Abstract
Matrix metalloproteinase (MMP)-13 has a pivotal, rate-limiting function in cartilage remodeling and degradation due to its specificity for cleaving type II collagen. The proximal MMP13 promoter contains evolutionarily conserved E26 transformation-specific sequence binding sites that are closely flanked by AP-1 and Runx2 binding motifs, and interplay among these and other factors has been implicated in regulation by stress and inflammatory signals. Here we report that ELF3 directly controls MMP13 promoter activity by targeting an E26 transformation-specific sequence binding site at position −78 bp and by cooperating with AP-1. In addition, ELF3 binding to the proximal MMP13 promoter is enhanced by IL-1β stimulation in chondrocytes, and the IL-1β-induced MMP13 expression is inhibited in primary human chondrocytes by siRNA-ELF3 knockdown and in chondrocytes from Elf3−/− mice. Further, we found that MEK/ERK signaling enhances ELF3-driven MMP13 transactivation and is required for IL-1β-induced ELF3 binding to the MMP13 promoter, as assessed by chromatin immunoprecipitation. Finally, we show that enhanced levels of ELF3 co-localize with MMP13 protein and activity in human osteoarthritic cartilage. These studies define a novel role for ELF3 as a procatabolic factor that may contribute to cartilage remodeling and degradation by regulating MMP13 gene transcription.
Keywords: Chondrocytes, Chromatin Immunoprecipitation (ChiP), Cytokine Action, Ets Family Transcription Factor, Gene Regulation, Matrix Metalloproteinase (MMP), Signal Transduction
Introduction
The matrix metalloproteinases (MMPs)3 are a family of enzymes that coordinately degrade components of the extracellular matrix in physiological/normal matrix remodeling processes (1) and in disease states wherein their aberrant and enhanced expression contributes to exacerbated matrix degradation (2, 3). Type II collagen is a major constituent of articular cartilage that contributes to its structural and functional properties by conferring tensile strength, and its degradation is the pivotal event that determines the irreversible progression of osteoarthritis (OA), in which articular cartilage is slowly and progressively destroyed (2). OA occurs in conjunction with changes in the synovium and subchondral bone that are associated with dysregulated chondrocyte physiology exemplified in part by the abnormal expression of catabolic and anabolic gene products (2). In this context, proinflammatory cytokines have been shown to trigger a diverse array of intracellular signaling pathways leading to the overexpression of a variety of matrix-degrading enzymes, including MMPs (2).
Because collagen degradation is mediated almost exclusively by MMPs, those with higher collagenolytic activity (collagenases) are the rate-limiting, major players in irreversible cartilage destruction (4), and MMP13 (collagenase 3) plays a very prominent role here. MMP13 preferentially and more potently cleaves type II collagen compared with other collagenases (5–7). Moreover, MMP13 levels and activity are enhanced in OA cartilage, associated with degenerative changes and co-localizing with MMP13-specific type II collagen cleavage products, inflammatory cytokines, and their receptors (4, 8). Further, in vivo evidence has shown OA changes in transgenic mice overexpressing constitutively active MMP13 (9), whereas Mmp13 knock-out mice are refractory to surgically induced OA (10). Therefore, a thorough understanding of how MMP13 is transcriptionally regulated by intracellular regulatory factors and how their activities are modulated by extracellular cues leading to aberrant MMP13 expression in OA cartilage is essential.
The E74-like factor 3 (ELF3), also known as ESE1, ESX, ERT, and JEN, is an epithelium-specific member of the E26 transformation-specific sequence (ETS) family of transcription factors (11). ETS transcription factors are trans-acting phosphoproteins that share a highly conserved DNA-binding winged helix-turn-helix domain (ETS domain), which specifically binds to ETS sequence motifs in the transcriptional control elements of target genes (12). ETS factors induce or repress transcription depending on their activation by mitogen-activated protein kinases (MAPKs) and by their association with other cofactors in a promoter context-specific manner (12, 13). ETS proteins mediate a variety of biological processes, including cell proliferation, differentiation, transformation, and tumor invasion, with the latter involving ECM remodeling by concerted modulation of MMPs and their inhibitors (14). ELF3 plays essential roles during epithelial cell differentiation (11, 15, 16), gut development (17), apoptosis (18, 19), and physiology of normal breast and breast cancer epithelial cells (20, 21). ELF3 activity within a given cell may depend on its expression levels and involve both nuclear transcriptional mechanisms and events independent of the nucleus (19). In addition, ELF3 is strongly induced in a variety of cell types by stress or inflammatory conditions dependent, at least in part, on canonical NF-κB (p65/p50) binding to a high affinity NF-κB site in the proximal ELF3 promoter (22, 23). ELF3 mediates inflammatory responses by regulating genes, such as nitric-oxide synthase 2 (NOS2), cyclooxygenase 2 (PTGS2/COX2), and angiopoietin-1 (22, 24–26), and thus may contribute as a procatabolic/antianabolic regulatory factor in inflammatory disease states, such as airway inflammation, OA, and rheumatoid arthritis (22, 23). In chondrocytes, ELF3 expression is induced in vitro by IL-1β, and we have previously reported that it accounts for the NF-κB-dependent and partially for the IL-1β-mediated repression of the COL2A1 promoter (27), thus making ELF3 a potential proinflammatory mediator in OA disease.
Herein, we provide evidence for a novel role of ELF3 as a regulatory component that drives the abnormal expression of MMP13 under proinflammatory conditions in chondrocytes. We show that ELF3 activates MMP13 transcription by binding to a conserved ETS site in its proximal promoter region and that ELF3 up-regulates MMP13 expression by acting in conjunction with MEK/ERK signaling and enhancing AP-1-driven MMP13 promoter activity in response to IL-1β stimulation. Importantly, dysregulated MMP13 expression and activity in OA articular cartilage is associated with enhanced levels of ELF3.
EXPERIMENTAL PROCEDURES
Cell Culture
Human primary chondrocytes were isolated, as described (28), from articular cartilage obtained from intact regions of femoral condyles of OA patients undergoing total knee replacement, with approval by the Institutional Review Board and patient consent. Immediately after surgery, the cells were isolated by sequential digestion with Pronase (Promega) and collagenase P (Promega), cultured to confluence in Dulbecco's modified Eagle's medium (DMEM)/F-12 containing 10% fetal bovine serum (FBS), and used for experimental purposes at passage one. Human immortalized T/C-28a2 chondrocytes were cultured in DMEM/Ham's F-12 containing 10% FBS, as described previously (27). Elf3 knock-out mice (17) were obtained from Dr. Melanie A. Pritchard (Monash University, Clayton, Victoria, Australia). Primary mouse chondrocytes were isolated as described (29) from 5–6-day-old wild type (C57BL/6) or Elf3 knock-out mouse articular cartilage. Passage 0 chondrocytes were seeded in DMEM/F-12 containing 10% FBS, allowed to reach confluence, trypsinized, and seeded in 6-well plates in DMEM/F-12 containing 10% FBS for experimental purposes. All experiments involving IL-1β and TNFα (R&D Systems) stimulation were performed in serum-free conditions after overnight starvation.
siRNA Transfection
Human primary or immortalized chondrocytes were transfected, as described previously (28). Briefly, 2.5 × 105 cells were seeded 24 h before transfection in 6-well plates in DMEM/F-12 containing 10% FBS. The non-targeting control siRNA (Dharmacon) or siRNAs against ELF3 (Dharmacon) were transfected at a final concentration of 50 nm using Lipofectamine PLUS reagents in serum-free medium. Transfection efficiency was assessed using non-targeting siRNA conjugated with rhodamine (Dharmacon), and knockdown efficacy was assessed by real-time PCR and Western blotting. At 72 h after transfection, cells were stimulated with IL-1β or vehicle for the indicated times.
Real-time Quantitative RT-PCR (RT-qPCR) Analysis
Total RNA was isolated using the RNeasy Plus minikit (Qiagen), and 150 ng were reverse transcribed, as described (28). Amplifications were carried out using SYBR Green I-based real-time PCR on the Opticon 2 real-time PCR detector system (Bio-Rad), as described (28), using the PCR primers and conditions indicated in supplemental Table S1. The data were calculated as the ratio of each gene to GAPDH, and HPRT1 was utilized as an additional housekeeping control.
Western Blot Analysis
Human and mouse primary chondrocytes and human immortalized chondrocytes were maintained in DMEM/F-12 medium containing 10% FBS. Before treatments with IL-1β, cells were incubated in serum-free medium overnight and challenged with the cytokine for the indicated times. To collect total cell lysates, cells were rinsed with cold PBS and lysed with 1× radioimmune precipitation assay buffer (Santa Cruz Biotechnology, Inc.) containing PMSF, proteinase inhibitors, and sodium orthovanadate, as indicated by the supplier. Protein content was determined using Coomassie Plus protein assay reagent (Pierce), and equal amounts of protein lysates were resolved in 10% Tris-HCl polyacrylamide gels under reducing conditions. Proteins were then transferred to polyvinylidine fluoride (PVDF) membranes using a semidry transfer system (Bio-Rad). Membranes were blocked with 5% nonfat milk in Tris-buffered saline with 0.1% Tween 20 for 1 h at room temperature and then incubated with primary antibodies against ELF3 (Abcam), phospho-p38, phospho-SAPK/JNK, phospho-ERK1/2, total p38, total SAPK/JNK, or total ERK1/2 (Cell Signaling); β-tubulin (Abcam) was used as loading control. After washing, the membrane was incubated with the corresponding secondary antibody conjugated with horseradish peroxidase. Signals were detected using enhanced chemiluminescence (Amersham Biosciences).
Plasmid Construction
The −1602/+20-MMP13-Luc promoter construct was described previously (30). The MMP13 promoter sequences spanning −1007/+20 and −273/+20 bp were prepared by PCR using the pCAT-MMP13 promoter construct as template as described (30). The sense primer for each construct included an artificial SacI site and the antisense primer included an artificial XhoI site. The constructs were cloned into the pGL2-Basic luciferase reporter gene vector (Promega).
