Background: Vibrio cholerae has a need for sialic acid transport and catabolism for colonization.
Results: The VC1777–1779 genes encode a functional tripartite ATP-independent periplasmic (TRAP) transporter for sialic acid in which the membrane subunits form a tight complex.
Conclusion: Vibrio cholerae encodes a functional sialic acid TRAP transporter.
Significance: This study reveals the route of sialic acid acquisition by an important human pathogen.
Keywords: Bacteria, Membrane Proteins, Reconstitution of Membrane Transporters, Sialic Acid, Transporters, TRAP Transporter, Vibrio cholerae
Abstract
Tripartite ATP-independent periplasmic (TRAP) transporters are widespread in bacteria but poorly characterized. They contain three subunits, a small membrane protein, a large membrane protein, and a substrate-binding protein (SBP). Although the function of the SBP is well established, the membrane components have only been studied in detail for the sialic acid TRAP transporter SiaPQM from Haemophilus influenzae, where the membrane proteins are genetically fused. Herein, we report the first in vitro characterization of a truly tripartite TRAP transporter, the SiaPQM system (VC1777–1779) from the human pathogen Vibrio cholerae. The active reconstituted transporter catalyzes unidirectional Na+-dependent sialic acid uptake having similar biochemical features to the orthologous system in H. influenzae. However, using this tripartite transporter, we demonstrate the tight association of the small, SiaQ, and large, SiaM, membrane proteins that form a 1:1 complex. Using reconstituted proteoliposomes containing particular combinations of the three subunits, we demonstrate biochemically that all three subunits are likely to be essential to form a functional TRAP transporter.
Introduction
Substrate-binding protein (SBP)2-dependent transporters are responsible for the movement of a plethora of important nutrients into bacterial cells. The most widely studied SBP-dependent transporters are the ATP-binding cassette transporters that utilize the binding and hydrolysis of ATP to power the translocation of the substrate across the membrane (1). However, SBPs are also used in another large and common family of transporters called the tripartite ATP-independent periplasmic (TRAP) transporters (2). The key difference between ATP-binding cassette and TRAP transporters is that although ATP-binding cassette transporters are primary transporters, TRAP transporters are secondary transporters that use electrochemical gradients across the membrane to energize uptake (3, 4). Much progress has been made recently in the understanding of the structure and function of SBPs from TRAP transporters (5–11), but our knowledge of the membrane components is much poorer. TRAP transporters contain two membrane components. The large (or “M”) subunit is predicted to have 12 transmembrane helices (TMHs) and is thought to form the translocation channel as it is a member of the ion transporter superfamily of secondary transporters (12). The small (or “Q”) subunit has four predicted TMHs, and the DctQ protein from Rhodobacter capsulatus has been experimentally determined to have an NINCIN topology (13). The function of the Q subunit is as yet unknown, but studies from the dctPQM C4-dicarboxylate TRAP transporter from R. capsulatus demonstrated genetically that the dctQ gene, like dctP and dctM, was essential for a functional DctPQM transporter (3).
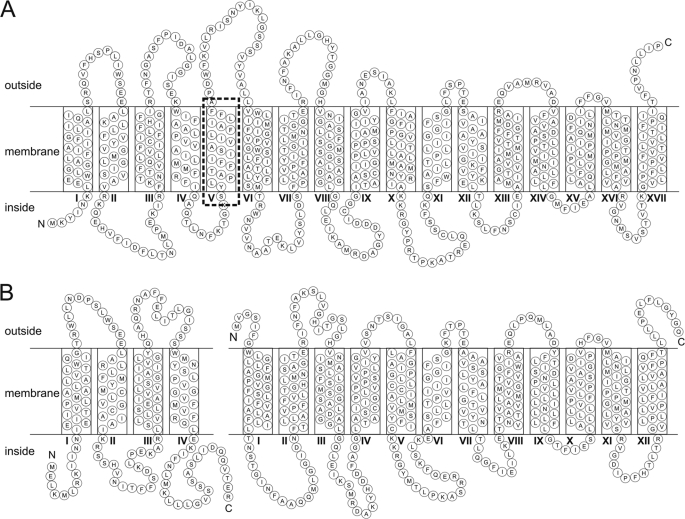
In a typical TRAP transporter operon, the Q subunit is encoded upstream of the M subunit, although there are now a few rare examples of the order being reversed (14). Often the Q and M genes are fused genetically into a longer gene, QM, and this is the case in the sialic acid-specific TRAP transporter, SiaPQM, from Haemophilus influenzae, where the membrane domains are encoded by the siaQM gene (also called siaT by other authors) (15, 16). In these fused systems, which are not uncommon, there is a predicted extra “linker helix” that is needed to connect the CIN end of the Q subunit with the predicted NOUT location of the M subunit (Fig. 1 and supplemental Fig. 1). Hence, the SiaQM protein from H. influenzae is predicted to have 17 TMHs (2).
FIGURE 1.
Topology models of HiSiaQM, VcSiaQ, and VcSiaM. Consensus topology models of HiSiaQM from H. influenzae (A) and VcSiaQ (left) and VcSiaM (right) from V. cholerae (B). To construct the consensus models, a multiple sequence alignment was performed on the integral membrane proteins from seven TRAP transporters previously predicted to be specific to Neu5Ac (15). Included in this alignment were three TRAP transporters whose integral membrane component was composed of two separate proteins and four fused membrane components. The boundaries of the TMHs for each protein were predicted using the algorithm TMHMM (36). The predicted transmembrane helix boundaries for each protein and the multiple sequence alignment were overlaid, allowing us to define the consensus boundaries of the transmembrane helices. The topology models were then constructed in CoralDRAW. The linker helix of HiSiaQM is indicated by a dashed box.
The SiaPQM system from H. influenzae (from now on referred to as HiSiaPQM) is the most comprehensively studied TRAP transporter to date, as it has been fully reconstituted into proteoliposomes and determined to function as a unidirectional Na+-dependent secondary transporter (2). This system was chosen for study partly due to the natural fusion of the membrane domains such that the reconstituted systems are effectively bipartite rather than tripartite. The structure of the SBP for this transporter, SiaP, is also known, which has been solved in ligand-free and ligand-bound forms (6, 11). The transporter is biologically important in H. influenzae as sialic acid is an important host-derived molecule that is needed for persistence in the host, and loss of the transporter causes defects in colonization (15, 17, 18).
More recently, an important role for sialic acid transport and catabolism has been identified in the human pathogen Vibrio cholerae, as the sialic acid catabolic gene nanA, which encodes a cytoplasmic enzyme that catalyzes the first step in sialic acid catabolism, has been shown to be important for colonization of the host (19). The nanA gene is carried on a pathogenicity island (VPI-2) present in toxigenic strains of V. cholerae (20, 21), which also encodes a secreted sialidase, that presumably releases the substrate (the common sialic acid N-acetylneuraminic acid) for use by the cell, while at the same time revealing binding sites for the cholera toxin (22). How sialic acid is taken into the cell has not been formally demonstrated, but it is likely to be through a TRAP transporter (VC1777–1779) encoded in the VPI-2, which is orthologous to HiSiaPQM (15, 19–21, 23, 24).
