Abstract
It is believed that 3.2 billion bp of the human genome harbor ∼35,000 protein-coding genes. On average, one could expect one gene per 300,000 nucleotides (nt). Although the distribution of the genes in the human genome is not random,it is rather surprising that a large number of genes overlap in the mammalian genomes. Thousands of overlapping genes were recently identified in the human and mouse genomes. However,the origin and evolution of overlapping genes are still unknown. We identified 1316 pairs of overlapping genes in humans and mice and studied their evolutionary patterns. It appears that these genes do not demonstrate greater than usual conservation. Studies of the gene structure and overlap pattern showed that only a small fraction of analyzed genes preserved exactly the same pattern in both organisms.
Overlapping genes occur frequently in viral and cellular prokaryotic genomes as well as in organelles such as mitochondria (Normark et al. 1983). Until recently, it was believed that they occur much less frequently in eukaryotic nuclear genomes. Although their presence in human and other species' genomes was reported previously (Williams and Fried 1986; Lazar et al. 1989; Miyajima et al. 1989; Burke et al. 1998; Cooper et al. 1998; Bachman et al. 1999; Shintani et al. 1999; Misener and Walker 2000; Morelli et al. 2000; Slavov et al. 2000; Zhuo et al. 2001), until lately very little was known about their frequency and genome-wide distribution. Recent reports show that overlapping genes occur relatively frequently in human and other mammalian genomes (Kiyosawa and Abe 2002; Lehner et al. 2002; Okazaki et al. 2002; Shendure and Church 2002; Yelin et al. 2003). Nevertheless, there is still little known about the origin, evolution, or cross-species conservation of overlapping genes.
Shintani et al. (1999)suggested that the overlap between two genes studied by them, ACAT2 and TCP1, arose during the transition from therapsid reptiles to mammals, and that the overlap could have happened in one of two ways. In one scenario, the rearrangement may have been accompanied by the loss of a part of the 3′-untranslated region (UTR), including the polyadenylation signal, from one gene. By chance, however, the 3′-UTR of the new neighbor on the opposite strand contained all the signals necessary for termination and transcription so that the translocated gene could continue to function. Alternatively, the two genes became neighbors through the rearrangement but at first did not overlap. Later, one of the genes lost its original polyadenylation signal, but was able to use a signal that happened to be present on the noncoding strand of the other gene. Keese and Gibbs (1992)suspect that overlapping genes arise as a result of overprinting—a process of generating new genes from preexisting nucleotide sequences. However, both studies were done based on a single pair of eukaryotic overlapping genes. The hypothesis by Shintani et al. (1999)can only be applied to those overlapping gene pairs in which the overlap occurs at the 3′-end and does not include coding sequences. The hypothesis by Keese and Gibbs needs to be confirmed by larger studies. Interestingly, in both studies the time of origin of the gene overlaps was estimated to take place after the divergence of mammals from birds.
As suggested by Miyata and Yasunaga (1978), the rate of evolution can be expected to be slower in overlapping genes. This is in agreement with a study by Lipman (1997) in which the higher rate of conservation of noncoding sequences of some genes is explained by the presence of antisense transcripts. However, there is not enough experimental evidence that higher conservation is a common feature of coding and noncoding overlapping genes. Shintani et al. (1999)found high 3′-UTR conservation in only one of two studied overlapping genes, and Svaren et al. (1997)found only one area with higher conservation in 3′-UTRs of overlapping Stat6 and Nab2 genes, and, even then, the authors expect this partial conservation to be due to some additional regulatory functions and not necessarily due to the overlap between the genes.
Here we report a study of 774 overlapping genes in human and 542 overlapping gene pairs in mouse as well as analysis of 778 human and mouse orthologous genes that, in at least one species, share exons with another gene.