The deletion mutant lacking the proximal −231/−39 bp sequence was generated by digestion of the −1602/+20 bp promoter construct with BbvCI and StuI (New England Biolabs) followed by blunt-ended ligation (Takara). Point mutants of the proximal ETS binding sites (EBS) (designated EBS-A, -B1, and -B2mut) within the MMP13 promoter were generated by two-step PCR mutagenesis using the wild type −273/+20-MMP13 construct as template. For the first PCR, common forward or reverse primers, containing XmaI and XhoI sites, respectively, were used in combination with the primers indicated in Table 1. The resulting PCR products were purified and utilized in the second PCR, performed with the common forward and reverse primers containing XmaI and XhoI restriction sites utilized for the first PCR. The resultant amplification products were purified and digested with SacI and XhoI (New England Biolabs) and transferred into the pGL2-Basic backbone treated with the same enzymes.
TABLE 1.
PCR mutagenesis primers
The mutated EBS sites are indicated in boldface type, with lowercase indicating the mutation site.
| Description | Primer sequences |
|---|---|
| Common forward (XmaI) | 5′-TAACATAACCCGGGTGTACTACTCTCTGCT-3′ |
| Common reverse (XhoI) | 5′-GCCAAGCTTACTTAGATCTCGAGTCTAGATTG-3′ |
| EBS-A mutant | Forward: 5′- CACACTCGGGAGatAAAAGAAAAAGTCGCC-3′ |
| Reverse: 5′-GGCGACTTTTTCTTTTatCTCCCGAGTGTG-3′ | |
| EBS-B1 mutant | Forward: 5′- TTCAAGTGACTAatAAGTGGAAACCTATCC-3′ |
| Reverse: 5′-GGATAGGTTTCCACTTatTAGTCACTTGAA-3′ | |
| EBS-B2 mutant | Forward: 5′-AAGTGACTAGGAAGTGtcAACCTATCCATA-3′ |
| Reverse: 5′-TATGGATAGGTTgaCACTTCCTAGTCACTT-3′ |
Expression vectors encoding ELF3, ELF5, EHF, RUNX2, c-Fos, and c-Jun have been described (27, 30). Vectors encoding JNK, ERK1/2, p38, MKP1, and the constitutively active MAP2K MKK7, MKK6, and MEK1 were purchased from Addgene and described elsewhere (30). All constructs were confirmed by DNA sequencing, performed at the Cornell University Life Sciences Core Laboratories Center.
Transfection and Reporter Assays
Transient transfection experiments were carried out in T/C-28a2 cells using Lipofectamine PLUSTM reagents (Invitrogen). Cells were seeded 24 h before transfection in 24-well tissue culture plates at 2.5 × 104 cells/cm2 in DMEM/F-12 containing 10% FBS. Transfections were carried out in serum-free and antibiotic-free medium using no more than 450 ng of plasmid DNA, including 350 ng of reporter constructs. Cell lysates were prepared by extraction with 100 μl of reporter lysis buffer (Promega), and the protein content was determined using the Coomassie Plus protein assay reagent. Unless specified otherwise, luciferase activities were normalized to the pRL-SV40 Renilla luciferase control vector (Promega).
Chromatin Immunoprecipitation (ChIP) Assay
The ChIP-IT Express Enzymatic Kit (Active Motif) was used to perform ChIP assays according to the manufacturer's instructions with minor modifications. Briefly, T/C-28a2 cells were plated on 150-mm dishes and either transfected with FLAG-tagged ELF3 expression vectors or incubated with 1 ng/ml IL-1β for 2 h, with or without a 45-min pretreatment with U0126 (2.5 μm; Sigma). Cross-linking was performed with 1% formaldehyde for 10 min at room temperature; nuclei were isolated and chromatin was enzymatically sheared for 8 min at 37 °C, resulting in chromatin fragments of 250–1000 bp. Chromatin was precleared by incubation with 25 μl of protein G magnetic beads and 5 μg of nonspecific (control) rabbit IgG (Cell Signaling) for 2 h at 4 °C with rotation. After preclearing and removal of the protein G magnetic beads, the lysates were incubated at 4 °C for 16 h with 5 μg of rabbit anti-ELF3 antibody (Orbigen), rabbit anti-FLAG antibody (Sigma), or normal rabbit IgG (Cell Signaling), and 10 μl of the precleared chromatin was stored to be used as assay input. After reverse cross-linking of the DNA-protein complexes, the DNA was subjected to PCR analysis using 5 μl of the eluted DNA and the following set of primers: 5′-CCCTCAAATTCTACCACAAACC-3′ (forward) and 5′-CAATGGTGAGTCATCACTTATGG-3′ (reverse), spanning from −157 to −38 bp of the proximal human MMP13 promoter. The PCR products were resolved on a 2.5% agarose gel. For real-time PCR analysis, the precipitated DNA was purified using DNA minicolumns (Qiagen), and the final DNA preparations were PCR-amplified using 2 μl of the purified DNA and the Opticon 2 real-time PCR detector system (Bio-Rad) utilizing the aforementioned primers. Primer efficiency was calculated utilizing serial dilutions of the pooled input DNA samples. For real-time PCR analysis, the CT of each sample was normalized to the CT of the input sample (10%). Specific GAPDH primers provided by the manufacturer (Active Motif) were used for assessing the ChIP quality of the digested chromatin and as negative controls for the precipitated DNA.
Histological and Immunohistochemical Analysis
Full thickness human articular cartilage samples, including cartilage and bone, were obtained with Institutional Review Board approval from the medial tibial plateaus of the knee joints of 10 OA patients undergoing total knee replacement. After fixation, decalcification, and paraffin embedding, sections of 6 μm were cut, deparaffinized in xylene, and rehydrated through an ethanol series. Every 10th section was collected for Safranin O/Fast green staining, and the OA stage was graded independently by two blinded observers according to the OARSI grading scale (31). For immunostaining, successive adjacent sections within each patient were deparaffinized and quenched for endogenous peroxidase activity. The sections were then blocked and incubated overnight with antibodies against ELF3 (Abcam), MMP13 (Chemicon), and C1,2C (IBEX). For negative controls, isotype-matched IgG (Santa Cruz Biotechnology, Inc.) was used in place of the primary antibodies. The sections were counterstained with methyl green, and the Vectastain ABC Elite kit (Vector Laboratories) was used as described by the manufacturer. Immunostainings were performed in at least three different donors per OARSI grade and repeated at least three times per donor.
Statistical Analysis
Data are reported as means ± standard error (S.E.) of at least three independent experiments. Statistical analysis was performed by analysis of variance followed by Student's t test with p values of <0.05 considered significant.
RESULTS
ELF3 Is a Novel Regulator of MMP13 Expression in Human and Murine Primary Chondrocytes
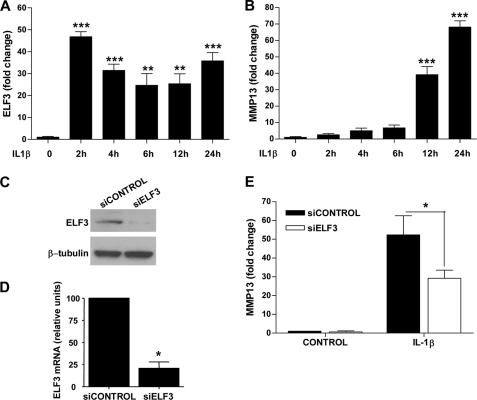
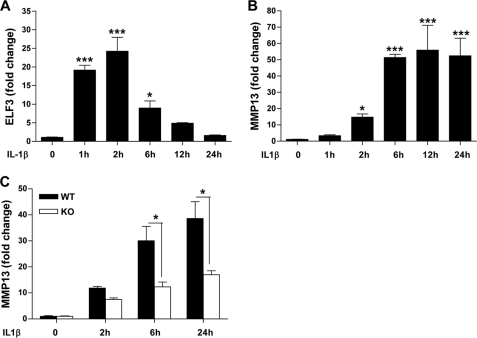
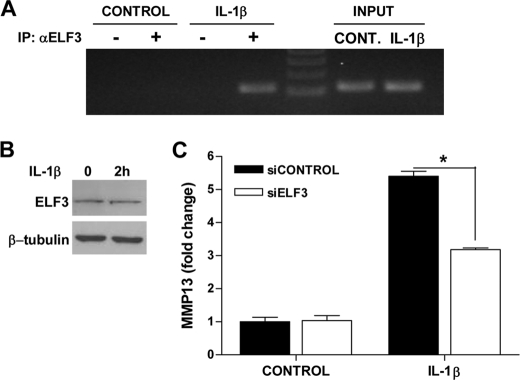
ELF3 expression is induced by inflammatory cytokines, including IL-1β, in different cell types in an NF-κB (p65/p50)-dependent manner (22, 23, 27), and in previous work, we showed that ELF3 mediates the antianabolic actions of IL-1β in chondrocytes by binding to the COL2A1 promoter and suppressing its activity (27). We initially screened for genes whose expression depended on ELF3 for their response to IL-1β by performing a TaqMan low density array screen of RNAs isolated from primary human chondrocytes transfected with siRNA-ELF3 or non-targeting siRNA oligonucleotides, which showed that MMP13 was among the genes that were differentially regulated by ELF3 in response to IL-1β stimulation (data not shown). Indeed, subsequent RT-qPCR analyses revealed that IL-1β rapidly induced the sustained expression of ELF3 in human primary OA articular chondrocytes, which was followed by the strong up-regulation of MMP13 mRNA (Fig. 1, A and B). Like that of MMP13, the IL-1β-dependent expression profiles of MMP1, -3, and -9 were also enhanced at later times, whereas COL2A1 mRNA was suppressed (supplemental Fig. S1). Importantly, IL-1β-induced MMP13 mRNA in primary human OA chondrocytes was significantly down-modulated by ELF3 knockdown (KD) (Fig. 1, C–E). In murine chondrocytes, IL-1β-induced Elf3 mRNA peaked at 2 h (declining thereafter), followed by the induction of Mmp13 mRNA peaking at 6 h and remaining stable up to 24 h (Fig. 2, A and B). Similar to our findings in human primary chondrocytes, IL-1β-induced Mmp13 mRNA was strongly although not completely reduced in Elf3−/− mouse chondrocytes compared with their wild type counterparts (Fig. 2C), clearly indicating that ELF3 is an important contributing factor for IL-1β-induced MMP13 expression in chondrocytes and also that other regulatory factors contribute to MMP13 expression independent of ELF3.