Although we have demonstrated previously the VcSiaP (VC1779) has identical ligand-binding properties to HiSiaP (4), we demonstrate here using purified and reconstituted proteins that VC1777–1779 encodes a functional sialic acid transporter with bioenergetic properties identical to the HiSiaPQM system. Significantly, the VcSiaPQM system differs from the HiSiaPQM system as Q and M membrane subunits are encoded by separate genes, and as such this is the first in vitro characterization of a truly tripartite TRAP transporter. We use the VcSiaPQM system to demonstrate that the Q and M subunits form a tight stoichiometric complex in the membrane and present the first biochemical data suggesting that all three subunits are essential for transporter function.
EXPERIMENTAL PROCEDURES
Expression Vector Construction
All proteins used in this work were overexpressed using a PBAD expression system that allows addition of decahistidine tags at either the N or C termini of the recombinant protein. The details of expression vector construction for the expression HiSiaQM have been described previously (4). N- and C-terminally tagged versions of VcSiaP (VC1779), VcSiaQ (VC1778), and VcSiaM (VC1777) were produced by PCR amplification of the genes using primer pairs (see supplemental Table 1 for primer sequences). The expression vector used to co-express the dicistronic VcsiaQM genes was produced by PCR amplification of the region containing both open reading frames with the native intergenic region (supplemental Table 1). All PCR products were cloned into both appropriate vectors using ligation-independent cloning to generate N- and C-terminally decahistidine-tagged constructs.
Protein Expression
The C-terminally tagged VcSiaQ, N-terminally tagged VcSiaM, and N-terminally tagged VcSiaQM were expressed essentially as described for N-terminally tagged HiSiaQM (4). Briefly, Escherichia coli MC1061 (pBADcVcSiaQ), MC1061 (pBADnVcSiaM), and MC1061 (pBADnVcSiaQM) were grown at 37 °C in Luria-Bertani broth (LB), supplemented with 0.5% (v/v) glycerol and 100 μg/ml ampicillin, until an A650 of 1 was reached. Each culture was subsequently induced by addition of 50 mg/liter l-arabinose and grown for a further 2 h. Cells were harvested by centrifugation at 4500 × g for 12 min at 4 °C and resuspended in 50 mm potassium phosphate, pH 7.8, supplemented with 20% glycerol (v/v) in a ratio of 35 ml/liter culture.
N-terminally tagged VcSiaP was overexpressed as described previously for N-terminally tagged HiSiaP (4). Briefly, E. coli MC1061/pBADnVcSiaP was grown at 37 °C in M9 minimal medium, supplemented with 100 μg/ml ampicillin, 0.2% (v/v) glycerol, 40 μg/ml l-leucine, 40 μg/ml l-isoleucine, 50 μg/ml 4-methyl-5-β-hydroxyethylthiazole, 2 μg/ml thiamine, 50 μg/ml nicotinic acid, and 1 mm MgSO4. When an A650 of 1 was reached, the culture was induced by addition of 50 mg/liter arabinose and grown for a further 2 h. Cells were harvested by centrifugation at 4500 × g for 12 min at 4 °C and resuspended in 50 mm potassium phosphate, pH 7.8, supplemented with 20% glycerol (v/v) in a ratio of 35 ml/liter culture.
Cytoplasm and Membrane Vesicle Preparation
The cytoplasm and membrane vesicles of the expressions strains were prepared by rupturing the cells using a Misonix sonicator 3000. All cell suspensions were ruptured in the presence of 50 mm potassium phosphate, pH 7.8, supplemented with 20% glycerol, 100 μg/ml DNase I (Sigma), and 1 mm MgCl2. The SBP-containing cytoplasmic fraction was separated from the insoluble fraction and unbroken cells by centrifugation at 45,000 × g for 30 min at 4 °C. The cytoplasm was then stored at −20 °C. The membrane vesicles were separated from cell debris and unbroken cells by centrifugation at 10,000 × g for 20 min at 4 °C. To collect the membrane vesicles, the resultant supernatant was centrifuged at 200,000 × g for 1 h at 4 °C. The membrane vesicles were resuspended in 50 mm potassium phosphate, pH 7.8, supplemented with 20% glycerol in a ratio of 1 ml/liter culture. The vesicles were then snap-frozen in liquid nitrogen and stored at −80 °C.
SBP Purification
VcSiaP was purified in a single step by using immobilized metal ion affinity chromatography (IMAC) with TALON Co2+-resin (Clontech). The TALON resin was equilibrated with wash buffer (WB) containing 50 mm potassium phosphate, pH 7.8, 200 mm NaCl, 20% glycerol, supplemented with 10 mm imidazole. The cytoplasmic fraction containing VcSiaP was supplemented with 200 mm NaCl, 10 mm imidazole and incubated with pre-equilibrated TALON resin for 1 h at room temperature. Impurities were eluted by washing the resin with WB supplemented with 10 mm imidazole. VcSiaP was eluted using WB supplemented with 500 mm imidazole. Purified VcSiaP was dialyzed overnight against 50 mm potassium phosphate, pH 7.8, 200 mm NaCl.
Membrane Protein Purification
VcSiaQM and NanT were purified using IMAC essentially as described previously for HiSiaQM (4). Membrane vesicles containing VcSiaQM were diluted 10-fold into WB supplemented with 10 mm imidazole and 0.5% (w/v) n-dodecyl-β-d-maltoside (DDM) and incubated on ice for 30 min. Material not solubilized by the DDM was removed by centrifugation at 200,000 × g for 20 min at 4 °C. The resultant supernatant was incubated for 1 h at 4 °C with Ni2+-nitrilotriacetic acid superflow resin (Qiagen, 0.3 ml, 1 ml of membrane vesicles) pre-equilibrated with WB supplemented with 10 mm imidazole and 0.05% DDM (w/v). The resin was distributed between disposable polystyrene columns (Pierce) and washed with 10 column volumes of WB supplemented with 40 mm imidazole and 0.05% DDM (w/v). The resin was then washed with 10 column volumes of WB supplemented with 40 mm imidazole and 0.15% n-decyl-β-d-maltoside (w/v). Protein was eluted by addition of WB supplemented with 500 mm imidazole and 0.15% n-decyl-β-d-maltoside (w/v). 5 mm EDTA was added to the elution fractions. DDM concentrations were determined using the methods of Urbani and Warne (25).
Membrane Reconstitution of VcSiaQM and NanT
The liposomes used in the reconstitution were a 3:1 (w/w) ratio of purified E. coli lipids and egg l-α-phosphatidylcholine (Avanti Polar Lipids) with a concentration of 20 mg of lipid/ml. Purification eluate containing 200 μg of VcSiaQM was diluted to 2 ml with 50 mm potassium phosphate, pH 7.8, 0.15% n-decyl-β-d-maltoside (w/v). The protein sample was mixed with 400 μl of liposomes and incubated on ice for 10 min. The protein/lipid mixture was rapidly diluted in 70 ml of 50 mm potassium phosphate, pH 7. Proteoliposomes were collected by centrifugation at 200,000 × g for 1.5 h at 4 °C. Proteoliposomes were resuspended in 50 mm potassium phosphate, pH 7, and snap-frozen. The proteoliposomes were then frozen and slowly thawed on ice five times.
Preparation of Proteoliposomes and Transport Assays
For use in the transport assays, proteoliposomes containing 400 μg of protein/ml were centrifuged for 20 min at 200,000 × g at 4 °C. The pellet of proteoliposomes was resuspended in lumenal buffer (100 mm potassium acetate, 20 mm potassium phosphate, pH 7, and 2 mm MgSO4, unless otherwise stated) and extruded through a 400-nm polycarbonate filter (Avestin Inc.). The extruded proteoliposomes were centrifuged for 20 min at 200,000 × g at 15 °C, and the resultant proteoliposomes pellet was resuspended in lumenal buffer to a concentration of 4 μg/μl protein.
