm of 1.0 was reached. Invertase activity was assayed in whole cells. Values are the averages of results obtained on four independent experiments. *, statistically significant differences between groups, p < 0.01.
Taken together, these results demonstrate that Lys6, Lys7, and/or Lys12 are critical amino acid residues in the Hxk2 NLS1 sequence. Moreover, because the nes2(Ala) mutation did not affect Hxk2 import because it does not decrease its nuclear accumulation, our data suggest that only the NLS1 motif is necessary for efficient nuclear import of Hxk2-GFP in vivo. Furthermore, the Hxk2nls1(Ala) mutant protein is catalytically functional because it confers glucose and fructose growth capacity to a triple sugar kinase mutant strain (Δhxk1Δhxk2Δglk1) (Fig. 4b), but did not restore the glucose repression capacity to a Δhxk1Δhxk2 double mutant strain (Fig. 4c). These data suggest that the Hxk2nls1(Ala) protein has lost the Hxk2 nuclear import function and therefore is defective in glucose repression signaling. These results are consistent with the idea that Kap60 and Kap95 proteins participate in Hxk2 nuclear import and prompted us to further define the possible role of these carriers in Hxk2 nuclear trafficking.
Hxk2 Interacts with Kap60 and Kap95
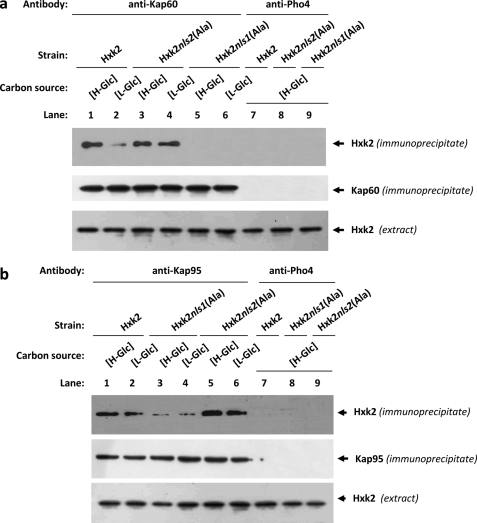
To test whether Hxk2 binds to Kap60 and Kap95 in vivo we have used an immunoprecipitation assay in cells expressing Hxk2, Hxk2nls1(Ala), or Hxk2nls2(Ala) as the only Hxk2 protein. Cell extracts from wild-type (Hxk2), FMY304 (Hxk2nls1(Ala)), or FMY305 (Hxk2nls2(Ala)) strains were immunoprecipitated with anti-Kap60 and anti-Kap95 antibodies. The resulting immunoprecipitates were assayed for the presence of Hxk2, Hxk2nls1(Ala), and Hxk2nls2(Ala) by immunoblot analysis with anti-Hxk2 antibodies. As shown in Fig. 5a, a specific Hxk2 signal was observed with samples immunoprecipitated with an anti-Kap60 antibody in the strains expressing wild-type Hxk2 and Hxk2nls2(Ala) mutant proteins; no signals were observed when the experiment was done using the strain expressing the Hxk2nls1(Ala) although, as can be seen in Fig. 5a, both Hxk2nls1(Ala) and Hxk2nls2(Ala) proteins are recognized by the anti-Hxk2 antibody. Moreover, because the interaction between wild-type Hxk2 and Kap60 proteins was stronger with samples from high-glucose grown cultures than from low-glucose cultures, our data suggest that the high-glucose condition increases the affinity of Hxk2 for the import receptor.
FIGURE 5.