RESULTS
Identification of Overlapping Genes
We used the NCBI human genome assembly Build 33 (April 2003)and the mouse genome assembly Build 30 (March 2003)as the sequence source for identification of overlapping genes. Out of 34,604 genes annotated in the human genome, we identified 774 pairs of overlapping genes, and of 33,936 analyzed genes in the mouse genome, we identified 578 pairs of overlapping genes. We focused on annotated genomic sequence genes only and did not include ESTs to get high-quality data for mouse-human comparison. As shown by other studies (Wolfsberg and Landsman 1997)as well as our work in the early stages of this study, EST sequences can be identified as overlapping because of chimeric sequences, mislabeling, and genomic sequence contamination. More than 10% of such identified, overlapping genes are artifacts (Yelin et al. 2003). Because chimeric sequences can also be found among annotated mRNAs (Lehner et al. 2002), we used genomic localization to confirm the presence of gene overlaps in our study. Our earlier studies showed that simple presence of regions of complementarities between mRNAs is not sufficient and can lead to false-positive results. Because we wanted to compare human and mouse protein-coding genes, we excluded from our search noncoding genes, which in the genome scale are involved in ∼75% of gene overlaps (Kiyosawa et al. 2003).
As shown in Table 1, among 774 overlapping protein coding genes in the human genome, 542 had overlapping exons. In 299 pairs of genes with overlapping exons, coding sequence was involved, and in 57 cases, coding sequences from both genes are coded by the same genomic fragment. From all human overlapping genes, 53% had tail-to-tail overlap (3′ to 3′), 30.23% showed head-to-head overlap (5′ to 5′), and 16.28% represented embedded genes. In the mouse genome, we found 578 pairs of protein-coding overlapping genes, and 455 of these pairs had overlapping exons. Of these, 232 pairs of genes with overlapping exons had coding sequence involved, and among these 31 pairs showed overlap between coding sequences of both genes. In mouse, 54.32% of genes overlapped at the 3′-ends, 36.51% of overlaps were head-to-head overlaps, and 9.17% of the gene pairs had one gene embedded into another. The fraction of gene pairs overlapping at the 5′-ends is significantly higher than previously reported by Shendure and Lehner, who found that only 5.53% (Shendure and Church 2002)and 15% (Lehner et al. 2002)of overlapping genes had head-to-head orientation. However, results similar to ours were presented by Yelin et al. (2003), who found that 31% of identified human overlapping genes overlap at the 5′-end. We also found 18 cases in the human genome and eight in mouse where one gene had exons overlapping with exons of not one but two different genes. These genes represent previously unreported triplets of overlapping genes. Table 2 lists all cases of such overlapping triplets. An example with three human overlapping genes— MUTYH, TOE1, and TESK2—is presented in Figure 1. The gene TOE1 has overlapping exons with MUTYH at the 5′-end and with TESK2 at the 3′-end. In the human genome we also found a segment with four exon overlapping genes: LOC338549, IDI2, HT009, and IDI1.
Table 1.
Frequency of Different Types of Overlaps Between Protein-Coding Genes in Human and Mouse Genomes
|
Human
|
Mouse
|
|||
|---|---|---|---|---|
| Overlapping genes | Genes with overlapping exons | Overlapping genes | Genes with overlapping exons | |
| Total | 774 | 542 | 578 | 455 |
| Embedded | 126 (16.28%) | 15 (2.77%) | 53 (9.17%) | 7 (1.54%) |
| Tail to tail | 414 (53.49%) | 360 (66.42%) | 314 (54.32%) | 280 (61.54%) |
| Head to head | 234 (30.23%) | 167 (30.81%) | 211 (36.51%) | 168 (36.92%) |
| Involving coding sequence | 299 (55.17%) | 232 (50.99%) | ||
| Coding–coding overlap | 57 (10.52%) | 31 (96.81%) | ||
Table 2.
Human and Mouse Genes Participating in Multiple Overlaps
| Human | ||||
|---|---|---|---|---|
| MUTYH | TOE1 | TESK2 | ||
| LOC348527 | PRKCZ | LOC199990 | ||
| MBDin | STAF65(gamma) | SLC4A1AP | ||
| AUP1 | PRSS25 | LOXL3 | ||
| STAB1 | FLJ12442 | LOC285395 | ||
| PKD2L2 | C5orf5 | LOC202047 | ||
| HOXA3 | LOC285943 | HOXA4 | ||
| LOC338549 | IDI2 | HT009 | ||
| IDI2 | HT009 | IDI1 | ||
| LOC283262 | LOC347872 | MGC5306 | ||
| HSPC134 | LOC338899 | LOC145553 | ||
| FLJ20190 | LOC145694 | KIF23 | ||
| SNN | LOC51061 | FLJ21777 | ||
| LOC339053 | MGC35048 | LOC283827 | ||
| DRG2 | LOC147229 | MYO15A | ||
| AKAP8 | LOC284430 | NAKAP95 | ||
| MCOLN1 | LOC147791 | NTE | ||
| LOC286522 | RPL10 | DNASE1L1 | ||
| Mouse
|
||||
| AW105885 | 2010003O02Rik | LOC230075 | ||
| 5830400N10Rik | LOC329840 | AI563590 | ||
| Pex1 | C030048B08Rik | 1700109H08Rik | ||
| Lass1 | Rent1 | B430202H16Rik | ||
| IIf3 | LOC330902 | Tgut-pending | ||
| Zfp278 | LOC327858 | LOC216505 | ||
| 1110031I02Rik | LOC238023 | MGC28978 | ||
| Vdac2 | 1810030M08Rik | A430057M04Rik | ||
| LOC277250 | 5830462I21Rik | MGC28827 | ||
Genes in the middle column have exons overlapping with exons of
other genes at both 3′- and 5′-ends.