FIGURE 1.
ELF3 regulates IL-1β-induced MMP13 gene expression in human primary articular chondrocytes. Chondrocytes were isolated from articular cartilage from the femoral condyles of five OA patients undergoing total knee replacement, and cultures at passage 1 were examined for the relative levels of (A) ELF3 and (B) MMP13 mRNA in response to IL-1β stimulation for 2–24 h. The same primary chondrocytes were transfected with 50 nm siRNA oligonucleotides against ELF3 (siELF3) or non-targeting siRNA (siCONTROL). Knockdown efficiency was assessed at 72 h by immunoblotting (C) and RT-qPCR (D) at 72 h post-transfection, the cells were stimulated with 1 ng/ml IL-1β for 24 h, and cellular MMP13 mRNA levels were analyzed by RT-qPCR (E). The values were normalized to GAPDH and are shown as mean ± S.E. (error bars). *, p < 0.05; **, p < 0.01; ***, p < 0.001; ELF3 Western blots were reprobed for β-tubulin as a loading reference control.
FIGURE 2.
Effects of ELF3 on MMP13 expression in mouse primary chondrocytes. Mouse primary chondrocytes were isolated from articular cartilage from wild-type 5–6-day-old C57BL/6 mice and incubated with 1 ng/ml IL-1β for the indicated times. IL-1β-induced ELF3 (A) and MMP13 (B) mRNA levels were analyzed by RT-qPCR. C, mouse primary chondrocytes isolated from articular cartilage from wild-type (WT) and Elf3 knock-out (KO) mice were incubated with 1 ng/ml IL-1β for the indicated times. Total RNAs were isolated, and MMP13 mRNA was analyzed by RT-qPCR. Each value was normalized to GAPDH in the same sample and shown as mean ± S.E. (error bars); *, p < 0.05; ***, p < 0.001.
We also investigated the contribution of ELF3 in induction of Mmp13 mRNA by TNFα in murine wild-type and Elf3−/− chondrocytes. Consistent with previous reports using chondrocyte cell lines and other cell types (22, 23), RT-qPCR analysis showed that stimulation with TNFα leads to increased Elf3 mRNA expression but not with the potency of IL-1β (supplemental Fig. S2A). TNFα-induced Mmp13 mRNA levels were suppressed in Elf3−/− mouse chondrocytes compared with their wild-type counterparts (supplemental Fig. S2B). This latter decrease in Mmp13 mRNA only reached statistical significance after 24 h of TNFα stimulation, indicating that other mediators are also involved in the induction of Mmp13 by TNFα in chondrocytes (32, 33).
Next, we explored the relative contribution of ELF3 to the IL-1β-induced expression of other important factors involved in cartilage degradative processes (supplemental Fig. S3). Consistent with reports showing that ELF3 acts as a transactivator of NOS2 and COX2/PTGS2 (25, 26), RT-qPCR analyses revealed that the IL-1β-induced Nos2 and Ptgs2 mRNA levels were significantly reduced in Elf3−/− compared with wild type mouse chondrocytes, whereas ELF3 did not impact the IL-1β-induction of Mmp2, -3, and -9 mRNA levels (supplemental Fig. S3).
ELF3 Protein Levels Are Enhanced in Human OA Cartilage and Co-localize with Increased MMP13 Protein Levels and Activity
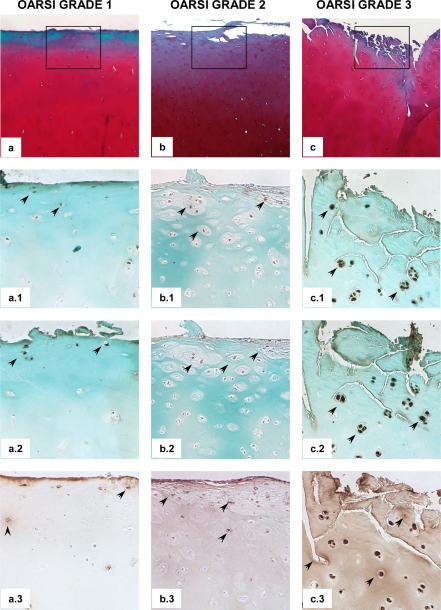
To further investigate their regulatory and spatial relationship in the context of OA disease, we analyzed the relative in situ levels of ELF3 and MMP13 proteins in different sections of human OA cartilage. Immunohistochemical analysis revealed enhanced ELF3 immunostaining in OA cartilage, in conjunction with increased MMP13 protein levels and activity, as judged by positive immunostaining for collagen type II cleavage epitopes with the C1,2C antibody, residing within areas of cartilage degradation (Fig. 3).
FIGURE 3.
ELF3 protein levels co-localize with increased MMP13 protein levels and activity in human OA cartilage. Sections of knee articular cartilage obtained from OA patients undergoing total knee replacement (n = 10) were Safranin O-stained (a, b, and c) and subjected to evaluation according to the OARSI cartilage OA histopathology grading system. Representative sections are shown. Successive sections were subjected to immunohistochemical staining using antibodies against ELF3 (a.1, b.1, and c.1), MMP13 (a.2, b.2, and c.2) and C1,2C (a.3, b.3, and c.3). Isotype-matched IgG was used as negative control (not shown). Squares indicate areas selected for higher magnification photomicrographs. The arrows indicate some of the areas with positive immunostaining. Note the more diffuse, matrix C1,2C-positive immunostaining with increased OARSI score. Original magnifications were 40× (a, b, and c) and 100× (a.1, b.1, c.1, a.2, b.2, c.2, a.3, b.3, and c.3).
ELF3 Activates MMP13 Promoter and Enhances AP-1-driven MMP13 Transactivation via a Highly Conserved Proximal ETS Binding Site
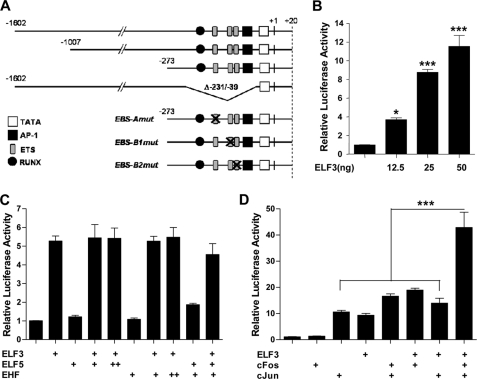
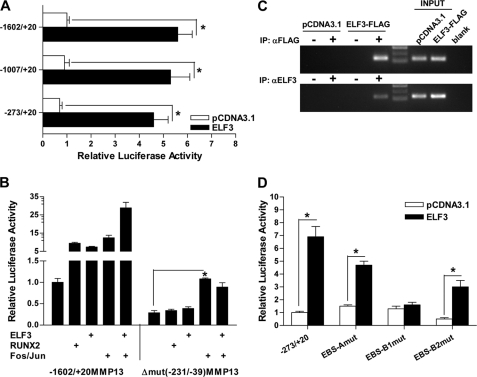
To determine if ELF3 activates MMP13 transcription, we used human MMP13 promoter-driven luciferase reporter constructs (depicted in Fig. 4A) in co-transfection experiments in T/C-28a2 human immortalized chondrocytes. Indeed, enforced ELF3 expression transactivated the −1602/+20 bp MMP13 reporter construct in these cells (Fig. 4B). However, the overexpression of two other ETS family members, ELF5 (ESE2) and EHF (ESE3), which share a high degree of homology with the ELF3 DNA binding domain (34, 35), did not transactivate the MMP13 promoter; nor did they enhance or interfere with ELF3-driven MMP13 transactivation (Fig. 4C).
FIGURE 4.
ELF3 transactivates MMP13 and enhances the AP-1-driven MMP13 activation. Schematic representation (not scaled) of the MMP13-luciferase reporter constructs utilized in this study (A). T/C-28a2 cells were co-transfected with 325 ng of the −1602/+20 bp MMP13 promoter and 12.5, 25, or 50 ng of expression vector encoding human ELF3 (B); 25 ng of expression vector encoding human ELF3, alone or in co-transfection with 25 (+) or 50 (++) ng of ELF5 or EHF expression vector (C); or 25 ng of each expression vector encoding ELF3, c-Fos, and c-Jun alone or together (D). Luciferase activities are shown as fold-change; protein input was used to normalize luciferase activities in D. *, p < 0.05; ***, p < 0.001. Error bars, S.E.
ETS transcription factors can work as co-factors of other transcriptional regulators (12, 13), and the MMP13 promoter contains evolutionarily conserved proximal EBS that are adjacent to an AP-1 motif, which may then interact in promoter transactivation (36). We therefore investigated if ELF3 can act in a collaborative fashion with c-Fos and c-Jun (AP-1) family members in the context of the human MMP13 promoter. As shown in Fig. 4D, c-Fos/c-Jun overexpression not only leads to the transactivation of the −1602/+20 bp MMP13 promoter but also co-enforced ELF3-driven MMP13 promoter activation. These results appear to explain the functional EBS-AP-1 interaction in transactivating the MMP13 promoter.
To more precisely define the ETS binding site(s) responsible for ELF3-driven MMP13 activation, we first used luciferase reporter constructs containing different sequences of the proximal human MMP13 promoter. ELF3 overexpression transactivated each of three MMP13 reporter constructs spanning −1602/+20, −1007/+20, and −273/+20 bp of the MMP13 promoter (Fig. 5A), suggesting that ELF3-driven MMP13 transactivation depends upon the EBS located within the −273/+20 bp proximal promoter region. We generated, therefore, a deletion construct of the −1602/+20 bp MMP13 promoter lacking the −231/−39 sequence (denoted as Δ−231/−39-MMP13 in Fig. 4A). Note that the Δ−231/−39-MMP13 construct, in addition to missing the EBS, also lacks the adjacent AP-1 and RUNX2 binding sites. As expected due to the absence of the proximal AP-1 site (37), the Δ−231/−39-MMP13 construct had reduced basal activity. Importantly, whereas overexpression of either c-Fos/c-Jun, RUNX2, or ELF3 transactivated the wild type −1602/+20-MMP13 promoter construct, deletion of the −231/−39 bp sequence completely abolished RUNX2- and ELF3-driven MMP13 promoter transactivation. Due to other remaining upstream AP-1 binding sites, the Δ−231/−39-MMP13 construct modestly responded to AP-1 (c-Fos/c-Jun) overexpression, but the positive effect of ELF3 on AP-1-induced MMP13 activation was absent (Fig. 5B), demonstrating that this proximal promoter region is responsible for ELF3-dependent EBS-AP-1 enhancement.