Interaction of Kap60 and Kap95 with Hxk2. In vivo co-immunoprecipitation of Kap60 (a) and Kap95 (b) with Hxk2, Hxk2nls1(Ala), and Hxk2nls2(Ala). The wild-type, FMY304, and FMY305 strains were grown in YEPD media until an A600 nm of 0.8 was reached and then shifted to high (H-Glc) and low (L-Glc) glucose conditions for 1 h. The cell extracts were immunoprecipitated with a polyclonal anti-Kap60 or anti-Kap95 antibodies (lanes 1–6), or a polyclonal antibody to Pho4 (lanes 7–9). Immunoprecipitates were separated by 12% SDS-PAGE, and co-precipitated Hxk2 variants were visualized on a Western blot with monoclonal anti-Hxk2 antibody. The level of immunoprecipitated Kap60 or Kap95 in the blotted samples was determined by using anti-Kap60 and anti-Kap95 antibodies, respectively. The level of Hxk2 present in the different extracts used in Fig. 5, a and b, was determined by Western blot using anti-Hxk2 antibody. The Western blots shown are representative of results obtained from four independent experiments.
As shown in Fig. 5b, a specific Hxk2 signal was observed, both at high- and low-glucose conditions, with samples immunoprecipitated with an anti-Kap95 antibody in strains expressing wild-type Hxk2, Hxk2nls1(Ala), or Hxk2nls2(Ala) mutant proteins. However, the signals observed both under high- and low-glucose conditions, with the Hxk2nls1(Ala) were much weaker than those observed with the wild-type Hxk2 or the Hxk2nls2(Ala) mutant proteins. Moreover, similar amounts of Kap60 and Kap95 proteins were detected in the immunoprecipitates when anti-Kap60 and anti-Kap95 antibody was, respectively, used. Similar amounts of the Hxk2 proteins were also detected in different protein extracts used by immunoblot analysis with an anti-Hxk2 antibody (Fig. 5, a and b). When an anti-Pho4 antibody was used to detect unspecific immunoprecipitation no signals were observed.
Thus, the Hxk2-Kap60 interaction is both glucose and NLS1 motif dependent. Whereas, the Hxk2-Kap95 interaction was detected both at high- and low-glucose conditions, which suggest that both proteins interact in a manner not regulated by glucose. Identical results were observed when the cell extracts were immunoprecipitated with an anti-Hxk2 antibody and the resulting immunoprecipitates were assayed for the presence of Kap60 or Kap95 proteins by immunoblot analysis with the corresponding antibodies (data not shown).
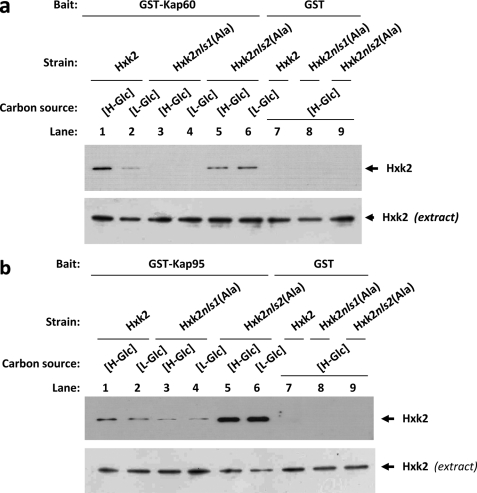
To confirm the in vitro interaction of Hxk2 with Kap60 and Kap95, we also employed the GST pulldown technique. We used crude protein extracts from Hxk2, Hxk2nls1(Ala), or Hxk2nls2(Ala) producing strains and bacterially produced GST-Kap60 and GST-Kap95 fusion proteins. As shown in Fig. 6a, a clear retention of the Hxk2 protein was observed, both at high- and low-glucose conditions, for the sample containing GST-Kap60 and crude extract from strains expressing the wild-type Hxk2 or Hxk2nls2(Ala) proteins. No retention was detected with extracts obtained from the Hxk2nls1(Ala) producing strain grown under both high- and low-glucose conditions. Thus the NLS1 motif of Hxk2 is required for the Hxk2-Kap60 interaction. These results also confirm that the interaction between Hxk2 and Kap60 proteins is stronger under high-glucose than under low-glucose conditions and suggest that both proteins interact in a glucose-dependent manner. As shown in Fig. 6b, when crude protein extracts from Hxk2, Hxk2nls1(Ala), or Hxk2nls2(Ala) producing strains and bacterially produced GST-Kap95 fusion protein were used in GST pulldown experiments, identical results to that shown in Fig. 5b were obtained. In this case, we also detected weaker signals with the Hxk2nls1(Ala) mutant protein than those observed with the wild-type Hxk2 or the Hxk2nls2(Ala) mutant protein. When a control with the GST protein in the reaction mixture was used, no signal of Hxk2, Hxk2nls1(Ala), or Hxk2nls2(Ala) was observed.