Figure 1.
Overlap between three human genes: MUTH, FLJ13949, and TESK2. Dark boxes represent coding sequence. Light boxes represent untranslated regions.
Identification of Human-Mouse Orthologs
We used NCBI HomoloGene data to identify mouse genes orthologous to overlapping human genes and vice versa. The 542 human gene pairs with overlapping exons could be divided into three categories—54 cases in which neither gene had a mouse homolog, 233 cases in which exactly one member of the gene pair had a mouse homolog, and 255 cases in which both members of the overlapping gene pair had mouse homologs annotated in HomoloGene. Among these 255 pairs with both mouse orthologs, in 95 cases genes were overlapping in both human and mouse, in 150 cases genes were overlapping in human but not in mouse although both mouse genes were mapped to the same contig, and in 10 cases identified homologs were in different contigs. The same search was performed using known mouse exon overlapping genes. For 455 pairs of these genes, in 36 cases neither of the two overlapping genes had an annotated human homolog, in 179 cases only one gene had a human homolog, and in 240 cases both homologs were found. Out of 240 cases in which both human homologs were identified, in 95 instances genes overlapped in both human and mouse (as expected from the above results), in 144 cases both human homologs were in the same contig but did not overlap, and in one case human homologs were on different contigs.
From the entire set of homologous genes, for further analysis, we selected only those where both homologs were found and both were on the same contig. The presence of the overlap in both organisms was not required. This gave us 389 pairs (778 genes)of human and mouse orthologous overlapping genes in which in at least one species, overlapping genes shared not only genomic location but also exonic sequence.
Human-Mouse Sequence Conservation
It was observed that a large fraction of the mammalian genes show high conservation of 5′- and 3′-untranslated regions. Lipman (1997)suggested that a subset of these cases can be explained by the existence of overlapping antisense transcripts. We analyzed the set of human and mouse orthologous genes overlapping in at least one of the organisms. We excluded any pair for which only one homolog was identified as well as all cases in which identified homologs were in different locations to eliminate possible cases of paralogous and not orthologous sequence assignment. After aligning all putative orthologous sequences, we additionally eliminated from our set all genes that had a similarity level between human and mouse coding sequences below 69%. Such low similarity between sequences may indicate that we may not have a true ortholog, and therefore we chose not to include them. The cutoff threshold of 69% was chosen based on the distribution of human-mouse ortholog similarities (W. Makalowski, unpubl.). We found 44 such cases, and these orthologs as well as their overlapping counterparts were eliminated from the set. We excluded all orthologous pairs when more than one homolog was annotated in HomoloGene. This gave us a set of 638 human-mouse orthologs overlapping with another gene in at least one organism.