FIGURE 5.
ELF3 binds to the proximal MMP13 promoter and transactivates MMP13 via an evolutionarily conserved proximal EBS. T/C-28a2 cells were co-transfected with 325 ng of luciferase reporter constructs spanning −1602/+20, −1007/+20, or −273/+20 bp of the MMP13 promoter along with 25 ng of ELF3 expression vector (A) or wild type −1602/+20 bp or Δ−231/−39 MMP13 reporter construct and 25 ng of expression vector encoding ELF3, RUNX2, c-Fos, or c-Jun (B). Luciferase activities in B were normalized to the protein input. C, T/C-28a2 cells were transfected with ELF3-FLAG or the empty pCDNA3.1 expression vectors for 18 h. Chromatin was cross-linked and enzymatically sheared, and after reverse cross-linking of the DNA-protein complexes, the precleared lysates were incubated overnight at 4 °C with antibodies against FLAG (+) or normal rabbit IgG (−) (top) or against ELF3 (+) or normal rabbit IgG (−) (bottom). The human MMP13 promoter region was PCR-amplified using primers spanning from −157 to −38 bp, and the PCR products were resolved on a 2.5% agarose gel. Data are representative of two independent experiments performed in duplicate. D, T/C-28a2 cells were co-transfected with 25 ng of the ELF3 expression vector and 325 ng of the wild-type −267/+27 bp MMP13 promoter sequence or sequences containing point mutations of the proximal A, B1, or B2 ETS binding sites. *, p < 0.05. Error bars, S.E. IP, immunoprecipitation.
Next, we investigated ELF3 binding in vitro and in vivo to the ETS binding sites contained within the −231/−39 bp proximal promoter sequence. Comparative analyses of DNA sequences from multiple species using the University of California Santa Cruz Genome Browser (Human Assembly, March 2006) confirmed the presence of the previously described evolutionarily conserved and functional RUNX2, AP-1, and −78 bp ETS/PEA3 (labeled as B1) binding sites in the proximal MMP13 promoter (37, 38). This analysis also identified a putative ETS site (B2) in tandem with the −78 bp EBS and another upstream EBS (A), which overlaps with a negative regulatory AG-rich element (AGRE) (39) that is less well conserved in different species (see supplemental Fig. S4A for further details). EMSA and antibody-mediated supershift analysis showed binding of in vitro-translated ELF3 to the EBS sequences present in the proximal MMP13 promoter region (supplemental Fig. S4B). We next assessed ChIP in T/C-28a2 cells, in which we overexpressed a FLAG-tagged ELF3 construct. Utilizing anti-FLAG or anti-ELF3-specific antibodies and PCR primers spanning the −157/−38 bp MMP13 promoter region, we detected the binding of ELF3 to the MMP13 proximal promoter in situ (Fig. 5C). These results are in accord with our interpretation that the EBS in the MMP13 proximal promoter region is responsible for its ELF3-driven transactivation.
To precisely identify the ELF3-responsive proximal site or sites, we generated point mutations of all three proximal EBS contained within the −231/−39 bp sequence. Reporter assays performed with the wild-type −273/+20 bp MMP13 promoter sequence and the EBS-MMP13 mutant constructs (Fig. 5D) indicated that mutation of the A site did not affect transactivation by ELF3. Compared with the wild-type construct, the EBS-A mutant had significantly (p = 0.007) increased basal promoter activity, consistent with previous reports showing inhibitory actions of the MMP13 proximal AGRE site (39, 40), which overlaps with the A site. Mutation of the B2 site, although decreasing the basal promoter activity, also did not affect the ELF3-driven MMP13 promoter transactivation. In contrast, the EBS-B1 mutation, corresponding to the evolutionarily conserved ETS binding site located at −78 bp, completely abolished ELF3-driven MMP13 promoter activity. In addition, the EBS-B1 mutation also disrupted ELF3 enhancement of AP-1-driven MMP13 transactivation without impeding AP-1-mediated MMP13 transactivation (supplemental Fig. S5). Taken together, these results show that ELF3 binds to and activates the proximal MMP13 promoter via a highly conserved proximal ETS binding site in a manner that may involve direct or indirect interactions with AP-1 family members.
Contribution of MEK/ERK Signaling to IL-1β-induced ELF3 Binding and Transactivation of MMP13 Promoter
ETS factors can be regulated by MAPK-mediated phosphorylation (12, 13), and the IL-1β induction of MMPs in chondrocytes relies, at least in part, on MAPK activation of different downstream transcription factors (2, 41). Consequently, the mechanism whereby ELF3 regulates MMP13 expression in response to IL-1β stimulation may involve not only increased ELF3 gene expression but also modulation of its activity. To begin to address this question, we first investigated whether IL-1β stimulation induced changes in ELF3 activity by analyzing ELF3 binding to the MMP13 promoter in ChIP assays performed with immortalized chondrocytes challenged with the cytokine for 2 h. As shown in Fig. 6A, IL-1β treatment of the cells increased endogenous ELF3 binding to the MMP13 proximal promoter without significantly modifying total ELF3 protein levels (Fig. 6B), indicating that IL-1β indeed modulates ELF3 activity in chondrocytes. IL-1β-induced ELF3 binding to the endogenous MMP13 promoter correlated with the enhanced level of MMP13 RNA in response to IL-1β in T/C-28a2 cells. In addition, ELF3 knockdown led to decreased MMP13 expression at 6 h after IL-1β stimulation (Fig. 6C), implicating ELF3 in the early activation of MMP13 transcription, in agreement with our results utilizing Elf3−/− mouse chondrocytes (Fig. 2C).
FIGURE 6.
IL-1β enhances the endogenous ELF3 binding to the proximal MMP13 promoter. A, after overnight incubation in serum-free medium, T/C-28a2 cells were incubated with 1 ng/ml IL-1β for 2 h. After stimulation, the chromatin was cross-linked and enzymatically sheared, and after reverse cross-linking of the DNA-protein complexes, the precleared lysates were incubated with antibodies against ELF3 (+) or normal IgG (−) overnight at 4 °C. The human MMP13 promoter region was PCR-amplified using primers spanning from −157 to −38 bp, and the PCR products were resolved on a 2.5% agarose gel. B, ELF3 protein levels were analyzed by Western blotting using cell lysates prepared from T/C-28a2 cells stimulated with vehicle or 1 ng/ml IL-1β for 2 h. C, T/C-28a2 cells were transfected with 50 nm siRNA oligonucleotides against ELF3 (siELF3) or non-targeting siRNA (siCONTROL). At 72 h post-transfection, cells were stimulated with 1 ng/ml IL-1β for 6 h. Total RNA was isolated, and MMP13 mRNA was analyzed by RT-qPCR. Each value was normalized to GAPDH in the same sample and shown as mean ± S.E. *, p < 0.05. IP, immunoprecipitation.
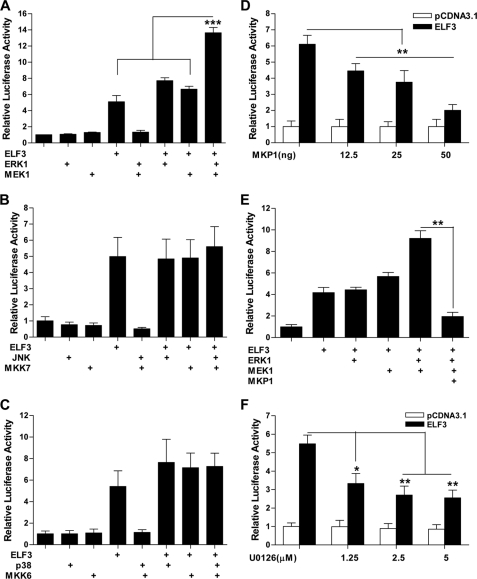
We next analyzed whether ELF3-driven MMP13 transactivation was modulated by MAPK signaling. We first performed luciferase reporter assays using cell extracts of immortalized chondrocytes co-transfected with the −1602/+20-MMP13 (Fig. 7) and −273/+20-MMP13 (not shown) reporter constructs and vector(s) expressing ELF3, p38, JNK, ERK-1, MKK-6, MKK-7, and MEK-1. As shown in Fig. 7A, MEK1/ERK1 overexpression significantly enhanced ELF3-driven MMP13 transactivation, whereas overexpressing MKK7/JNK (Fig. 7B) or MKK6/p38 (Fig. 7C) did not. Furthermore, enforced, concentration-dependent expression of MAPK phosphatase 1 (MKP1) reduced both ELF3 activation of MMP13 (Fig. 7D) and the MEK1/ERK1 enhancement of ELF3-1-driven MMP13 promoter activation (Fig. 7E). Moreover, U0126, a MEK1/2-specific pharmacological inhibitor, dose-dependently decreased MMP13 promoter activity induced by ELF3 overexpression (Fig. 7F), further indicating that MEK/ERK signaling contributes to ELF3-driven MMP13 transcription in chondrocytes.
FIGURE 7.
MEK/ERK overexpression enhances the ELF3-driven MMP13 promoter activation. T/C-28a2 cells were co-transfected with 325 ng of the −1602/+20 bp MMP13 promoter, 25 ng of ELF3 expression vector, and (A) 25 ng of each expression vector encoding ERK1 and MEK1; (B) 25 ng of each expression vector encoding JNK and MKK7; (C) 25 ng of each expression vector encoding p38 and MKK6; (D) increasing amounts (0, 12.5 ng, 25 ng, and 50 ng) of expression vector encoding MKP1, and (E) 25 ng of expression vector encoding MEK1 or ERK1 and 50 ng of expression vector encoding MKP1; or (C) treated with vehicle (DMSO) or the indicated concentrations of U0126 at 4 h after transfection, followed by 20 h of incubation after addition of the inhibitor. Luciferase activities are shown as fold-change with untreated controls set at 1.0. *, p < 0.05; **, p < 0.01; ***, p < 0.001. Error bars, S.E.