FIGURE 6.
GST pulldown assays of the interaction of Kap60 and Kap95 with Hxk2. The GST-Kap60 (a) and GST-Kap95 (b) fusion proteins were purified on glutathione-Sepharose columns. Equal amounts of GST-Kap60 and GST-Kap95 were incubated with cell extracts from wild-type, FMY304, and FMY305 strains. The yeast strains were grown in YEPD media until an A600 nm of 0.8 was reached and then shifted to low (L-Glc) glucose conditions for 1 h. After exhaustive washing the proteins were separated by 12% SDS-PAGE, and retained Hxk2 variants were visualized on a Western blot with polyclonal anti-Hxk2 antibody (lanes 1–6). For the control samples, GST protein was also incubated with the high-glucose (H-Glc) cell extracts, but no signals were detected (lanes 7–9). The level of Hxk2 present in the different extracts used in a and b was determined by Western blot using anti-Hxk2 antibody. The Western blots shown are representative of results obtained from four independent experiments.
We conclude that consistent with the inability of the Hxk2nes2(Ala)nls1(Ala) mutant to enter the nucleus in vivo (Fig. 4a), the mutants lacking the NLS1 were unable to bind to the Kap60 importin (Figs. 5a and 6a). Thus, these results demonstrate that the Hxk2-Kap60 interaction crucially depends on the identified NLS1 sequence of Hxk2. Moreover, because the interaction between the wild-type Hxk2 and Kap60 was systematically stronger with samples from high-glucose grown cultures than from low-glucose cultures, our data suggests that the high-glucose condition increases the affinity of Hxk2 for the import receptor.
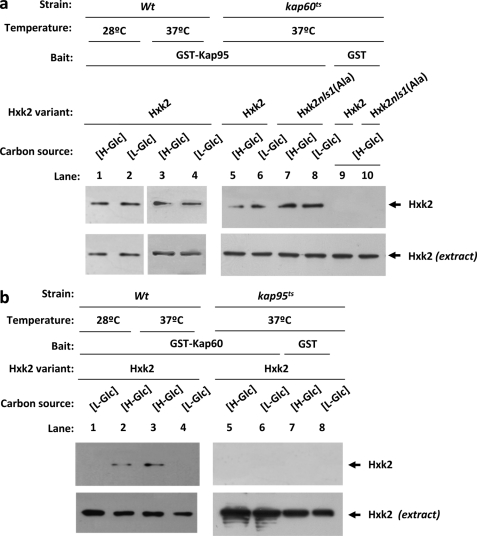
The Interaction between Hxk2 and Kap60 Is Altered in kap95ts Mutants
Whereas our data suggest that both Kap60 and Kap95 functions are required for efficient import of an Hxk2 cargo to the nucleus, these data do not address whether the role of Kap60 or Kap95 is necessary for the importin-Hxk2 interaction. To determine whether the kap95ts and kap60ts mutations alter the ability of Hxk2 to interact, respectively, with Kap60 and Kap95, we employed the GST pulldown technique. First, we used crude protein extracts from kap60ts mutant strains producing Hxk2 or Hxk2nls1(Ala) proteins and bacterially produced GST-Kap95 fusion protein, to determine the Hxk2-Kap95 interaction in the absence of a Kap60 functional protein. As shown in Fig. 7a, similar interaction patterns between Hxk2, Hxk2nls1(Ala), and Kap95 proteins were observed in kap60ts mutant cells at the restrictive temperature and in wild-type cells at both 28 and 37 °C. Second, we used crude protein extracts from kap95ts mutant strain producing the Hxk2 protein and bacterially produced GST-Kap60 fusion protein, to determine the Hxk2-Kap60 interaction in the absence of a Kap95 functional protein. Interestingly, in the absence of the Kap95 functional protein the Hxk2-Kap60 interaction is completely abolished at both high- and low-glucose conditions (Fig. 7b). These results suggest that the Kap95 protein performs an important role in the Hxk2-Kap60 interaction, which is also dependent on the NLS1 motif of Hxk2 and glucose levels in the culture medium.