For all transport assays performed, the proteoliposome suspension was diluted 50-fold into the following buffers to form the required electrochemical gradients. An inwardly directed Na+ (ΔμNa+) gradient was formed by dilution into buffer containing 100 mm sodium acetate, 20 mm sodium PIPES, pH 7, and 2 mm MgSO4. A Na+ gradient in combination with membrane potential (ΔμNa+ + ΔΨ) was formed by dilution into buffer containing 100 mm sodium acetate, 20 mm sodium PIPES, pH 7, 2 mm MgSO4, and 2 μm valinomycin. A Na+ gradient in combination with a pH gradient (ΔμNa+ + ΔpH) was formed by dilution into buffer containing 120 mm sodium PIPES, pH 7, and 2 mm MgSO4. A pH gradient (ΔpH) alone was formed by dilution into buffer containing 120 mm potassium phosphate, pH 7, and MgSO4. A membrane potential alone (ΔΨ) was formed by dilution into buffer containing 100 mm N-methylglucamine acetate, pH 7, 20 mm N-methylglucamine phosphate, pH 7, 2 mm MgSO4, and 2 μm valinomycin. All transport assays, unless indicated otherwise, contained 5 μm VcSiaP (except for the NanT transport assays) and 5 μm [14C]Neu5Ac. The cold chase assays were performed by application of a ΔμNa+ + ΔΨ followed by the addition of 1 mm unlabeled Neu5Ac at 100 s. The substrate counterflow assay was performed by dilution of proteoliposomes prepared by extrusion with an altered lumenal buffer containing 100 mm potassium acetate, 20 mm potassium phosphate, pH 7, 2 mm MgSO4 and 1 mm unlabeled Neu5Ac into the same buffer without the unlabeled Neu5Ac and with 5 μm [14C]Neu5Ac.
Transport assays were performed by incubating 600 μl of the appropriate buffer plus 5 μm [14C]Neu5Ac for 1 min with stirring at 30 °C in a water bath. 12 μl of proteoliposomes was added to the 600-μl buffer, and 50-μl samples were taken at regular intervals. Samples were incubated with cold wash buffer (the reaction buffer with the addition of 1 mm unlabeled Neu5Ac) for 10 s before rapid filtration with 0.45-μm pore diameter cellulose nitrate membranes (Millipore). The filters were washed with 2 ml of ice-cold 50 mm potassium phosphate, pH 7, and radioactivity associated with the filters was determined using liquid scintillation counting. All transport assays were performed in triplicate.
SEC-MALLS
SEC-MALLS was conducted using Wyatt HELEOS-II 18 light scattering and rEX refractive index detectors coupled to a Shimadzu LC-20 HPLC system with SPD20A UV detector. IMAC-purified protein samples were loaded onto a Superdex 200 column (10/300 GL, GE Healthcare) equilibrated with 50 mm potassium phosphate, pH 7.8, 200 mm NaCl, 0.05% (w/v) DDM, run at 0.5 ml/min. For data analysis, protein was taken to have a refractive index increment (dn/dc) of 0.186 ml/g and DDM of 0.143 ml/g (26). The ratio of DDM to protein was determined by a colorimetric assay of the DDM maltoside (25) as 219 ± 63 DDM per VcSiaQM and 235 ± 93 DDM per HiSiaQM. These ratios were used to calculate a composite dn/dc of 0.159 ml/g for HiSiaQM-DDM and VcSiaQM-DDM complexes (27). This dn/dc value was then used in the Wyatt Astra software to calculate molar masses for the complex particles.
Sedimentation Equilibrium Analytical Ultracentrifugation (AUC)
The sedimentation equilibrium experiments were performed on a Beckman Optima XL/1 analytical ultracentrifuge. All experiments were performed at 20 °C. Absorbance scans (280 nm) were taken every 4 h until equilibrium was achieved. Absorbance scans were conducted in step mode with 10 replicates per data point. Scan data were analyzed using the Beckman Origin software to give buoyant masses (uncorrected for partial specific volume or solvent density). Assuming an integral number of SiaQM molecules per particle, the number of detergent molecules required to achieve this buoyant mass was calculated (using a partial specific volume of 0.83 ml/g for DDM). The results were consistent with a monomer plus 244 DDM or a dimer plus 26 DDM molecules; the latter was discarded as unrealistic.
RESULTS
Co-expression of siaQ and siaM Is Essential for Accumulation of High Levels of V. cholerae SiaQM
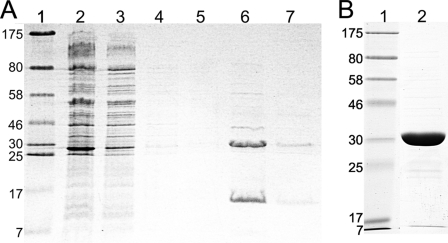
For in vitro characterization of VcSiaPQM, we first attempted to express the membrane subunits separately as either C- or N-terminally decahistidine-tagged proteins. The best expressing variants, a C-terminal tagged VcSiaQ and an N-terminal tagged for VcSiaM, could be expressed and purified using IMAC, but expression levels were very low (0.06 and 0.08 mg/liter, respectively). Using the same expression system, we generated an N-terminally decahistidine-tagged siaQ that was co-expressed with siaM, containing the native intergenic region between siaQ and siaM. Here, the expressed protein accumulated to higher levels than the individually expressed subunits and could be solubilized in DDM and purified using IMAC. When analyzed on SDS-PAGE (Fig. 2A), we observed both VcSiaQ (22 kDa) and VcSiaM (45 kDa) in the elution fraction. Both proteins migrated to positions corresponding to masses smaller than predicted, ∼15 and ∼30 kDa, respectively; however, this is a common observation for integral membrane proteins (27). Given that the histidine tag was on the VcSiaQ subunit only, the presence of VcSiaM in the elution fraction suggests that it is forming a protein complex with VcSiaQ. The identities of the two protein bands were confirmed using MALDI-tandem mass spectrometry as VcSiaQ and VcSiaM (data not shown). This co-expressed protein provided a combined yield of ∼0.4 mg/liter culture and was stable enough in DDM to make them amenable to further study.
FIGURE 2.
Expression and purification of the components of VcSiaPQM. A, Coomassie-stained SDS-polyacrylamide gel of the purification of the VcSiaQM complex (His-tagged on the N terminus of VcSiaQ) using IMAC. Lane 1, molecular weight ladder; lane 2, total membrane vesicles; lane 3, column flow-through; lane 4, 40 mm imidazole washing step; lanes 5–7, 500 mm imidazole elution steps. B, Coomassie-stained SDS-polyacrylamide gel of N-terminally tagged VcSiaP purified in one IMAC step. Lane 1, molecular mass marker with the sizes of the proteins indicated in kDa; lane 2, IMAC-purified VcSiaP.
To enable the reconstitution of the complete VcSiaPQM transporter we also expressed an N-terminally decahistidine-tagged version of VcSiaP, which purified to apparent homogeneity using IMAC producing a yield of 9.1 mg/liter culture (Fig. 2B). This binds the common sialic acid Neu5Ac with a high affinity of 0.11 ± 0.03 μm (data not shown), identical to that which we measured previously with a C-terminally hexahistidine-tagged version of the protein (4).