We compared untranslated regions and coding sequences of the human and mouse orthologs. At first we analyzed the entire set of 638 orthologs, regardless of category of the overlap. We compared separately all 5′-UTRs, coding sequences, and 3′-UTRs. From our comparison we discarded all alignments that were shorter than 30 bp. The median value of the CDS identity between human and mouse genes was 85.67%, for 3′-UTRs 63.89%, and for 5′-UTRs 61.54%. These values do not differ significantly when compared with the reported results for all known humanmouse orthologs (Makalowski and Boguski 1998)because all mean values lie within one standard deviation (Table 3). Although overlapping genes in our data set overall did not demonstrate greater than average conservation, we expected a higher level of similarity when coding regions were involved. We, therefore, partitioned our sequences based on the type of overlap and calculated identity statistics for each set. Our main focus was on the comparison of coding sequence when coding sequence from both genes was involved in the overlap, comparison of 5′-UTRs when 5′-UTRs were overlapping with the coding sequence of another gene, and 3′-UTRs when they were overlapping with the coding region of another gene. We analyzed these values when the above overlap was observed in at least one organism as well as when the particular type of overlap was observed in both organisms. In neither case did we observe significantly higher conservation of coding sequence or untranslated regions. Table 3 presents the results of our analysis. One can see that mean and median values do not differ significantly either between categories or when compared with overall human-mouse sequence identities. In all cases, the mean and median values obtained in our study lie within the range of one standard deviation reported for all human-mouse orthologs by Makalowski and Boguski (1998).
Table 3.
Summary of Sequence Similarities Between Untranslated Regions and Coding Sequences of Human and Mouse Orthologous Overlapping Genes
|
5′-UTR
|
CDS
|
3′-UTR
|
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Overlap | no. of seq. | median | mean | std | no. of seq. | median | mean | std | no. of seq. | median | mean | std | ||||||||||||
| Sequences that have overlaps in at least one of the species | ||||||||||||||||||||||||
| Coding—coding | 15 | 66.67 | 67.11 | 12.46 | 34 | 84.48 | 84.9 | 4.54 | 30 | 61.03 | 60.7 | 15.98 | ||||||||||||
| 3UTR_Coding | 102 | 60.96 | 61.75 | 15.09 | 185 | 85.36 | 84.66 | 4.95 | 165 | 63.23 | 63.35 | 13.52 | ||||||||||||
| 5UTR_Coding | 38 | 60.72 | 61.92 | 15.42 | 67 | 84.34 | 84 | 4.63 | 60 | 59.49 | 62.57 | 13.04 | ||||||||||||
| 5UTR_5UTR | 61 | 63.53 | 63.06 | 13.74 | 91 | 84.75 | 84.46 | 5.11 | 81 | 61.27 | 62.74 | 12.11 | ||||||||||||
| 3UTR_3UTR | 281 | 61.01 | 61.67 | 14.45 | 483 | 85.95 | 85.12 | 5.17 | 440 | 64.83 | 64.68 | 13.07 | ||||||||||||
| Sequences that have overlaps in both of the species | ||||||||||||||||||||||||
| Coding—coding | 0 | 0 | 0 | N/A | 0 | 0 | 0 | N/A | 0 | 0 | 0 | N/A | ||||||||||||
| 3UTR_Coding | 20 | 55.76 | 58.08 | 15.92 | 34 | 84.9 | 84.2 | 4.51 | 33 | 63.41 | 62.5 | 12.11 | ||||||||||||
| 5UTR_Coding | 1 | 72.7 | 72.7 | N/A | 2 | 88.97 | 88.97 | 2.89 | 2 | 69.48 | 69.48 | 5.79 | ||||||||||||
| 5UTR_5UTR | 3 | 72.52 | 69.53 | 5.33 | 4 | 86.71 | 85.3 | 6.04 | 4 | 69.48 | 69.94 | 10.97 | ||||||||||||
| 3UTR_3UTR | 74 | 60.7 | 61.26 | 15.41 | 124 | 85.34 | 85.02 | 4.8 | 120 | 69.06 | 68.27 | 11.47 | ||||||||||||
| Large scale human—mouse comparison (Makalowski and Boguski 1998) | ||||||||||||||||||||||||
| 871 | 67 | 69.7 | 12.9 | 1,138 | 89 | 86.4 | 12.3 | 938 | 69.4 | 71 | 12.2 | |||||||||||||
In bold are values that we expected to be significantly higher because of the overlap with another gene coding sequence and therefore be under higher selection against mutations.