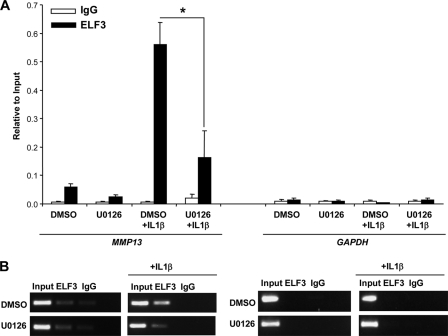
Because IL-1β treatment induced ELF3 binding to the proximal MMP13 promoter, and MEK/ERK signaling enhanced ELF3-driven MMP13 transactivation, we next investigated if IL-1β-induced ELF3 binding to the MMP13 promoter required MEK/ERK signaling. Depending upon the structural features of each ETS factor, along with cellular and DNA sequence-specific effects, modulation of ETS transcriptional activity in response to MAPKs can involve changes in subcellular localization, unmasking of their DNA binding domains, or differential interactions with transcriptional co-activators/repressors (12, 13). To determine the effect of IL-1β-dependent phosphorylation of ERK1/2, p38, and SAPK/JNK on ELF3 function, we employed the MEK1/2-specific inhibitor U0126 in T/C-28a2 cells. In agreement with previous reports in human primary (42) and immortalized chondrocytes (43), p38, SAPK/JNK, or ERK1/2 phosphorylation was induced by IL-1β treatment, and pretreatment with 2.5 μm U0126 inhibited both basal and IL-1β-induced ERK1/2 phosphorylation without affecting p38 or SAPK/JNK phosphorylation (supplemental Fig. S6). We analyzed IL-1β-induced ELF3 binding to the MMP13 promoter in ChIP assays performed in T/C-28a2 cells pretreated with vehicle (DMSO) or 2.5 μm U0126 for 45 min and stimulated for 2 h with 1 ng/ml of IL-1β. ChIP analysis revealed that the IL-1β-induced ELF3 binding to the proximal MMP13 promoter was significantly reduced by U0126-mediated inhibition of MEK/ERK signaling in human chondrocytes (Fig. 8).
FIGURE 8.
MEK/ERK signaling is required for the IL-1β-induced ELF3 binding to the proximal MMP13 promoter. After overnight incubation in serum-free medium, T/C-28a2 cells were pretreated with vehicle (DMSO) or 2.5 μm U0126 for 45 min before incubation with 1 ng/ml IL-1β in the presence of inhibitor. At 2 h after IL-1β stimulation, chromatin was cross-linked and enzymatically sheared, and after reverse cross-linking of the DNA-protein complexes, the precleared lysates were incubated with antibodies against ELF3 or normal IgG overnight at 4 °C. The ELF3 binding to the human MMP13 promoter region was analyzed using primers spanning from −157 to −38 bp of the promoter region by quantitative PCR (A) and conventional PCR (B), with PCR products resolved on a 2.5% agarose gel. GAPDH gene-specific primers were used as a negative control. *, p < 0.05. Error bars, S.E.
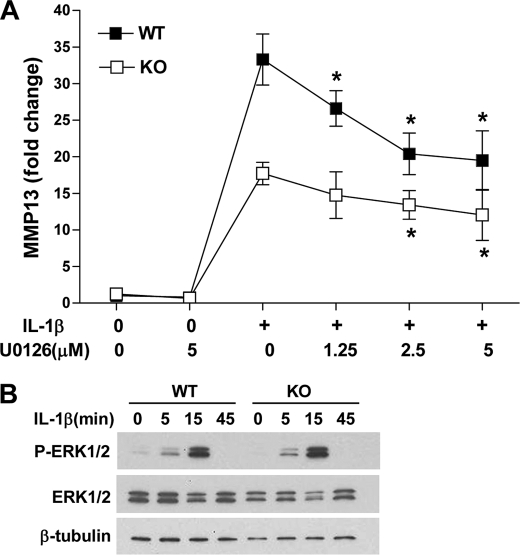
Finally, we explored whether MEK/ERK signaling requires ELF3 to drive MMP13 gene expression. Wild type and Elf3−/− mouse primary chondrocytes were stimulated with IL-1β with or without U0126 pretreatment (Fig. 9A). RT-qPCR analysis revealed that blocking MEK/ERK signaling significantly suppressed IL-1β-induced levels of Mmp13 mRNA in both wild type and Elf3−/− chondrocytes. Previous reports (44, 45) have shown that ELF3 overexpression or knockdown in mammary epithelium-derived cells leads to enhanced or reduced MAPK activity, respectively. However, wild-type and Elf3−/− chondrocytes did not show a significant change in IL-1β-induced ERK1/2 phosphorylation (Fig. 9B). We conclude from these results that MEK/ERK signaling also acts independently of ELF3 to induce MMP13 gene expression in response to IL-1β.
FIGURE 9.
Effects of ELF3 on MEK/ERK-dependent MMP13 expression in mouse primary chondrocytes. Mouse primary chondrocytes were isolated from articular cartilage from wild type (WT) C57BL/6 and Elf3 knock-out (KO) 5–6-day-old mice. A, after overnight incubation in serum-free medium, cells were pretreated with vehicle (DMSO) or the indicated concentrations of U0126 for 45 min before incubation with 1 ng/ml IL-1β in the presence of the inhibitor. At 24 h after IL-1β stimulation, total RNAs were isolated, and MMP13 mRNA was analyzed by RT-qPCR. Each value was normalized to GAPDH in the same sample and shown as mean ± S.E. (error bars). B, phospho-ERK1/2 (P-ERK1/2) and total ERK1/2 (ERK1/2) protein levels were analyzed by Western blotting using cell lysates prepared from wild type and Elf3 knock-out primary chondrocytes stimulated with 1 ng/ml IL-1β for the indicated times after overnight incubation in serum-free conditions. Western blots were reprobed for β-tubulin as a loading reference control. *, p < 0.05 versus the IL-1β-stimulated controls.
DISCUSSION
Abnormal Stress-related Induction and Activation of MMP13 in Chondrocytes Fuels OA Disease Phenotype
Among the different MMPs abnormally expressed by OA chondrocytes, special attention has been focused on MMP13 due to its potent ability to cleave type II collagen (5). Evidence has shown increased MMP13 expression and activity in OA cartilage (7), co-localization of MMP13-specific type II collagen cleavage products with cytokines and their receptors (4, 8), OA-like changes associated with overexpression of constitutively active MMP13 (9) and protection of the Mmp13 knock-out mice against surgically induced OA (10). Consequently, numerous efforts have been directed toward a complete understanding of the mechanisms leading to the dysregulated expression of MMP13 in OA chondrocytes. In response to a variety of stress and inflammatory insults, an array of signaling pathways becomes abnormally activated in metabolically dormant articular chondrocytes. For instance, RAS/RAF/MEK/ERK signaling has been shown to control DDR2 receptor-driven up-regulation of MMP13 in OA disease (46). Moreover, stress-mediated activation of the NF-κB and MAPK pathways drives the induction of downstream transcriptional effectors with direct impact on chondrocyte physiology (2, 41), including the inhibition of anabolic genes (27, 47, 48) and the exacerbated expression of matrix degradative enzymes, as found in OA disease (49–52). In addition, because OA chondrocytes themselves produce proinflammatory factors, including IL-1β, a positive feedback loop is generated, which contributes to the dysregulation of chondrocyte functions and exacerbation of cartilage erosion and loss of function (8, 53).
MMP13 Transcription Is Subject to Effects of Multiple DNA Binding Activators
Proinflammatory stimuli, including IL-1β and TNFα, transactivate MMP promoters, depending on the presence and availability of AP-1 and ETS elements (2, 41). Similar to other MMP genes, the MMP13 promoter contains conserved ETS factor binding sites, which are adjacent to AP-1 binding sites (38). Different studies have shown that the proximal AP-1 site is crucial for both basal and cytokine-induced MMP13 transcription in response to c-Fos/c-Jun heterodimers or c-Jun/c-Jun homodimers, which can cooperate with ETS factors to activate transcription (36, 38, 54). In chondrocytes, the NF-κB (p65/p50) members (55, 56), RUNX2 (37), HIF2α (57), C/EBPβ (58), and c-Fos/c-Jun (59) have each been implicated in the mechanisms of cytokine-induced MMP13 transcription (37, 54, 56, 57, 59–63). Activation of JNK, p38, and ERK1/2 signaling mediates MMP transcription by activating AP-1 (56) and RUNX2 (63) in addition to ETS factors, including ELK1 (64). ERK1/2 phosphorylation is increased in vivo in OA cartilage, and the MEK/ERK-induced phosphorylation of ELK1 mediates FGF-2-induced MMP13 expression by enhancing ELK1 transcriptional activation (64). Furthermore, ETS1 overexpression leads to increased MMP13 expression both in vitro and in vivo (65), and IFNα stimulation of primary cultures of hepatic stellate cells induces PEA3 engagement with Jak1 and Stat1 and subsequent MMP13 expression (66). These findings indicate that the induction of MMP13 by different ETS factors is stimulus- and cell type-dependent.