VcSiaPQM Transporter Containing Co-expressed SiaQM Is Active as a Na+-driven Secondary Transporter for Sialic Acid
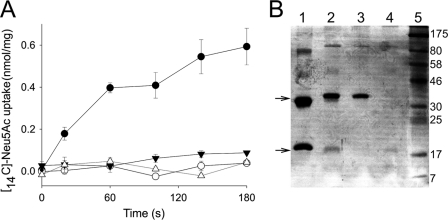
To determine whether the purified SiaQ and SiaM proteins are active and assess the properties of the resulting VcSiaPQM transporter, we reconstituted the IMAC-purified SiaQ and SiaM into proteoliposomes, using a dilution method (28), as used previously for HiSiaQM (4). To determine the energetic requirements of transport by VcSiaPQM, different electrochemical gradients across the membrane were systematically varied, and the resulting effects on [14C]Neu5Ac uptake in the presence of VcSiaP were monitored. As was observed for HiSiaPQM, the application of an inwardly directed sodium gradient (ΔμNa+) to VcSiaQM-containing proteoliposomes resulted in rapid uptake of [14C]Neu5Ac into the lumen of the proteoliposomes where it accumulated ∼11-fold over 6 min (Fig. 3A). The presence of either a pH gradient alone (ΔpH, increasing internal pH from 7 to 8.7) or a membrane potential alone (ΔΨ, −40 mV) was insufficient to support transport by VcSiaPQM (Fig. 3A). In the presence of ΔμNa+, the addition of ΔpH did not alter the rate of uptake, indicating no direct role for protons in the transport activity of VcSiaPQM. The highest transport activity was observed in the presence of a ΔμNa+ in combination with a ΔΨ (Fig. 3A). This stimulation of transport activity by inclusion of a ΔΨ suggests that transport of VcSiaPQM is an electrogenic process. As Neu5Ac has a single negative charge at physiological pH (pKa = 2.6), this implies that Neu5Ac is co-transported with at least two Na+ ions. These data demonstrate the VcSiaPQM system is a sialic acid transporter, which has similar properties to the known bipartite counterpart from H. influenzae (2).
FIGURE 3.
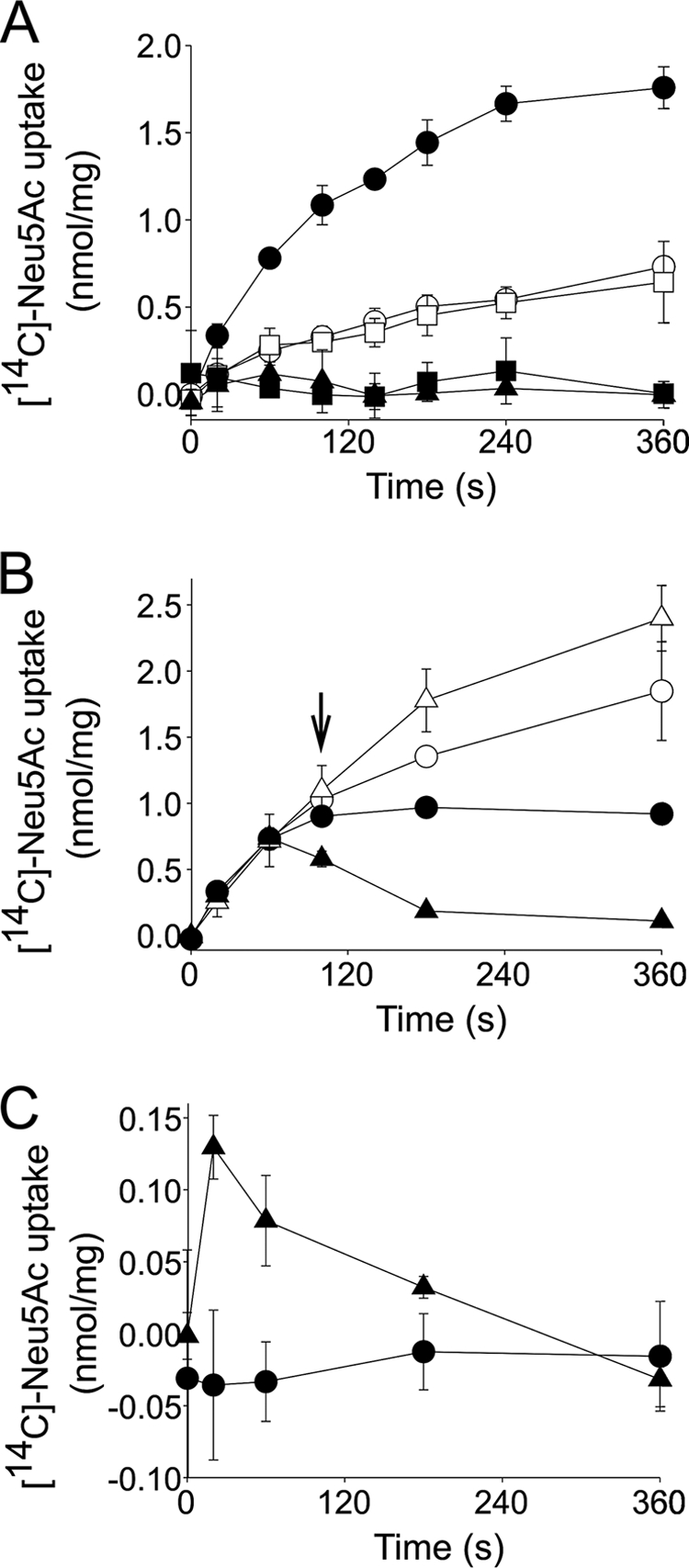
Energetic characterization of VcSiaPQM. A, transport of 5 μm [14C]Neu5Ac into the lumen of VcSiaQM-containing proteoliposomes in the presence of 5 μm VcSiaP and the following electrochemical gradients: ΔμNa+ΔΨ (closed circles), ΔμNa alone (open circles), ΔμNa+ΔpH (open squares), ΔpH alone (closed squares), and ΔΨ alone (closed triangles). B, cold chase assay with unlabeled Neu5Ac. Transport of 5 μm [14C]Neu5Ac into the lumen of proteoliposomes containing VcSiaQM (circles, also in the presence of 5 μm VcSiaP) with an applied ΔμNa + ΔΨ or NanT (triangles) with an applied ΔpH. At 100 s (arrow), either 1 mm unlabeled Neu5Ac (closed symbols) or an equivalent volume of distilled H2O (open symbols) was added to the reaction. C, solute counterflow activity of proteoliposomes prepared via extrusion containing lumenal buffer with 1 mm unlabeled Neu5A and either VcSiaQM with 5 μm VcSiaP (circles) or NanT (triangles).
VcSiaPQM Functions as a Unidirectional Secondary Transporter
Characterization of HiSiaPQM revealed that this TRAP transporter has unusual properties that distinguish it from other secondary carriers, in that it appears to function unidirectionally (4). To assess whether this interesting property was also a feature of a second TRAP transporter, we repeated these experiments with VcSiaPQM and also included purified and reconstituted NanT, the E. coli K12 sialic acid transporter from the major facilitator superfamily as a control (29–31). Using membrane-reconstituted protein, we demonstrated that NanT is a ΔpH-driven transporter and that ΔμNa+ plays no part in Neu5Ac transport (supplemental Fig. 2).