Conservation of Gene Structure and Overlap Pattern
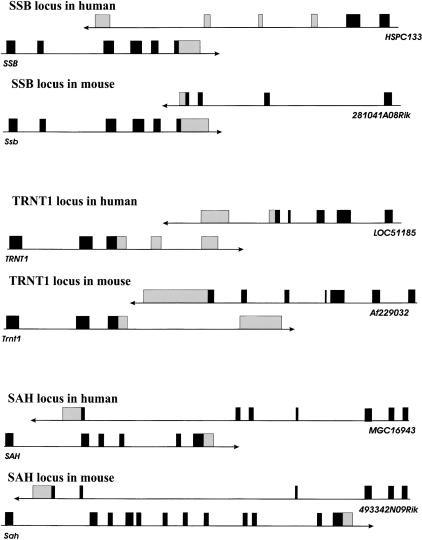
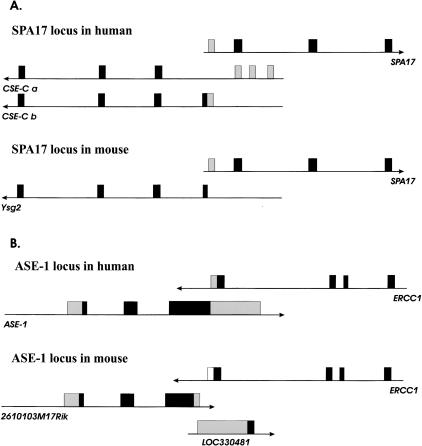
Besides sequence similarity, we also studied the conservation of the gene structure and overlap pattern. As mentioned above, only in 95 out of 255 cases were mouse homologs of overlapping human genes involved in overlaps in the mouse genome. A significant fraction of these 95 pairs show different overlap patterns in the two genomes. This is in contrast to the data presented by Shendure and Church (2002), where in all cases of human and mouse orthologous overlapping genes, the pattern of overlap was the same in both organisms. In many cases, lack of similarity could be explained by unfinished sequencing of one or both UTRs. For example, in the human gene SRR, the 3′-UTR sequence overlaps with the 3′-UTR and coding region of FLJ10534. In mouse, the 3′-UTR of Srr does not overlap with a coding region of the gene LOC193029, but only overlaps with the 3′-UTR. However, the 3′-UTR of LOC193029 overlaps with both a coding region and the 3′-UTR of Srr. Although there is a reversed pattern of overlaps, it is apparent that the 3′-UTRs of human FLJ10534 and mouse Srr are very short, and, if extended, they would overlap with coding sequences of the gene sharing the same locus. Based on this observation, we can expect that the 3′-UTRs of both genes should overlap with both the 3′-UTR and coding sequence of another gene if they were fully sequenced. Figure 2 shows a few examples of genes in which the overlap pattern is different in human and mouse and cannot be explained by the missing ends of 3′- or 5′-UTRs. One explanation of the above discrepancies in genome structure and overlap pattern could be that we are observing different splice variants of one or both genes in each organism. For example, Figure 3A shows that the 5′-UTR of SPA17 overlaps with the 5′-UTR of one CSE-C splice variant (variant b), and that the second exon of SPA17, containing coding sequence, overlaps with the third noncoding exon of CSE-C variant a. Therefore, depending on which splice variant we look at, we see a different overlap pattern. In mouse, only one splice variant, with a missing 5′-UTR, is annotated on the map, and there is no overlap between the orthologous genes. Problems with different overlap patterns could also be caused by incorrect annotation. An example of such a case is the ASE-1 gene (Fig. 3B). In human, the 3′-UTR of ASE-1 overlaps with the 3′-UTR and coding sequence of ERCC1. In mouse, 2610103M17Rik, the ortholog of ASE-1, does not overlap with Ercc1. However, there is another gene placed on the mouse genome map between Ercc1 and 2610103M17Rik, LOC330481. This gene overlaps at the 3′-end with Ercc1 and at the 5′-end with 2610103M17Rik. It seems likely that LOC330481 is a part of the 2610103M17Rik 3′-UTR and not a separate gene, and therefore both are parts of the mouse ASE-1 ortholog. If this is true, the pattern of overlaps in human and mouse does not differ in this case.
Figure 2.
Examples of genes with different patterns of overlap in human and mouse. Dark boxes represent coding sequence. Light boxes represent untranslated regions.
Figure 3.
Examples of different patterns of overlap in human and mouse caused by different splice variants or annotation error. Dark boxes represent coding sequence. Light boxes represent untranslated regions.