FIGURE 7.
GST pulldown assays of the interaction of Kap60 and Kap95 with Hxk2 in the absence of functional Kap95 and Kap60 proteins, respectively. The GST-Kap95 and GST-Kap60 fusion proteins were purified on glutathione-Sepharose columns. a, equal amounts of GST-Kap95 were incubated with cell extracts from wild-type, JCY1410 (kap60ts), and FMY306 (kap60tsHxk2nls1(Ala)) strains. b, equal amounts of GST-Kap60 were incubated with cell extracts from wild-type and JCY1407 (kap95ts) strains. The yeast strains were grown in YEPD media until an A600 nm of 0.8 was reached and then shifted to low (L-Glc) glucose conditions for 1 h. The wild-type strain was grown at 28 °C and the mutant cells were grown at 25 °C (permissive temperature) and then shifted to 37 °C (nonpermissive temperature) for 5 h. After exhaustive washing the proteins were separated by 12% SDS-PAGE, and the retained Hxk2 variants were visualized on a Western blot with polyclonal anti-Hxk2 antibody. For the control samples, GST protein was also incubated with the high-glucose (H-Glc) cell extracts, but no signals were detected. The level of Hxk2 present in the different extracts used in a and b was determined by Western blot using anti-Hxk2 antibody. The Western blots shown are representative of results obtained from four independent experiments.
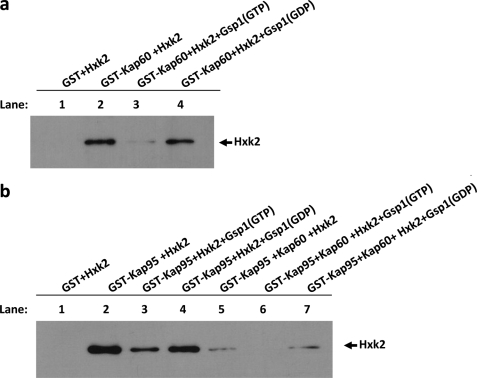
“In Vitro” Formation of the Import Complex
In recent years, a great deal of evidence has supported the idea that Kap95 and Kap60 bind cooperatively to Gsp1(GDP) and its import cargo, leading to the formation of a tetrameric transport complex in the cytoplasm. Once in the nucleus, GDP/GTP exchange induced by guanine nucleotide exchange factors promotes the import complex dissociation, thereby liberating cargo and the carriers that can return to the cytoplasm for another round of import. Thus the classical importin-α/β pathway uses the Gsp1-GTP/GDP cycle to control transport directionality (27). To reconstruct the import complex in vitro and study how the different factors affect the Hxk2-Kap60 and -Kap95 interaction affinity, we performed GST pulldown experiments with purified proteins. We used GST-Hxk2, GST-Gsp1, GST-Kap95, and GST-Kap60 fusion proteins from E. coli lysates. The GST-fused proteins were immobilized with glutathione-Sepharose and purified with thrombin. First, we immobilized GST-Kap60 on glutathione-Sepharose beads and analyzed the binding of Hxk2 to Kap60. Our results indicate that purified Hxk2 has a strong affinity for Kap60 (Fig. 8a, lane 2). Moreover, as expected for an importin-type receptor, the complex formation between Kap60 and Hxk2 is regulated by the guanine nucleotide-binding protein Gsp1. Our data indicate that the Hxk2-Kap60 complex is formed with equal readiness in the presence or absence of Gsp1-GDP (Fig. 8a, lanes 2 and 4). However, the presence of Gsp1-GTP facilitates the dissociation of the complex between Hxk2 and Kap60 (Fig. 8a, lane 3). Second, we studied the interaction of Hxk2 with Kap95, as can be seen in Fig. 8b, purified Hxk2 has a strong affinity for Kap95 in the absence or presence of Gsp1(GDP) (lanes 2 and 4). However, the presence of Gsp1(GTP) decreases the affinity of Hxk2 for Kap95 (lane 3).