The directionality of VcSiaPQM transport was investigated using substrate exchange and substrate counterflow experiments in vitro. In the substrate exchange experiments, [14C]Neu5Ac was accumulated in VcSiaQM or NanT-containing proteoliposomes energized by a ΔμNa+ + ΔΨ or ΔpH, respectively. At 100 s, 1 mm unlabeled Neu5Ac (200-fold excess) was added, which resulted in rapid efflux of the preaccumulated [14C]Neu5Ac from NanT-containing proteoliposomes, as expected for a conventional secondary transporter (Fig. 3B). Addition of excess unlabeled substrate to VcSiaPQM-containing proteoliposomes resulted in a cessation of transport activity but no efflux of preaccumulated [14C]Neu5Ac (Fig. 3B), indicating that VcSiaPQM cannot perform this substrate exchange phenotype characteristic of secondary carriers. In both cases, the addition of distilled H2O resulted in continued transport of [14C]Neu5Ac into the proteoliposomes (Fig. 3B).
The abilities of VcSiaPQM and NanT to facilitate substrate counterflow activity were also assessed. This assay measures the ability of a transporter to exchange lumenal unlabeled substrate present at a high concentration with external radiolabeled substrate present at a relatively low concentration (32, 33). This exchange of substrate is facilitated by the accessibility of the substrate-binding site alternating from one side of the membrane to the other. This results in rapid but transient accumulation of the radiolabeled substrate followed by apparent efflux as the concentration on both sides of the membrane equilibrates. VcSiaPQM- and NanT-containing proteoliposomes were loaded with 1 mm unlabeled Neu5Ac by extrusion and diluted into a solution containing 5 μm [14C]Neu5Ac. NanT-containing proteoliposomes had distinct counterflow activity under these conditions, whereas VcSiaPQM-containing proteoliposomes did not (Fig. 3C). Under these experimental conditions, these findings suggest that like HiSiaQM, VcSiaPQM is a unidirectional secondary transporter. This is unlike the control MFS protein NanT, which has the ability to transport Neu5Ac in a reversible manner.
Failure of Separately Expressed Membrane Domains to Form a Functional TRAP Transporter
Although the essential role of the SBP for TRAP transporter function is well established both in vivo and in vitro (2, 3, 15), the essential role of both membrane subunits has only been demonstrated genetically (3, 15). Using the tripartite VcSiaPQM system, we sought to determine whether each individual subunit of the transporter was necessary and/or sufficient to catalyze transport.
Transport of [14C]Neu5Ac into proteoliposomes prepared to produce a ΔμNa+ + ΔΨ was assessed using a range of differently prepared and reconstituted SiaPQM subunits. First, we compared SiaQM co-expressed and reconstituted to SiaQ and SiaM expressed and purified separately, mixed, and reconstituted together. Assays were performed in the presence of VcSiaP. Although we could measure uptake of [14C]Neu5Ac into proteoliposome containing co-expressed SiaQM (Fig. 4A), we could not detect uptake for the separately expressed and mixed SiaQM (Fig. 4A). Also, no uptake was observed when VcSiaQ or VcSiaM was reconstituted alone either in the presence of absence of VcSiaP, which suggests that they also are not functional individually (Fig. 4A) (data not shown). We confirmed the proteins present in the proteoliposomes on silver-stained SDS-polyacrylamide gels (Fig. 4B) and also by Western blot (data not shown), but we cannot rule out that the His tags used for purification of these proteins might interfere with their function. As mentioned previously, the expression of VcSiaQ and VcSiaM alone reduced their stability and also resulted in diminished reconstitution efficacy (Fig. 4B), although there is clearly a protein present in each of the different conditions. We note that the SiaM from the co-expressed protein migrates slightly further than the singly expressed sample, likely as the single expressed protein has an additional N-terminal decahistidine tag. These data suggest that none of the three subunits expressed alone can catalyze transport, consistent with all the subunits being essential for TRAP transporter function and that co-expression of VcSiaQ and VcSiaM is essential for function.
FIGURE 4.
Requirements of different transporter subunits for transporter function. A, transport of 5 μm [14C]Neu5Ac into the lumen of proteoliposomes containing VcSiaQM (co-expressed, closed circle), VcSiaQ and SiaM (separately expressed, open circles), VcSiaQ alone (closed inverted triangles), and VcSiaM alone (open triangles). All transport assays were performed with ΔμNa + ΔΨ applied and in the presence of 5 μm VcSiaP. B, silver-stained SDS-polyacrylamide gel of the proteoliposomes containing different combinations of reconstituted VcSiaQ and VcSiaM. Lane 1, molecular weight ladder; lane 2, VcSiaQ and VcSiaM co-expressed, co-purified, and reconstituted; lane 3, VcSiaQ and VcSiaM, separately expressed, purified, mixed, and then reconstituted; lane 4, VcSiaM reconstituted alone; lane 5, VcSiaQ reconstituted alone. The top and bottom arrows indicate the position of the bands for VcSiaM and VcSiaQ, respectively.
VcSiaQ and VcSiaM Form a Complex in Solution
During purification of co-expressed SiaQM, it was clear that the two proteins were associated during IMAC (Fig. 2A). We analyzed this interaction further using size-exclusion chromatography, and here we included HiSiaQM as a control. The co-expressed IMAC-purified VcSiaQM eluted as a single large peak in the chromatogram indicating the presence of a single protein or protein complex (Fig. 5A). SDS-PAGE analysis of the samples across the peak revealed that, despite their large difference in molecular weight, VcSiaQ and VcSiaM shared an identical elution profile (Fig. 5B), indicating that the proteins are forming a complex. As a control, we also performed an identical procedure with HiSiaQM, the fused QM protein, which elutes slightly earlier than VcSiaQM due to the extra mass due to the presence of the linker helix (Fig. 5A) and on SDS-PAGE yields a single band that corresponds to the HiSiaQM protein (Fig. 5C).
FIGURE 5.
Size-exclusion chromatography of VcSiaQM and HiSiaQM. A, SEC trace of IMAC-purified VcSiaQM (black line) and HiSiaQM (gray line) showing absorbance (A280) versus retention time. B and C, Coomassie-stained SDS-polyacrylamide gels of the fractions taken across the SEC peaks of VcSiaQM and HiSiaQM, respectively.
To investigate further the nature of the interaction between the SiaQ and SiaM, we studied the naturally fused HiSiaQM and introduced a Factor Xa protease cleavage site into the solvent-exposed loop predicted to follow helix 5 (cutting at residue 200), producing the mutant HiSiaQMFxa200. This mutant protein was produced with a similar yield to wild-type HiSiaQM and was stable in DDM as determined by SEC analysis (data not shown). After incubation at room temperature for 3 h in the presence or absence of Factor Xa (supplemental Methods), SEC was used to determine whether the two domains of HiSiaQMFxa200, when no longer physically attached, would remain in complex. Although protein aggregation was apparent in both the presence and absence of Factor Xa, a peak in the SEC trace was observed for both samples at a retention volume of ∼11.9 ml (supplemental Fig. 3A). SDS-PAGE analysis of the fractions across these peaks revealed that in the absence of Factor Xa, only full-length HiSiaQMFxa200 eluted (supplemental Fig. 2B), although in the presence of Factor Xa, HiSiaQMFxa200 was partially digested resulting in both cleaved and noncleaved protein in the sample (supplemental Fig. 3B). Significantly, the elution pattern of the cleaved protein was identical to the uncleaved, suggesting that the two polypeptides remained packed into a tight complex despite no longer being covalently linked.