DISCUSSION
We identified >774 gene pairs sharing a locus in the human genome and 542 in the mouse genome. It was expected that overlapping genes, especially those in which coding sequences are involved, would be more conserved between species than nonoverlapping genes (Lipman 1997; Yelin et al. 2003). This is mostly because a mutation in the overlapping region would cause changes in both genes, and the selection against these mutations should therefore be stronger. This hypothesis does not hold in our data. The identity between overlapping human and mouse orthologs was not higher than average. Even when we limited the comparison to genes in which a particular overlap was observed in both species, we did not notice any significant difference in conservation of coding sequences or untranslated regions. We also noticed that the pattern of overlap can be different across species. The lack of higher conservation rates and differing overlap patterns raises interesting questions about the evolution of overlapping genes. The hypothesis of overlapping genes origin suggested by Shintani et al. (1999)cannot fully explain these phenomena because it is related to 3′-end overlaps only and does not consider 5′-overlaps at all. Overprinting as the origin of overlapping genes, as suggested by Keese and Gibbs (1992), may provide a better explanation, but more detailed studies would be required. However, because overprinting generates novel genes, the hypothesis may help in deciphering some observations. Keese and Gibbs suggest that overprinting took place after the divergence of mammals from birds, and that overlapping genes represent young, phylogenetically restricted genes encoding proteins with diverse functions, and are therefore specialized to the present life-style of the organism in which they are found. This could explain why we could not find orthologs of human overlapping genes in Caenorhabditis elegans. The lineage-specific nature of the overlapping genes, as suggested by Shendure and Church, may explain only partial intersection between the human and mouse gene sets. We observed the lack of many mouse orthologs of human overlapping genes and many cases in which only one ortholog was found. Although we suspect that sometimes these orthologs have simply not been identified yet, this may not explain cases in which one ortholog was found but none of the neighboring, closely localized genes matched the other gene. Of course, at this point we cannot exclude the possibility that in cases like the above we have paralogs and not orthologs. Additionally, the overlap is not always repeated in both human and mouse, and the structure of genes involved in the overlap sometimes differs as well. This could mean that overlaps can be generated by multiple mechanisms, including overprinting, as suggested by Keese and Gibbs, or rearrangements or loss of parts of 3′-UTRs and use of neighboring gene signals, as suggested by Shintani et al. In our data set, we can find cases supporting each of these mechanisms, as well as implying that overlaps between some genes could be specific to mammals, and others could have happened after separation of mouse and human. This observation is made based on the fact that, in some cases, the lack of an overlap in one of the species cannot be explained by unfinished sequencing, alternative splice variants, or wrong annotations, as well as the fact that homologs of overlapping genes were sometimes found on different contigs or even different chromosomes. We excluded all such cases from our human-mouse comparison because of the possibility of paralogy. In many instances, those were the only homologous sequences identified. If these are orthologous genes, in addition to rearrangement, some other mechanism would be required to explain divergence, such as duplication of the overlapping gene region followed by loss of one gene from each duplicate. However, further and more detailed studies including more lineages are required to confirm any of the above hypotheses.
METHODS
Sequence Data
We analyzed assembled and annotated human and mouse genomic sequences from GenBank to identify overlapping genes. We used NCBI's Build 33 release of the human genome (April 2003)and Build 30 of the mouse genome (March 2003).
Identification of the Overlapping Genes
The GenBank files contain annotations for both gene and mRNA features. However, the location of the gene tag is not very well defined and is not consistent. The gene feature location may or may not include the promoter region and regulatory elements. Therefore, in the interest of consistency, we defined the gene locations in terms of the mRNA location, which is represented by the set of intervals on the genomic region, referring to the particular mRNA exons.
Identification of Human/Mouse Orthologs and Their Authentication
Mouse orthologs of human overlapping genes were identified by searching the HomoloGene database at NCBI. We searched the HomoloGene database for mouse orthologs of every human overlapping gene we identified, and vice versa. All cases in which no ortholog was identified for either of the two overlapping genes were excluded from further search. We also excluded all cases in which genes were not located on the same contig to avoid the analysis of paralogous and not orthologous genes as well as genes in which more than one homolog was annotated in the HomoloGene database. Furthermore, all orthologous pairs that had coding sequence identity below 69% were eliminated to avoid the possibility of analysis of paralogs. The 69% cutoff level was chosen based on the distribution of coding sequences identities of human and mouse orthologs (W. Makalowski, unpubl.). We also excluded overlapping counterparts of genes with low identity to be consistent with the requirement of including only those genes for which both orthologs, for a given overlapping gene pair, were identified.