FIGURE 8.
The import Hxk2-Kap60-Kap95 complex formation. GST pulldown assays of the effect of Gsp1 on the Hxk2-Kap60 (a) and Hxk2-Kap95 (b) interaction stability. GST-Hxk2, GST-Gsp1, and GST-Kap60 fusion proteins were purified on glutathione-Sepharose columns and incubated with thrombin to isolate Hxk2, Gsp1, and Kap60 proteins, respectively. The Gsp1 protein was loaded with GTP to generate Gsp1(GTP), or with GDP to generate Gsp1(GDP). The Hxk2 protein was incubated with purified GST-Kap60 or GST-Kap95 bound to glutathione-Sepharose beads in the presence of Gsp1(GTP) and Gsp1(GDP) in the absence or presence of Kap60 purified protein (lanes 5–7). The beads were washed extensively. Co-precipitated proteins were resolved by 12% SDS-PAGE and visualized on a Western blot with polyclonal anti-Hxk2 antibody.
Furthermore, when we tried to reconstruct the quaternary import complex by using GSP-Kap95 immobilized with glutathione-Sepharose and Hxk2, Gsp1(GTP/GDP), and Kap60 purified proteins, we obtained interesting results representing the whole quaternary import complex reconstructed in vitro. Our data demonstrate that the Kap95-Hxk2 interaction in the presence of Kap60 is still regulated by the Gsp1-GTP/GDP cycle. The Hxk2-Kap95 interaction signal is less intense in the presence or absence of Gsp1-GDP (Fig. 8b, lanes 5 and 7), but it is absolutely absent in the presence of Gsp1-GTP. This experiment suggests that a quaternary complex between Kap95, Kap60, Gsp1, and Hxk2 is formed in a cooperative way in vitro. Moreover, the complex formation is controlled by the GDP/GTP-bound state of Gsp1, because loading of Gsp1 with GTP disrupts the formation of the import complex.
DISCUSSION
In the glucose-repression signaling pathway, two major mediators are the proteins, Mig1 and Hxk2. In high-glucose growth conditions, nuclear Hxk2 interacts both with Mig1 and the Snf1 subunit of the Snf1 protein kinase complex to establish the repression state of several genes. Hxk2 interacts strongly with Mig1 in high glucose-grown cells and previous findings implicate serine 311 of Mig1 as a critical residue for this interaction (13). In low-glucose grown cells, the Hxk2 interaction with Mig1 is abolished and a transient increase in interaction between Snf1 and Mig1 was detected (28). This interaction pattern potentially stimulates Mig1 phosphorylation by the Snf1 kinase at serine 311 and this phosphorylation results in most Mig1 being exported from the nucleus to the cytoplasm. Simultaneously, Hxk2 was no longer retained in the nucleus and accumulates in the cytoplasm too. Thus, Hxk2 shuttles between the nucleus and cytoplasm in response to glucose availability. Recently, the mechanism of nuclear Hxk2 export has been reported (15), but the Hxk2 nuclear import mechanism remains to be established.