ELF3 Is Novel Activator of MMP13 Transcription in Stressed Chondrocytes
Our results show for the first time that ELF3 is among the primary transcription factors that activate MMP13 transcription in response to IL-1β. Importantly, ELF3 deficiency, both by knockdown in vitro and by Elf3 knock-out in vivo, resulted in significantly reduced MMP13 expression in response to IL-1β and TNFα. ELF3 overexpression transactivated the MMP13 promoter, dependent on its −78 bp (B1) evolutionarily conserved ETS site, and ELF3 acted in conjunction with AP-1 (c-Fos/c-Jun) to drive MMP13 transcription, which helps to explain the previously described molecular interplay of these cis-acting elements and their trans-acting binding factors (14, 36). In addition, IL-1β stimulation enhanced ELF3 binding to the MMP13 proximal promoter, and MEK/ERK signaling both enhanced ELF3-driven activation of MMP13 transcription and participated in IL-1β-induced ELF3 binding to the promoter. However, it remains to be determined if the ELF3/AP-1 functional interplay uncovered here involves either direct or indirect interactions with AP-1 family members. Moreover, ELF3/AP-1 interaction at the MMP13 proximal promoter probably represents an important nexus for the recruitment of other critical transcriptional co-activators and chromatin-modifying proteins, which now also warrant further investigation. Although our results collectively show that ELF3 is an important new contributing factor for the control of MMP13 transcription in response to proinflammatory stimuli in chondrocytes, the delay in MMP13 mRNA induction in response to IL-1β or TNFα stimulation indicates that other factors, which are activated after ELF3, must work in conjunction with ELF3 to drive MMP13 transcription in response to stress-related stimuli.
ELF3 Also Contributes to Transcriptional Activation of Proinflammatory Mediators in Stressed Chondrocytes
ELF3 is also known to mediate stress-related, proinflammatory reactions involving the repression of anabolic genes (27) and the activation of inflammatory response genes, such as NOS2 and PTGS2/COX2 (25, 26). ELF3 has also been reported to modulate the transcription of proinflammatory cytokines, such as IL-6 (23). ELF3 may also activate (20, 67) or repress (68) the transcription of MMPs. However, previous reports have shown no transactivating effect of ELF3 overexpression on a rat MMP3 promoter construct, whereas it strongly activated a reporter construct with a multimerized stromelysin EBS (20). Indeed, our experiments confirmed that ELF3 contributes to the IL-1β-induced expression of NOS2 and PTGS2/COX2 in human chondrocytes (supplemental Fig. S3). Thus, ELF3 could mediate stress/inflammatory responses that drive MMP transcription via the direct transcriptional activation of the relevant promoters and also by other indirect mechanisms.
ELF3 Activity Is Post-translationally Modulated by MEK/ERK Signaling
An array of post-translational modifications, including phosphorylation, glycosylation, and sumoylation, can regulate ETS activity, resulting in loss of repression, increased activation, increased or decreased stability, alterations in protein-protein interaction, nuclear translocation, or enhanced DNA binding (12, 13). Previous reports have demonstrated that ELF3 stability and activity are modulated by phosphorylation (44) and that stable ELF3 overexpression leads to MAPK activation (45). However, to the best of our knowledge, our results provide evidence of the first functional link between MEK/ERK signaling and ELF3 activation. Initial comparison of the ELF3 structure with that of ETS1 revealed little conservation within the PNT domain (11), which contains ERK2 docking sites in both ETS1 and -2 (69), and removal of the PNT domain did not affect the ability of ELF3 to transactivate the type II TGF-β receptor promoter (67). However, potential phosphoacceptor sites for p38, JNK, ERK, and dual specificity kinases have been identified in the ELF3 structure (11). Our results in overexpressing the relevant constitutively active MAP2K along with the relevant MAPK show that the ELF3 activity is enhanced by MEK/ERK overexpression. In accord with the latter observation, MKP1 overexpression shows that the potency of ELF3 in transactivating MMP13 also depends upon phosphorylation events. Moreover, the pharmacological inhibition of MEK/ERK in both reporter and ChIP assays reinforces the contribution of MEK/ERK to ELF3 activity, in accord with ELF3 being a downstream effector of MEK/ERK-mediated MMP13 expression in chondrocytes. However, the precise mechanisms by which MEK/ERK modulate ELF3 activity, whether by favoring specific protein-protein interactions, increasing DNA binding affinity, or modifying protein stability, remain unclear and require further investigation.
Linkage of Abnormal Levels of ELF3 with MMP13 Activity in OA Disease
Because ELF3 expression is low in most non-epithelial tissues, including cartilage, and we do not detect Elf3 mRNA in the developing cartilage of mouse embryos by in situ hybridization analysis,4 ELF3 is unlikely to participate in normal cartilage homeostasis. Moreover, little is known regarding in vivo contributions of ELF3 to the development and function of different tissues due to the severe defects in the gastric epithelium of Elf3 knock-out mice that compromise their survival (17) and thus impede a detailed analysis over time. In that regard, the in vivo analysis of the intrinsic roles of ELF3 in normal cartilage homeostasis or in driving MMP13 expression during chondrocyte hypertrophy and endochondral ossification processes would require the use of cartilage-targeted, conditional Elf3 knock-out mice, which would be necessary to determine if the roles of ELF3 in controlling MMP13 expression are cell- or context-specific processes. However, in both OA and RA, ELF3 expression is up-regulated in synovial tissues (22). Our results suggest that ELF3 may serve as a novel sensor in cartilage pathology, responding to and mediating mechanical stress and proinflammatory insults and acting as a contributing factor to OA development and/or progression by inducing MMP13 transcription, among other downstream targets. Indeed, our results showing increased ELF3 expression in response to inflammatory stimuli in vitro as well as increased ELF3 levels correlating with increased MMP13 protein and activity in areas of cartilage degradation in human OA are indicative of a detrimental role of ELF3. The latter fits nicely with its in vitro contribution to IL-1β-induced MMP13 enhancement and COL2A1 repression (27) in articular chondrocytes. This concept is also consistent with reports showing that abnormal ELF3 expression alters normal physiology in different scenarios, resulting in breast tumorigenesis (19, 20), contributing to airway inflammation (23), or mediating angiogenesis in the setting of inflammation (24).
In summary, we have shown co-localization of ELF3 protein with MMP13 and MMP13 cleavage products in OA cartilage, a functional role for ELF3 in IL-1β-induced MMP13 expression in chondrocytes, and involvement of the MEK/ERK signaling in both enhancing ELF3-driven MMP13 transactivation and ELF3 binding to the MMP13 proximal promoter in response to IL-1β. Given the aforementioned results, we speculate that ELF3 is one of the MEK/ERK downstream effectors that induces aberrant expression and activity of MMP13, when chondrocytes and other cell types respond to inflammation and stress. Modulation of the MEK/ERK/ELF3 axis may be a contributing factor to OA development and/or progression, and its role in determining MMP13 expression and activity in this and other disease contexts now merits further investigation.
Supplementary Material
Acknowledgment
We are grateful to Dr. Thomas P. Sculco (Hospital for Special Surgery) for providing human cartilage specimens.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-AG022021, R21-AR054887, and RC4 AR060546 (to M. B. G.). This work was also supported by an Arthritis Foundation postdoctoral fellowship (to M. O.) and an Osteoarthritis Research Society international scholarship (to E. O.).

This article contains supplemental Table S1 and Figs. S1–S6.
D. A. Plumb, M. Otero, C. L. Dragomir, and M. B. Goldring, unpublished observations.
- MMP
- matrix metalloproteinase
- OA
- osteoarthritis
- ETS
- E26 transformation-specific sequence
- OARSI
- Osteoarthritis Research Society International
- RT-qPCR
- real-time quantitative RT-PCR
- EBS
- ETS binding site(s)
- AGRE
- AG-rich element.
REFERENCES
- 1. Inada M., Wang Y., Byrne M. H., Rahman M. U., Miyaura C., López-Otín C., Krane S. M. (2004) Critical roles for collagenase-3 (Mmp13) in development of growth plate cartilage and in endochondral ossification. Proc. Natl. Acad. Sci. U.S.A. 101, 17192–17197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Goldring M. B., Otero M., Plumb D. A., Dragomir C., Favero M., El Hachem K., Hashimoto K., Roach H. I., Olivotto E., Borzì R. M., Marcu K. B. (2011) Roles of inflammatory and anabolic cytokines in cartilage metabolism. Signals and multiple effectors converge upon MMP-13 regulation in osteoarthritis. Eur. Cell Mater. 21, 202–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Murphy G., Nagase H. (2008) Reappraising metalloproteinases in rheumatoid arthritis and osteoarthritis. Destruction or repair? Nat. Clin. Pract. Rheumatol. 4, 128–135 [DOI] [PubMed] [Google Scholar]