Stoichiometry of SiaQ, SiaM, and SiaQM Fusions
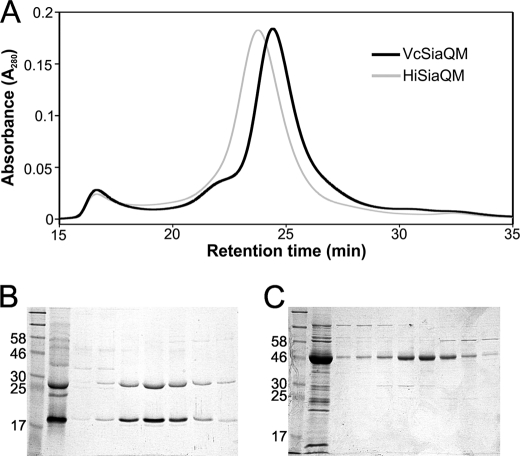
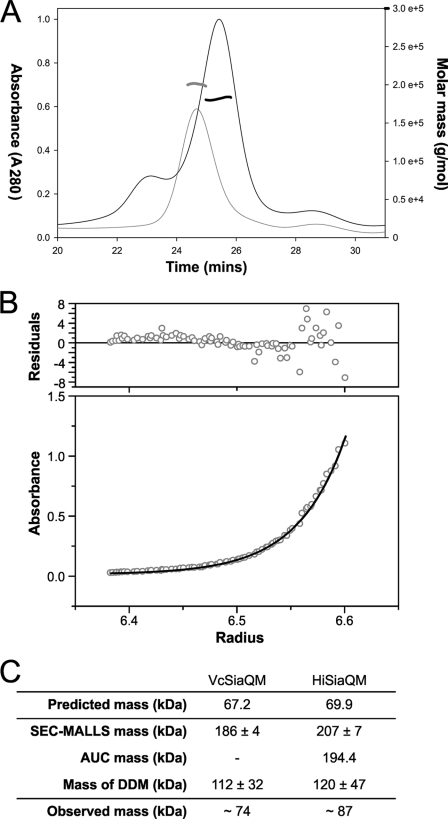
Using the separately encoded SiaQ and SiaM subunits of the V. cholerae transporter, we wished to determine experimentally the stoichiometry of the two components in the purified transporter. For this, we first used SEC combined with SEC-MALLS. Individually, VcSiaQM and HiSiaQM were extracted from the membrane using DDM and purified using IMAC, as described previously. The molecular weight of each protein-DDM complex was assessed using SEC-MALLS, revealing masses of 186 ± 4 and 207 ± 7 kDa for VcSiaQM and HiSiaQM, respectively (Fig. 6A).
FIGURE 6.
Molecular masses of HiSiaQM and VcSiaQM determined by AUC and SEC-MALLS. A, SEC-MALLS trace of VcSiaQM (black lines) and HiSiaQM (gray lines) showing the relative absorbance at A280 trace normalized to 1 (thin lines) and the molar masses of the species present in the samples (thick lines). B, representative dataset from the sedimentation equilibrium experiment performed on HiSiaQM using a Beckman Optima XL/1 analytical ultracentrifuge at 20 °C. C, table summarizing the data obtained for the masses of VcSiaQM and HiSiaQM using AUC, SEC-MALLS, and the colorimetric maltoside assay.
As a separate measure of the size of the protein-detergent complex, we used sedimentation equilibrium AUC experiments to corroborate the mass ranges obtained from SEC-MALLS. Unfortunately, VcSiaQM was too unstable over the 24-h period of the experiment to obtain a reliable molecular weight from AUC; however. the HiSiaQM-DDM complex was found to be 194.4 kDa using AUC, which is in good agreement with the SEC-MALLS data (Fig. 6, B and C).
To determine the mass of the detergent component of the complex, we used the colorimetric assay that measures the maltoside components of the DDM (Fig. 6C) (25). For VcSiaQM, the DDM assay revealed that there were 219 ± 63 DDM molecules per 1:1 complex of VcSiaQM, giving a detergent mass component of 112 ± 32 kDa. Subtracting this from the mass of the complex from SEC-MALLS leaves a mass of ∼74 kDa, which is only consistent with a single molecule of SiaQ and a single molecule of SiaM in the complex, which would have a mass of 67 kDa. Similarly, for HiSiaQM there were 235 ± 93 DDM molecules in the micelle giving a mass contribution of 120 ± 47 kDa. Subtracting these values from the masses of the whole complex gives a mass for HiSiaQM of around ∼87 kDa, which is only consistent with the protein being a monomer in the detergent micelle as its predicted mass of 69.9 kDa. Although there is considerable variation in the DDM assay, the masses suggest a likely 1:1 stoichiometry of the SiaQ and SiaM subunits in VcSiaQM as seen in the naturally fused HiSiaQM.
DISCUSSION
In this work, we have described the in vitro characterization of the sialic acid-specific TRAP transporter, VcSiaPQM, from V. cholerae. This TRAP transporter is of particular interest as it is a tripartite orthologue of the bipartite H. influenzae SiaPQM. The in vitro characterization of a tripartite TRAP transporter offers a number of advantages over bipartite systems, most prominently, the ability to express each protein separately to determine their individual characteristics and contributions. This is particularly important as the functions of the two integral membrane proteins have not been explicitly demonstrated.
The catabolism of sialic acid transport in V. cholerae is known to be important, and a strain lacking the first catabolic gene nanA has a defect in early colonization of the mouse intestine (19). The siaPQM genes (called dctPWQ in Ref. 19) are encoded within the Vibrio pathogenicity island 2 (VPI-2), which is present in strains of V. cholerae that can cause epidemic cholera outbreaks. The siaP gene is specifically induced by sialic acid as are related genes in the VPI-2 that also include the catabolic genes for sialic acid (19), and there is evidence that these genes are expressed in vivo (34). There is a strong correlation between the ability to grow on sialic acid and the presence of VPI-2 (19), which suggests that SiaPQM is the sole sialic acid transport in this bacterium. Although we have previously demonstrated that VcSiaP can bind sialic acid (4), this is the first characterization of VcSiaPQM as a functional sialic acid transporter.
Of note, the orthologous transporter genes in the related marine pathogen Vibrio vulnificus ATCC 29307 are also part of a pathogenicity island, and the adjacent catabolic gene, nanA, is important for virulence in a mouse model of infection (35), and it is probable that these genes encode a functional sialic acid transporter in this bacterium.
Biochemically, we found the VcSiaPQM is indistinguishable from HiSiaPQM, which confirmed that TRAP transporters can be Na+-dependent and that they have unique properties relating to their directionality that are conferred by the presence of the SBP. The only small difference we observed was that in HiSiaPQM transport decreased about 2-fold when a ΔpH was added in addition to ΔμNa+ (4) This was not seen with VcSiaPQM (Fig. 3A), which supports our previous interpretation that this effect was due to the added ΔpH affecting the protein conformation of HiSiaQM rather than being required for the transport cycle per se. Presumably differences in the cytoplasmic face of VcSiaQM versus HiSiaQM make the latter more susceptible to pH changes. The Na+ dependence we observed fits well with our previous observation of a propensity for TRAP transporters to be used preferentially by marine-dwelling bacteria (14), and this is also not surprising for V. cholerae, which can live in sea water where the salt concentration is ∼600 mm. The question of whether all TRAP transporters are Na+-dependent will require characterization of further, more diverse, members of the TRAP transporter family. This is also true for our repeated observation of the unidirectional movement of substrate, as judged from the cold chase assay and substrate counterflow assay. Importantly, in this study we included the NanT protein from E. coli as a control, which we show here for the first time in vitro is a sialic acid transporter that has typical properties of a major facilitator superfamily protein.