This study shows that nuclear import of Hxk2 is not mediated by importins Nmd5 and Msn5, because deletion of both nonessential proteins does not abolish the import of Hxk2 to the nucleus. Although Mig1 interacts with Hxk2 in the nucleus, Mig1 does not need to form a complex with Hxk2 to enter the nucleus because Hxk2 is found in the nucleus in an xpo1–1Δmig1 double mutant (13, 29). In this work, we determined that functional Kap60 and Kap95 proteins are required for Hxk2 to enter the nucleus. Moreover, the Lys6, Lys7, and Lys12 amino acids of Hxk2 (NLS1 motif) were also required for Hxk2 to enter the nucleus, and the change of these lysines to alanines reduced the invertase repression capacity of Hxk2nls1(Ala), because the protein is essentially excluded from the nucleus. Several NLS-cargoes transported by the Kap60/Kap95 system have been identified so far (30); however, only a few studies have shown which sequences within the transported protein-mediated interaction and rigorous evidence for a direct interaction between α-importin Kap60, β-importin Kap95, and cargo is usually not provided. We demonstrate in this study that Hxk2 associates with both Kap60 and Kap95 in protein immunoprecipitation and pulldown experiments from yeast cell lysate. We also demonstrate that diminution of either Kap60 or Kap95 function prevented Hxk2 from entering the nucleus.
The Hxk2 protein directly interacts with α-importin Kap60 and the β-importin Kap95 both at high- and low-glucose concentrations; however, a strong signal was consistently detected in samples from cells grown in high-glucose medium and no or very weak interaction was observed between Kap60 and Hxk2 proteins under low-glucose growth conditions. Moreover, no interaction was observed between Kap60 and the Hxk2nls1(Ala) mutant protein, in neither high- nor low-glucose conditions, and this indicates that the NLS1 motif is necessary for interaction between Kap60 and Hxk2. The interaction affinity between Hxk2 and Kap95 is similar under high- and low-glucose growth conditions. However, although the NLS1 motif of Hxk2 is not required for the Hxk2nls1(Ala)-Kap95 interaction, the Kap95 protein is essential for interaction between Hxk2 and the α-importin Kap60.
A potentially interesting observation also made in this report is the role of the NLS2 motif of Hxk2 in Kap60 and Kap95 interactions. Although interaction of the wild-type Hxk2 with Kap60 is glucose regulated, the interaction between Hxk2nls2(Ala) and Kap60 is constitutive. Moreover, the NLS2 mutation increases the affinity between Hxk2 and Kap95 as shown Fig. 6b. Thus, although the NLS2 motif of Hxk2 in not required for nuclear import it may be an important tool to help in the understanding of the regulation of the Hxk2 nucleocytoplasmic shuttling.
Our data suggest that Hxk2 probably enters the nucleus via the canonical route of binding the α-importin Kap60 through the N-terminal basic amino acid NLS on Hxk2. To achieve this interaction the presence of β-importin Kap95 that is essential for recruitment of the Hxk2 protein in the correct orientation to the ternary complex is necessary. Then, the tripartite protein complex could be recognized by the nuclear pore for transport into the nucleus. Moreover, we have reconstructed from bacterially produced proteins the Hxk2 import complex in vitro. Our GST pulldown assay data, obtained by using the purified proteins, suggest that the Hxk2-Kap60-Kap95 ternary complex is stable in the presence of Gsp1(GDP) as happens in the cytoplasm. However, the complex is disrupted in the presence of high levels of Gsp1(GTP), a condition that occurs in the nucleus. Thus, efficient dissociation of the Hxk2-Kap60-Kap95 complex is mediated primarily by Gsp1(GTP), which displaces Hxk2 from the α/β-importin receptor. These experiments, performed using purified proteins from bacteria, also exclude the possibility that some unknown protein(s) in the yeast extract could mediate the interaction between the Hxk2 protein and the Kap60 or Kap95 proteins.
The current study has identified a new pathway for Hxk2 import into the nucleus. Hxk2 directly binds the α- and β-importins, and together they form a complex to allow transit of Hxk2 through the nuclear pore. Kap95 facilitates recognition of the Hxk2 NLS1 motif by Kap60 and both importins are essential for Hxk2 nuclear import. In addition, we found that Kap60 and Kap95 form a stable complex with Hxk2 in the cytoplasm where the level of Gsp1(GTP) is low. After translocation into the nucleus where Gsp1(GTP) is high, this complex is dissociated via a mechanism that includes binding of Gsp1(GTP) to the α/β-importins complex. It has been shown recently that Xpo1 is the export receptor for Hxk2 (15). Here, we show that Kap60/Kap95 serve as the import receptors. This work completes the mechanisms by which Hxk2 enters and exits from the nucleus in S. cerevisiae. The regulatory mechanisms that operate in the nuclear import-export cycle of Hxk2 remain to be determined.