- 4. Billinghurst R. C., Dahlberg L., Ionescu M., Reiner A., Bourne R., Rorabeck C., Mitchell P., Hambor J., Diekmann O., Tschesche H., Chen J., Van Wart H., Poole A. R. (1997) Enhanced cleavage of type II collagen by collagenases in osteoarthritic articular cartilage. J. Clin. Invest. 99, 1534–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Knäuper V., López-Otin C., Smith B., Knight G., Murphy G. (1996) Biochemical characterization of human collagenase-3. J. Biol. Chem. 271, 1544–1550 [DOI] [PubMed] [Google Scholar]
- 6. Mitchell P. G., Magna H. A., Reeves L. M., Lopresti-Morrow L. L., Yocum S. A., Rosner P. J., Geoghegan K. F., Hambor J. E. (1996) Cloning, expression, and type II collagenolytic activity of matrix metalloproteinase-13 from human osteoarthritic cartilage. J. Clin. Invest. 97, 761–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reboul P., Pelletier J. P., Tardif G., Cloutier J. M., Martel-Pelletier J. (1996) The new collagenase, collagenase-3, is expressed and synthesized by human chondrocytes but not by synoviocytes. A role in osteoarthritis. J. Clin. Invest. 97, 2011–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tetlow L. C., Adlam D. J., Woolley D. E. (2001) Matrix metalloproteinase and proinflammatory cytokine production by chondrocytes of human osteoarthritic cartilage. Associations with degenerative changes. Arthritis Rheum. 44, 585–594 [DOI] [PubMed] [Google Scholar]
- 9. Neuhold L. A., Killar L., Zhao W., Sung M. L., Warner L., Kulik J., Turner J., Wu W., Billinghurst C., Meijers T., Poole A. R., Babij P., DeGennaro L. J. (2001) Postnatal expression in hyaline cartilage of constitutively active human collagenase-3 (MMP-13) induces osteoarthritis in mice. J. Clin. Invest. 107, 35–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Little C. B., Barai A., Burkhardt D., Smith S. M., Fosang A. J., Werb Z., Shah M., Thompson E. W. (2009) Matrix metalloproteinase 13-deficient mice are resistant to osteoarthritic cartilage erosion but not chondrocyte hypertrophy or osteophyte development. Arthritis Rheum. 60, 3723–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oettgen P., Alani R. M., Barcinski M. A., Brown L., Akbarali Y., Boltax J., Kunsch C., Munger K., Libermann T. A. (1997) Isolation and characterization of a novel epithelium-specific transcription factor, ESE-1, a member of the ets family. Mol. Cell. Biol. 17, 4419–4433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sharrocks A. D. (2001) The ETS-domain transcription factor family. Nat. Rev. Mol. Cell Biol. 2, 827–837 [DOI] [PubMed] [Google Scholar]
- 13. Tootle T. L., Rebay I. (2005) Post-translational modifications influence transcription factor activity. A view from the ETS superfamily. BioEssays 27, 285–298 [DOI] [PubMed] [Google Scholar]
- 14. Trojanowska M. (2000) Ets factors and regulation of the extracellular matrix. Oncogene 19, 6464–6471 [DOI] [PubMed] [Google Scholar]
- 15. Cabral A., Fischer D. F., Vermeij W. P., Backendorf C. (2003) Distinct functional interactions of human Skn-1 isoforms with Ese-1 during keratinocyte terminal differentiation. J. Biol. Chem. 278, 17792–17799 [DOI] [PubMed] [Google Scholar]
- 16. Reddy S. P., Vuong H., Adiseshaiah P. (2003) Interplay between proximal and distal promoter elements is required for squamous differentiation marker induction in the bronchial epithelium. Role for ESE-1, Sp1, and AP-1 proteins. J. Biol. Chem. 278, 21378–21387 [DOI] [PubMed] [Google Scholar]
- 17. Ng A. Y., Waring P., Ristevski S., Wang C., Wilson T., Pritchard M., Hertzog P., Kola I. (2002) Inactivation of the transcription factor Elf3 in mice results in dysmorphogenesis and altered differentiation of intestinal epithelium. Gastroenterology 122, 1455–1466 [DOI] [PubMed] [Google Scholar]
- 18. Lee S. H., Bahn J. H., Choi C. K., Whitlock N. C., English A. E., Safe S., Baek S. J. (2008) ESE-1/EGR-1 pathway plays a role in tolfenamic acid-induced apoptosis in colorectal cancer cells. Mol. Cancer Ther. 7, 3739–3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Prescott J. D., Koto K. S., Singh M., Gutierrez-Hartmann A. (2004) The ETS transcription factor ESE-1 transforms MCF-12A human mammary epithelial cells via a novel cytoplasmic mechanism. Mol. Cell. Biol. 24, 5548–5564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eckel K. L., Tentler J. J., Cappetta G. J., Diamond S. E., Gutierrez-Hartmann A. (2003) The epithelial-specific ETS transcription factor ESX/ESE-1/Elf-3 modulates breast cancer-associated gene expression. DNA Cell Biol. 22, 79–94 [DOI] [PubMed] [Google Scholar]
- 21. Neve R., Chang C. H., Scott G. K., Wong A., Friis R. R., Hynes N. E., Benz C. C. (1998) The epithelium-specific ets transcription factor ESX is associated with mammary gland development and involution. FASEB J. 12, 1541–1550 [DOI] [PubMed] [Google Scholar]
- 22. Grall F., Gu X., Tan L., Cho J. Y., Inan M. S., Pettit A. R., Thamrongsak U., Choy B. K., Manning C., Akbarali Y., Zerbini L., Rudders S., Goldring S. R., Gravallese E. M., Oettgen P., Goldring M. B., Libermann T. A. (2003) Responses to the proinflammatory cytokines interleukin-1 and tumor necrosis factor α in cells derived from rheumatoid synovium and other joint tissues involve nuclear factor κB-mediated induction of the Ets transcription factor ESE-1. Arthritis Rheum. 48, 1249–1260 [DOI] [PubMed] [Google Scholar]
- 23. Wu J., Duan R., Cao H., Field D., Newnham C. M., Koehler D. R., Zamel N., Pritchard M. A., Hertzog P., Post M., Tanswell A. K., Hu J. (2008) Regulation of epithelium-specific Ets-like factors ESE-1 and ESE-3 in airway epithelial cells. Potential roles in airway inflammation. Cell Res. 18, 649–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brown C., Gaspar J., Pettit A., Lee R., Gu X., Wang H., Manning C., Voland C., Goldring S. R., Goldring M. B., Libermann T. A., Gravallese E. M., Oettgen P. (2004) ESE-1 is a novel transcriptional mediator of angiopoietin-1 expression in the setting of inflammation. J. Biol. Chem. 279, 12794–12803 [DOI] [PubMed] [Google Scholar]
- 25. Grall F. T., Prall W. C., Wei W., Gu X., Cho J. Y., Choy B. K., Zerbini L. F., Inan M. S., Goldring S. R., Gravallese E. M., Goldring M. B., Oettgen P., Libermann T. A. (2005) The Ets transcription factor ESE-1 mediates induction of the COX-2 gene by LPS in monocytes. FEBS J. 272, 1676–1687 [DOI] [PubMed] [Google Scholar]
- 26. Rudders S., Gaspar J., Madore R., Voland C., Grall F., Patel A., Pellacani A., Perrella M. A., Libermann T. A., Oettgen P. (2001) ESE-1 is a novel transcriptional mediator of inflammation that interacts with NF-κB to regulate the inducible nitric-oxide synthase gene. J. Biol. Chem. 276, 3302–3309 [DOI] [PubMed] [Google Scholar]
- 27. Peng H., Tan L., Osaki M., Zhan Y., Ijiri K., Tsuchimochi K., Otero M., Wang H., Choy B. K., Grall F. T., Gu X., Libermann T. A., Oettgen P., Goldring M. B. (2008) ESE-1 is a potent repressor of type II collagen gene (COL2A1) transcription in human chondrocytes. J. Cell. Physiol. 215, 562–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ijiri K., Zerbini L. F., Peng H., Otu H. H., Tsuchimochi K., Otero M., Dragomir C., Walsh N., Bierbaum B. E., Mattingly D., van Flandern G., Komiya S., Aigner T., Libermann T. A., Goldring M. B. (2008) Differential expression of GADD45β in normal and osteoarthritic cartilage. Potential role in homeostasis of articular chondrocytes. Arthritis Rheum. 58, 2075–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gosset M., Berenbaum F., Thirion S., Jacques C. (2008) Primary culture and phenotyping of murine chondrocytes. Nat. Protoc. 3, 1253–1260 [DOI] [PubMed] [Google Scholar]
- 30. Tsuchimochi K., Otero M., Dragomir C. L., Plumb D. A., Zerbini L. F., Libermann T. A., Marcu K. B., Komiya S., Ijiri K., Goldring M. B. (2010) GADD45β enhances Col10a1 transcription via the MTK1/MKK3/6/p38 axis and activation of C/EBPβ-TAD4 in terminally differentiating chondrocytes. J. Biol. Chem. 285, 8395–8407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pritzker K. P., Gay S., Jimenez S. A., Ostergaard K., Pelletier J. P., Revell P. A., Salter D., van den Berg W. B. (2006) Osteoarthritis cartilage histopathology. Grading and staging. Osteoarthr. Cartil. 14, 13–29 [DOI] [PubMed] [Google Scholar]
- 32. Joosten L. A., Helsen M. M., Saxne T., van De Loo F. A., Heinegard D., van Den Berg W. B. (1999) IL-1 αβ blockade prevents cartilage and bone destruction in murine type II collagen-induced arthritis, whereas TNF-α blockade only ameliorates joint inflammation. J. Immunol. 163, 5049–5055 [PubMed] [Google Scholar]
- 33. Zwerina J., Redlich K., Polzer K., Joosten L., Krönke G., Distler J., Hess A., Pundt N., Pap T., Hoffmann O., Gasser J., Scheinecker C., Smolen J. S., van den Berg W., Schett G. (2007) TNF-induced structural joint damage is mediated by IL-1. Proc. Natl. Acad. Sci. U.S.A. 104, 11742–11747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kas K., Finger E., Grall F., Gu X., Akbarali Y., Boltax J., Weiss A., Oettgen P., Kapeller R., Libermann T. A. (2000) ESE-3, a novel member of an epithelium-specific ets transcription factor subfamily, demonstrates different target gene specificity from ESE-1. J. Biol. Chem. 275, 2986–2998 [DOI] [PubMed] [Google Scholar]
- 35. Oettgen P., Kas K., Dube A., Gu X., Grall F., Thamrongsak U., Akbarali Y., Finger E., Boltax J., Endress G., Munger K., Kunsch C., Libermann T. A. (1999) Characterization of ESE-2, a novel ESE-1-related Ets transcription factor that is restricted to glandular epithelium and differentiated keratinocytes. J. Biol. Chem. 274, 29439–29452 [DOI] [PubMed] [Google Scholar]
- 36. Tardif G., Reboul P., Dupuis M., Geng C., Duval N., Pelletier J. P., Martel-Pelletier J. (2001) Transforming growth factor-β-induced collagenase-3 production in human osteoarthritic chondrocytes is triggered by Smad proteins. Cooperation between activator protein-1 and PEA-3 binding sites. J. Rheumatol. 28, 1631–1639 [PubMed] [Google Scholar]