The formation of a complex between the small and large subunits was a primary reason to study the VcSiaPQM transporter, and indeed tagging with the small subunit resulted in co-purification of VcSiaM. This complex was stable and could also be observed on subsequent size-exclusion chromatography. The formation of the complex between the two membrane domains was already suggested by the genetic fusion of the two genes seen in the H. influenzae system, but these data demonstrated biochemically that they form a tight complex. By using the HiSiaQM protein, we were able to split the protein after expression and purification and found that the split SiaQ and SiaM still formed a stable complex. The stabilization of the individual proteins was also observed during our initial expression trials where the individually expressed proteins did not accumulate to high levels, whereas the co-expressed proteins did. The failure of the separately expressed and mixed VcSiaQ and VcSiaM to form a function transporter in the presence of VcSiaP suggests that the proteins need to fold at the same time in the membrane and have an intimate interaction that cannot be formed if they have folded separately; however, we cannot rule out here that the His tag on the separately expressed subunits might interfere with complex formation.
It is worth noting that the levels of separately expressed and purified VcSiaM were greater than those for VcSiaQ, which perhaps relates to the 12-transmembrane subunits having ancestral function as a stand-alone transporter and hence being more stable. We have also observed similar increases in stability and yield during work on the E. coli TRAP transporter YiaMNO, when the yield of YiaN (the large subunit) was greatly increased when co-expressed with YiaM (the small subunit).3 These biochemical data suggest that the small subunit is an important final component of the transporter and that complex formation directly after translation is likely important for the function of the SiaQM complex as separately purified and mixed proteins are not active.
The determination of the molecular weight of the purified protein complex, accounting for the amount of bound detergent in the micelle, yields a mass that was consistent with a 1:1 stoichiometry for the two subunits, which is what is likely occurring in the TRAP transporters where the membrane domains are naturally fused. The relative stoichiometry of the SBP with the membrane domains is assumed to be 1:1:1, but this has not been proven formally. Interestingly, we have recently observed novel genetic fusions between the large membrane subunit and the SBP, with the SBP being C-terminal to the large membrane subunit. This genetic organization is seen in a limited number of species of Acidaminococcus and Ruminococcus but are clearly suggestive of a 1:1 stoichiometry of the large membrane domain to the SBP.4 Hence, our current working models of TRAP transporters assume a 1:1:1 stoichiometry of the three subunits in the active transporter, although the SBP is present in the periplasm at a large excess over the membrane domains in Gram-negative bacteria.
It is also interesting, given this recent observation of fusion between the equivalents of SiaM and SiaP, that this might indicate that SiaM is the site of interaction with SiaP, and these two subunits might be all that are needed for function. However, in our experiments we have never been able to observe transporter function unless all three subunits are present, and in fact only if SiaQMs have been co-expressed, presumably forming a complex during or immediately after insertion into the cytoplasmic membrane. Given that SiaM is distantly related to DcuC, which is a conventional secondary transporter with no associated ancillary proteins, it is perhaps significant that no transporter activity could be observed for SiaM alone or with SiaP added, even though the protein was clearly present at high levels in the proteoliposomes (Fig. 4B); however, it was not possible to ascertain whether, in the absence of SiaQ, SiaM is properly folded and able to function. Regardless, this observation provides further evidence for the need for SiaQ in the final transporter for function. What the role of SiaQ is in the transporter cycle is not yet clear; it shares no sequence similarity to any other known protein family. It has been suggested that the small subunit could act as a chaperone for the large subunit to ensure it is inserted and/or folded in the membrane properly or it could act as a “landing pad” for the SBP during the transport cycle (4); its true function awaits experimental determination.
In conclusion, we demonstrate that the VPI-2-encoded TRAP transporter SiaPQM is a functional sialic acid transporter and has similar properties to that of another human pathogen H. influenzae. By characterizing this truly tripartite transporter, we have been able to demonstrate for the first time biochemically that the membrane subunits form a 1:1 complex and that all three components of the transporter are required for in vitro function.
Supplementary Material
Acknowledgment
We thank Rosie Dixon for computational analysis of novel gene arrangement of the TRAP transporter genes.
This work was supported by Biotechnology and Biological Sciences Research Council Grant BB/F014759/1 (to G. H. T.).

This article contains supplemental Methods, Figs. 1–3, and Table 1.
C. Mulligan and G. H. Thomas, unpublished data.
R. Dixon and G. Thomas, unpublished data.
- SBP
- substrate-binding protein
- TRAP
- tripartite ATP-independent periplasmic
- DDM
- n-dodecyl-β-d-maltoside
- IMAC
- immobilized metal affinity chromatography
- SEC
- size-exclusion chromatography
- AUC
- analytical ultracentrifugation
- TMH
- transmembrane helix
- Neu5Ac
- N-acetylneuraminic acid
- SEC-MALLS
- size-exclusion chromatography coupled to multiangle laser light scattering.
REFERENCES
- 1. Davidson A. L., Dassa E., Orelle C., Chen J. (2008) Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 72, 317–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mulligan C., Fischer M., Thomas G. H. (2011) Tripartite ATP-independent periplasmic (TRAP) transporters in bacteria and archaea. FEMS Microbiol. Rev. 35, 68–86 [DOI] [PubMed] [Google Scholar]
- 3. Forward J. A., Behrendt M. C., Wyborn N. R., Cross R., Kelly D. J. (1997) TRAP transporters. A new family of periplasmic solute transport systems encoded by the dctPQM genes of Rhodobacter capsulatus and by homologs in diverse Gram-negative bacteria. J. Bacteriol. 179, 5482–5493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mulligan C., Geertsma E. R., Severi E., Kelly D. J., Poolman B., Thomas G. H. (2009) The substrate-binding protein imposes directionality on an electrochemical sodium gradient-driven TRAP transporter. Proc. Natl. Acad. Sci. U.S.A. 106, 1778–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fischer M., Zhang Q. Y., Hubbard R. E., Thomas G. H. (2010) Caught in a TRAP. Substrate-binding proteins in secondary transport. Trends Microbiol. 18, 471–478 [DOI] [PubMed] [Google Scholar]
- 6. Müller A., Severi E., Mulligan C., Watts A. G., Kelly D. J., Wilson K. S., Wilkinson A. J., Thomas G. H. (2006) Conservation of structure and mechanism in primary and secondary transporters exemplified by SiaP, a sialic acid-binding virulence factor from Haemophilus influenzae. J. Biol. Chem. 281, 22212–22222 [DOI] [PubMed] [Google Scholar]