Acknowledgments
We are grateful to following people for generously providing yeast strains and plasmids: E. Hurt for the pGEX4T3-GSP1 plasmid, M. P. Rout for pGEX-Kap95 and pGEX-Kap60 plasmids, and J. C. Igual for kap95ts and kap60ts strains.
This work was supported by Grants BFU2007-66063-C02-02 and BFU2010-19628-C02-01 from the Ministerio de Ciencia e Innovación (Spain).
- NLS
- nuclear localization sequence
- Hxk2
- hexokinase 2.
REFERENCES
- 1. Radu A., Blobel G., Moore M. S. (1995) Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc. Natl. Acad. Sci. U.S.A. 92, 1769–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Görlich D., Prehn S., Laskey R. A., Hartmann E. (1994) Isolation of a protein that is essential for the first step of nuclear protein import. Cell 79, 767–778 [DOI] [PubMed] [Google Scholar]
- 3. Stade K., Ford C. S., Guthrie C., Weis K. (1997) Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90, 1041–1050 [DOI] [PubMed] [Google Scholar]
- 4. Weis K. (2003) Regulating access to the genome. Nucleocytoplasmic transport throughout the cell cycle. Cell 112, 441–451 [DOI] [PubMed] [Google Scholar]
- 5. Kalab P., Weis K., Heald R. (2002) Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science 295, 2452–2456 [DOI] [PubMed] [Google Scholar]
- 6. Smith A. E., Slepchenko B. M., Schaff J. C., Loew L. M., Macara I. G. (2002) Systems analysis of Ran transport. Science 295, 488–491 [DOI] [PubMed] [Google Scholar]
- 7. Cook A., Bono F., Jinek M., Conti E. (2007) Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 76, 647–671 [DOI] [PubMed] [Google Scholar]
- 8. Kalderon D., Roberts B. L., Richardson W. D., Smith A. E. (1984) A short amino acid sequence able to specify nuclear location. Cell 39, 499–509 [DOI] [PubMed] [Google Scholar]
- 9. Görlich D., Kostka S., Kraft R., Dingwall C., Laskey R. A., Hartmann E., Prehn S. (1995) Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr. Biol. 5, 383–392 [DOI] [PubMed] [Google Scholar]
- 10. Herrero P., Galíndez J., Ruiz N., Martínez-Campa C., Moreno F. (1995) Transcriptional regulation of the Saccharomyces cerevisiae HXK1, HXK2, and GLK1 genes. Yeast 11, 137–144 [DOI] [PubMed] [Google Scholar]
- 11. Randez-Gil F., Herrero P., Sanz P., Prieto J. A., Moreno F. (1998) Hexokinase PII has a double cytosolic-nuclear localization in Saccharomyces cerevisiae. FEBS Lett. 425, 475–478 [DOI] [PubMed] [Google Scholar]
- 12. Ahuatzi D., Herrero P., de la Cera T., Moreno F. (2004) The glucose-regulated nuclear localization of hexokinase 2 in Saccharomyces cerevisiae is Mig1-dependent. J. Biol. Chem. 279, 14440–14446 [DOI] [PubMed] [Google Scholar]
- 13. Ahuatzi D., Riera A., Peláez R., Herrero P., Moreno F. (2007) Hxk2 regulates the phosphorylation state of Mig1 and therefore its nucleocytoplasmic distribution. J. Biol. Chem. 282, 4485–4493 [DOI] [PubMed] [Google Scholar]
- 14. Moreno F., Ahuatzi D., Riera A., Palomino C. A., Herrero P. (2005) Glucose sensing through the Hxk2-dependent signaling pathway. Biochem. Soc. Trans. 33, 265–268 [DOI] [PubMed] [Google Scholar]
- 15. Peláez R., Herrero P., Moreno F. (2009) Nuclear export of the yeast hexokinase 2 protein requires the Xpo1 (Crm1)-dependent pathway. J. Biol. Chem. 284, 20548–20555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wallis J. W., Chrebet G., Brodsky G., Rolfe M., Rothstein R. (1989) A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell 58, 409–419 [DOI] [PubMed] [Google Scholar]