- 37. Mengshol J. A., Vincenti M. P., Brinckerhoff C. E. (2001) IL-1 induces collagenase-3 (MMP-13) promoter activity in stably transfected chondrocytic cells. Requirement for Runx-2 and activation by p38 MAPK and JNK pathways. Nucleic Acids Res. 29, 4361–4372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pendás A. M., Balbín M., Llano E., Jiménez M. G., López-Otín C. (1997) Structural analysis and promoter characterization of the human collagenase-3 gene (MMP13). Genomics 40, 222–233 [DOI] [PubMed] [Google Scholar]
- 39. Benderdour M., Tardif G., Pelletier J. P., Dupuis M., Geng C., Martel-Pelletier J. (2002) A novel negative regulatory element in the human collagenase-3 proximal promoter region. Biochem. Biophys. Res. Commun. 291, 1151–1159 [DOI] [PubMed] [Google Scholar]
- 40. Fan Z., Tardif G., Boileau C., Bidwell J. P., Geng C., Hum D., Watson A., Pelletier J. P., Lavigne M., Martel-Pelletier J. (2006) Identification in human osteoarthritic chondrocytes of proteins binding to the novel regulatory site AGRE in the human matrix metalloprotease 13 proximal promoter. Arthritis Rheum. 54, 2471–2480 [DOI] [PubMed] [Google Scholar]
- 41. Yan C., Boyd D. D. (2007) Regulation of matrix metalloproteinase gene expression. J. Cell. Physiol. 211, 19–26 [DOI] [PubMed] [Google Scholar]
- 42. Geng Y., Valbracht J., Lotz M. (1996) Selective activation of the mitogen-activated protein kinase subgroups c-Jun NH2 terminal kinase and p38 by IL-1 and TNF in human articular chondrocytes. J. Clin. Invest. 98, 2425–2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nieminen R., Leinonen S., Lahti A., Vuolteenaho K., Jalonen U., Kankaanranta H., Goldring M. B., Moilanen E. (2005) Inhibitors of mitogen-activated protein kinases down-regulate COX-2 expression in human chondrocytes. Mediators Inflamm. 2005, 249–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Manavathi B., Rayala S. K., Kumar R. (2007) Phosphorylation-dependent regulation of stability and transforming potential of ETS transcriptional factor ESE-1 by p21-activated kinase 1. J. Biol. Chem. 282, 19820–19830 [DOI] [PubMed] [Google Scholar]
- 45. Schedin P. J., Eckel-Mahan K. L., McDaniel S. M., Prescott J. D., Brodsky K. S., Tentler J. J., Gutierrez-Hartmann A. (2004) ESX induces transformation and functional epithelial to mesenchymal transition in MCF-12A mammary epithelial cells. Oncogene 23, 1766–1779 [DOI] [PubMed] [Google Scholar]
- 46. Xu L., Peng H., Wu D., Hu K., Goldring M. B., Olsen B. R., Li Y. (2005) Activation of the discoidin domain receptor 2 induces expression of matrix metalloproteinase 13 associated with osteoarthritis in mice. J. Biol. Chem. 280, 548–555 [DOI] [PubMed] [Google Scholar]
- 47. Okazaki K., Li J., Yu H., Fukui N., Sandell L. J. (2002) CCAAT/enhancer-binding proteins β and δ mediate the repression of gene transcription of cartilage-derived retinoic acid-sensitive protein induced by interleukin-1 β. J. Biol. Chem. 277, 31526–31533 [DOI] [PubMed] [Google Scholar]
- 48. Tan L., Peng H., Osaki M., Choy B. K., Auron P. E., Sandell L. J., Goldring M. B. (2003) Egr-1 mediates transcriptional repression of COL2A1 promoter activity by interleukin-1β. J. Biol. Chem. 278, 17688–17700 [DOI] [PubMed] [Google Scholar]
- 49. Bau B., Gebhard P. M., Haag J., Knorr T., Bartnik E., Aigner T. (2002) Relative messenger RNA expression profiling of collagenases and aggrecanases in human articular chondrocytes in vivo and in vitro. Arthritis Rheum. 46, 2648–2657 [DOI] [PubMed] [Google Scholar]
- 50. Freemont A. J., Hampson V., Tilman R., Goupille P., Taiwo Y., Hoyland J. A. (1997) Gene expression of matrix metalloproteinases 1, 3, and 9 by chondrocytes in osteoarthritic human knee articular cartilage is zone- and grade-specific. Ann. Rheum. Dis. 56, 542–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Koshy P. J., Lundy C. J., Rowan A. D., Porter S., Edwards D. R., Hogan A., Clark I. M., Cawston T. E. (2002) The modulation of matrix metalloproteinase and ADAM gene expression in human chondrocytes by interleukin-1 and oncostatin M. A time course study using real-time quantitative reverse transcription-polymerase chain reaction. Arthritis Rheum. 46, 961–967 [DOI] [PubMed] [Google Scholar]
- 52. Rogerson F. M., Chung Y. M., Deutscher M. E., Last K., Fosang A. J. (2010) Cytokine-induced increases in ADAMTS-4 messenger RNA expression do not lead to increased aggrecanase activity in ADAMTS-5-deficient mice. Arthritis Rheum. 62, 3365–3373 [DOI] [PubMed] [Google Scholar]
- 53. Melchiorri C., Meliconi R., Frizziero L., Silvestri T., Pulsatelli L., Mazzetti I., Borzì R. M., Uguccioni M., Facchini A. (1998) Enhanced and coordinated in vivo expression of inflammatory cytokines and nitric-oxide synthase by chondrocytes from patients with osteoarthritis. Arthritis Rheum. 41, 2165–2174 [DOI] [PubMed] [Google Scholar]
- 54. Benderdour M., Tardif G., Pelletier J. P., Di Battista J. A., Reboul P., Ranger P., Martel-Pelletier J. (2002) Interleukin 17 (IL-17) induces collagenase-3 production in human osteoarthritic chondrocytes via AP-1-dependent activation. Differential activation of AP-1 members by IL-17 and IL-1β. J. Rheumatol. 29, 1262–1272 [PubMed] [Google Scholar]
- 55. Liacini A., Sylvester J., Li W. Q., Zafarullah M. (2002) Inhibition of interleukin-1-stimulated MAP kinases, activating protein-1 (AP-1), and nuclear factor κB (NF-κB) transcription factors down-regulates matrix metalloproteinase gene expression in articular chondrocytes. Matrix Biol. 21, 251–262 [DOI] [PubMed] [Google Scholar]
- 56. Mengshol J. A., Vincenti M. P., Coon C. I., Barchowsky A., Brinckerhoff C. E. (2000) Interleukin-1 induction of collagenase 3 (matrix metalloproteinase 13) gene expression in chondrocytes requires p38, c-Jun N-terminal kinase, and nuclear factor κB. Differential regulation of collagenase 1 and collagenase 3. Arthritis Rheum. 43, 801–811 [DOI] [PubMed] [Google Scholar]
- 57. Yang S., Kim J., Ryu J. H., Oh H., Chun C. H., Kim B. J., Min B. H., Chun J. S. (2010) Hypoxia-inducible factor-2α is a catabolic regulator of osteoarthritic cartilage destruction. Nat. Med. 16, 687–693 [DOI] [PubMed] [Google Scholar]
- 58. Hayashida M., Okazaki K., Fukushi J., Sakamoto A., Iwamoto Y. (2009) CCAAT/enhancer-binding protein β mediates expression of matrix metalloproteinase 13 in human articular chondrocytes in inflammatory arthritis. Arthritis Rheum. 60, 708–716 [DOI] [PubMed] [Google Scholar]
- 59. Litherland G. J., Elias M. S., Hui W., Macdonald C. D., Catterall J. B., Barter M. J., Farren M. J., Jefferson M., Rowan A. D. (2010) Protein kinase C isoforms ζ and ι mediate collagenase expression and cartilage destruction via STAT3- and ERK-dependent c-fos induction. J. Biol. Chem. 285, 22414–22425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Borden P., Solymar D., Sucharczuk A., Lindman B., Cannon P., Heller R. A. (1996) Cytokine control of interstitial collagenase and collagenase-3 gene expression in human chondrocytes. J. Biol. Chem. 271, 23577–23581 [DOI] [PubMed] [Google Scholar]
- 61. Goldring M. B., Birkhead J. R., Suen L. F., Yamin R., Mizuno S., Glowacki J., Arbiser J. L., Apperley J. F. (1994) Interleukin-1 β-modulated gene expression in immortalized human chondrocytes. J. Clin. Invest. 94, 2307–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lo Y. Y., Conquer J. A., Grinstein S., Cruz T. F. (1998) Interleukin-1 β induction of c-fos and collagenase expression in articular chondrocytes. Involvement of reactive oxygen species. J. Cell. Biochem. 69, 19–29 [DOI] [PubMed] [Google Scholar]
- 63. Pei Y., Harvey A., Yu X. P., Chandrasekhar S., Thirunavukkarasu K. (2006) Differential regulation of cytokine-induced MMP-1 and MMP-13 expression by p38 kinase inhibitors in human chondrosarcoma cells. Potential role of Runx2 in mediating p38 effects. Osteoarthr. Cartil. 14, 749–758 [DOI] [PubMed] [Google Scholar]
- 64. Muddasani P., Norman J. C., Ellman M., van Wijnen A. J., Im H. J. (2007) Basic fibroblast growth factor activates the MAPK and NFκB pathways that converge on Elk-1 to control production of matrix metalloproteinase-13 by human adult articular chondrocytes. J. Biol. Chem. 282, 31409–31421 [DOI] [PubMed] [Google Scholar]
- 65. Nagarajan P., Parikh N., Garrett-Sinha L. A., Sinha S. (2009) Ets1 induces dysplastic changes when expressed in terminally-differentiating squamous epidermal cells. PLoS One 4, e4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Díaz-Sanjuán T., García-Ruiz I., Rodríguez-Juan C., Muñoz-Yagüe T., Solís-Muñoz P., Solís-Herruzo J. A. (2009) Interferon α increases metalloproteinase-13 gene expression through a polyomavirus enhancer activator 3-dependent pathway in hepatic stellate cells. J. Hepatol. 50, 128–139 [DOI] [PubMed] [Google Scholar]
- 67. Kopp J. L., Wilder P. J., Desler M., Kinarsky L., Rizzino A. (2007) Different domains of the transcription factor ELF3 are required in a promoter-specific manner and multiple domains control its binding to DNA. J. Biol. Chem. 282, 3027–3041 [DOI] [PubMed] [Google Scholar]
- 68. Iwai S., Amekawa S., Yomogida K., Sumi T., Nakazawa M., Yura Y., Nishimune Y., Nozaki M. (2008) ESE-1 inhibits the invasion of oral squamous cell carcinoma in conjunction with MMP-9 suppression. Oral Dis. 14, 144–149 [DOI] [PubMed] [Google Scholar]
- 69. Seidel J. J., Graves B. J. (2002) An ERK2 docking site in the Pointed domain distinguishes a subset of ETS transcription factors. Genes Dev. 16, 127–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.