- 7. Rucktooa P., Antoine R., Herrou J., Huvent I., Locht C., Jacob-Dubuisson F., Villeret V., Bompard C. (2007) Crystal structures of two Bordetella pertussis periplasmic receptors contribute to defining a novel pyroglutamic acid-binding DctP subfamily. J. Mol. Biol. 370, 93–106 [DOI] [PubMed] [Google Scholar]
- 8. Kuhlmann S. I., Terwisscha van Scheltinga A. C., Bienert R., Kunte H. J., Ziegler C. (2008) 1.55 Å structure of the ectoine-binding protein TeaA of the osmoregulated TRAP-transporter TeaABC from Halomonas elongata. Biochemistry 47, 9475–9485 [DOI] [PubMed] [Google Scholar]
- 9. Gonin S., Arnoux P., Pierru B., Lavergne J., Alonso B., Sabaty M., Pignol D. (2007) Crystal structures of an extracytoplasmic solute receptor from a TRAP transporter in its open and closed forms reveal a helix-swapped dimer requiring a cation for α-keto acid binding. BMC Struct. Biol. 7, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lecher J., Pittelkow M., Zobel S., Bursy J., Bönig T., Smits S. H., Schmitt L., Bremer E. (2009) The crystal structure of UehA in complex with ectoine-A comparison with other TRAP-T-binding proteins. J. Mol. Biol. 389, 58–73 [DOI] [PubMed] [Google Scholar]
- 11. Johnston J. W., Coussens N. P., Allen S., Houtman J. C., Turner K. H., Zaleski A., Ramaswamy S., Gibson B. W., Apicella M. A. (2008) Characterization of the N-acetyl-5-neuraminic acid-binding site of the extracytoplasmic solute receptor (SiaP) of nontypeable Haemophilus influenzae strain 2019. J. Biol. Chem. 283, 855–865 [DOI] [PubMed] [Google Scholar]
- 12. Rabus R., Jack D. L., Kelly D. J., Saier M. H., Jr. (1999) TRAP transporters. An ancient family of extracytoplasmic solute-receptor-dependent secondary active transporters. Microbiology 145, 3431–3445 [DOI] [PubMed] [Google Scholar]
- 13. Wyborn N. R., Alderson J., Andrews S. C., Kelly D. J. (2001) Topological analysis of DctQ, the small integral membrane protein of the C4-dicarboxylate TRAP transporter of Rhodobacter capsulatus. FEMS Microbiol. Lett. 194, 13–17 [DOI] [PubMed] [Google Scholar]
- 14. Mulligan C., Kelly D. J., Thomas G. H. (2007) Tripartite ATP-independent periplasmic transporters. Application of a relational database for genome-wide analysis of transporter gene frequency and organization. J. Mol. Microbiol. Biotechnol. 12, 218–226 [DOI] [PubMed] [Google Scholar]
- 15. Severi E., Randle G., Kivlin P., Whitfield K., Young R., Moxon R., Kelly D., Hood D., Thomas G. H. (2005) Sialic acid transport in Haemophilus influenzae is essential for lipopolysaccharide sialylation and serum resistance and is dependent on a novel tripartite ATP-independent periplasmic transporter. Mol. Microbiol. 58, 1173–1185 [DOI] [PubMed] [Google Scholar]
- 16. Allen S., Zaleski A., Johnston J. W., Gibson B. W., Apicella M. A. (2005) Novel sialic acid transporter of Haemophilus influenzae. Infect. Immun. 73, 5291–5300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jenkins G. A., Figueira M., Kumar G. A., Sweetman W. A., Makepeace K., Pelton S. I., Moxon R., Hood D. W. (2010) Sialic acid-mediated transcriptional modulation of a highly conserved sialometabolism gene cluster in Haemophilus influenzae and its effect on virulence. BMC Microbiol. 10, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bouchet V., Hood D. W., Li J., Brisson J. R., Randle G. A., Martin A., Li Z., Goldstein R., Schweda E. K., Pelton S. I., Richards J. C., Moxon E. R. (2003) Host-derived sialic acid is incorporated into Haemophilus influenzae lipopolysaccharide and is a major virulence factor in experimental otitis media. Proc. Natl. Acad. Sci. U.S.A. 100, 8898–8903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Almagro-Moreno S., Boyd E. F. (2009) Sialic acid catabolism confers a competitive advantage to pathogenic Vibrio cholerae in the mouse intestine. Infect. Immun. 77, 3807–3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jermyn W. S., Boyd E. F. (2005) Molecular evolution of Vibrio pathogenicity island-2 (VPI-2). Mosaic structure among Vibrio cholerae and Vibrio mimicus natural isolates. Microbiology 151, 311–322 [DOI] [PubMed] [Google Scholar]
- 21. Jermyn W. S., Boyd E. F. (2002) Characterization of a novel Vibrio pathogenicity island (VPI-2) encoding neuraminidase (nanH) among toxigenic Vibrio cholerae isolates. Microbiology 148, 3681–3693 [DOI] [PubMed] [Google Scholar]
- 22. Rohmer L., Hocquet D., Miller S. I. (2011) Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol. 19, 341–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vimr E. R., Kalivoda K. A., Deszo E. L., Steenbergen S. M. (2004) Diversity of microbial sialic acid metabolism. Microbiol. Mol. Biol. Rev. 68, 132–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Almagro-Moreno S., Boyd E. F. (2009) Insights into the evolution of sialic acid catabolism among bacteria. BMC Evol. Biol. 9, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Urbani A., Warne T. (2005) A colorimetric determination for glycosidic and bile salt-based detergents. Applications in membrane protein research. Anal. Biochem. 336, 117–124 [DOI] [PubMed] [Google Scholar]
- 26. Slotboom D. J., Duurkens R. H., Olieman K., Erkens G. B. (2008) Static light scattering to characterize membrane proteins in detergent solution. Methods 46, 73–82 [DOI] [PubMed] [Google Scholar]
- 27. Ward A., O'Reilly J., Rutherford N. G., Ferguson S. M., Hoyle C. K., Palmer S. L., Clough J. L., Venter H., Xie H., Litherland G. J., Martin G. E., Wood J. M., Roberts P. E., Groves M. A., Liang W. J., Steel A., McKeown B. J., Henderson P. J. (1999) Expression of prokaryotic membrane transport proteins in Escherichia coli. Biochem. Soc. Trans. 27, 893–899 [DOI] [PubMed] [Google Scholar]
- 28. Baron C., Thompson T. E. (1975) Solubilization of bacterial membrane proteins using alkyl glucosides and dioctanoyl phosphatidylcholine. Biochim. Biophys. Acta 382, 276–285 [DOI] [PubMed] [Google Scholar]
- 29. Vimr E. R., Troy F. A. (1985) Identification of an inducible catabolic system for sialic acids (nan) in Escherichia coli. J. Bacteriol. 164, 845–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martinez J., Steenbergen S., Vimr E. (1995) Derived structure of the putative sialic acid transporter from Escherichia coli predicts a novel sugar permease domain. J. Bacteriol. 177, 6005–6010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Severi E., Hosie A. H., Hawkhead J. A., Thomas G. H. (2010) Characterization of a novel sialic acid transporter of the sodium solute symporter (SSS) family and in vivo comparison with known bacterial sialic acid transporters. FEMS Microbiol. Lett. 304, 47–54 [DOI] [PubMed] [Google Scholar]
- 32. Poolman B., Konings W. N. (1993) Secondary solute transport in bacteria. Biochim. Biophys. Acta 1183, 5–39 [DOI] [PubMed] [Google Scholar]
- 33. Kaczorowski G. J., Kaback H. R. (1979) Mechanism of lactose translocation in membrane vesicles from Escherichia coli. 1. Effect of pH on efflux, exchange, and counterflow. Biochemistry 18, 3691–3697 [DOI] [PubMed] [Google Scholar]
- 34. Nielsen A. T., Dolganov N. A., Otto G., Miller M. C., Wu C. Y., Schoolnik G. K. (2006) RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog. 2, e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jeong H. G., Oh M. H., Kim B. S., Lee M. Y., Han H. J., Choi S. H. (2009) The capability of catabolic utilization of N-acetylneuraminic acid, a sialic acid, is essential for Vibrio vulnificus pathogenesis. Infect. Immun. 77, 3209–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden Markov model. Application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.