- 17. Ma H., Botstein D. (1986) Effects of null mutations in the hexokinase genes of Saccharomyces cerevisiae on catabolite repression. Mol. Cell. Biol. 6, 4046–4052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Taberner F. J., Quilis I., Igual J. C. (2009) Spatial regulation of the start repressor Whi5. Cell Cycle 8, 3010–3018 [PubMed] [Google Scholar]
- 19. Kadowaki H., Kadowaki T., Wondisford F. E., Taylor S. I. (1989) Use of polymerase chain reaction catalyzed by Taq DNA polymerase for site-specific mutagenesis. Gene 76, 161–166 [DOI] [PubMed] [Google Scholar]
- 20. Künzler M., Gerstberger T., Stutz F., Bischoff F. R., Hurt E. (2000) Yeast Ran-binding protein 1 (Yrb1) shuttles between the nucleus and cytoplasm and is exported from the nucleus via a CRM1 (XPO1)-dependent pathway. Mol. Cell. Biol. 20, 4295–4308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leslie D. M., Timney B., Rout M. P., Aitchison J. D. (2006) Studying nuclear protein import in yeast. Methods 39, 291–308 [DOI] [PubMed] [Google Scholar]
- 22. Gascón S., Lampen J. O. (1968) Purification of the internal invertase of yeast. J. Biol. Chem. 243, 1567–1572 [PubMed] [Google Scholar]
- 23. DeVit M. J., Johnston M. (1999) The nuclear exportin Msn5 is required for nuclear export of the Mig1 glucose repressor of Saccharomyces cerevisiae. Curr. Biol. 9, 1231–1241 [DOI] [PubMed] [Google Scholar]
- 24. Polizotto R. S., Cyert M. S. (2001) Calcineurin-dependent nuclear import of the transcription factor Crz1p requires Nmd5p. J. Cell Biol. 154, 951–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ferrigno P., Posas F., Koepp D., Saito H., Silver P. A. (1998) Regulated nucleo/cytoplasmic exchange of HOG1 MAPK requires the importin β homologs NMD5 and XPO1. EMBO J. 17, 5606–5614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kosugi S., Hasebe M., Matsumura N., Takashima H., Miyamoto-Sato E., Tomita M., Yanagawa H. (2009) Six classes of nuclear localization signals specific to different binding grooves of importin α. J. Biol. Chem. 284, 478–485 [DOI] [PubMed] [Google Scholar]
- 27. Quan X., Yu J., Bussey H., Stochaj U. (2007) The localization of nuclear exporters of the importin-β family is regulated by Snf1 kinase, nutrient supply, and stress. Biochim. Biophys. Acta 1773, 1052–1061 [DOI] [PubMed] [Google Scholar]
- 28. Treitel M. A., Kuchin S., Carlson M. (1998) Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol. Cell. Biol. 18, 6273–6280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tomás-Cobos L., Sanz P. (2002) Active Snf1 protein kinase inhibits expression of the Saccharomyces cerevisiae HXT1 glucose transporter gene. Biochem. J. 368, 657–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lange A., Mills R. E., Lange C. J., Stewart M., Devine S. E., Corbett A. H. (2007) Classical nuclear localization signals. Definition, function, and interaction with importin α. J. Biol. Chem. 282, 5101–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Belinchón M. M., Gancedo J. M. (2007) Different signalling pathways mediate glucose induction of SUC2, HXT1, and pyruvate decarboxylase in yeast. FEMS Yeast Res. 7, 40–47 [DOI] [PubMed] [Google Scholar]