Human-Mouse Comparison
We downloaded mRNA sequences of all orthologous human and mouse genes. We used GenBank annotations to separate coding sequence and untranslated regions for similarity search. All sequences were aligned using map software (Huang 1994) with the following parameters: a mismatch penalty, -3; match, 10; gap opening penalty, 50; gap extension penalty, 5; longest penalized gap, 10. Then, identities (excluding gaps)between coding sequences, 5′-UTRs, and 3′-UTRs were calculated. We discarded from our comparison all alignments shorter than 30 bp.
Overlapping Genes Database
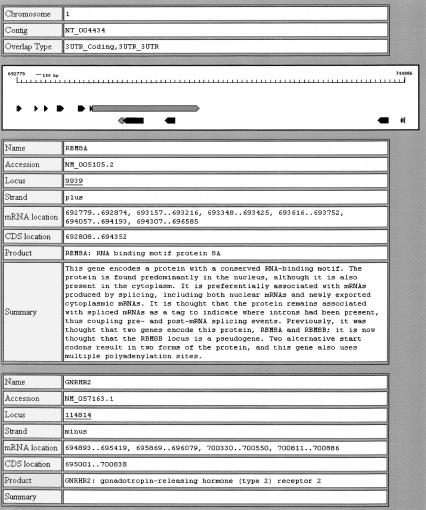
All overlap information that could be inferred from the annotated GenBank files for Build 33 of the human genome sequence and Build 30 of the mouse genome sequence is stored in a MySQL database. A Web-based graphical user interface to the data is available at http://posnania.cbio.psu.edu/research/overlapping_genes.html. The interface allows users to search for known overlapping genes by name, accession number, or LocusLink id. A second form allows users to browse the set of overlaps by overlap type (Coding-Coding, 3′-UTR-Coding, etc). Overlaps are displayed in a pictorial format with colors being used to distinguish untranslated regions from coding regions. Information retrieved dynamically from LocusLink is used to summarize the genes participating in the overlaps (see example in Fig. 4).
Figure 4.
The screenshot from the overlapping genes database presenting overlap details.
Acknowledgments
We thank Alex Richter for critical reading of the manuscript and Jaroslaw Bryko for his help in preliminary analysis.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1590904.
Footnotes
[Supplemental material is available online at www.genome.org and http://posnania.cbio.psu.edu/research/overlapping_genes.html.]
References
- Bachman, N.J., Wu, W., Schmidt, T.R., Grossman, L.I., and Lomax, M.I. 1999. The 5′ region of the COX4 gene contains a novel overlapping gene, NOC4. Mamm. Genome 10: 506-512. [DOI] [PubMed] [Google Scholar]
- Burke, J., Wang, H., Hide, W., and Davison, D.B. 1998. Alternative gene form discovery and candidate gene selection from gene indexing projects. Genome Res. 8: 276-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, P.R., Smilinich, N.J., Day, C.D., Nowak, N.J., Reid, L.H., Pearsall, R.S., Reece, M., Prawitt, D., Landers, J., Housman, D.E., et al. 1998. Divergently transcribed overlapping genes expressed in liver and kidney and located in the 11p15.5 imprinted domain. Genomics 49: 38-51. [DOI] [PubMed] [Google Scholar]
- Huang, X. 1994. On global sequence alignment. Comput. Appl. Biosci. 10: 227-235. [DOI] [PubMed] [Google Scholar]
- Keese, P.K. and Gibbs, A. 1992. Origins of genes: “Big bang” or continuous creation? Proc. Natl. Acad. Sci. 89: 9489-9493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosawa, H. and Abe, K. 2002. Speculations on the role of natural antisense transcripts in mammalian X chromosome evolution. Cytogenet. Genome Res. 99: 151-156. [DOI] [PubMed] [Google Scholar]
- Kiyosawa, H., Yamanaka, I., Osato, N., Kondo, S., and Hayashizaki, Y. 2003. Antisense transcripts with FANTOM2 clone set and their implications for gene regulation. Genome Res. 13: 1324-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar, M.A., Hodin, R.A., Darling, D.S., and Chin, W.W. 1989. A novel member of the thyroid/steroid hormone receptor family is encoded by the opposite strand of the rat c-erbA α transcriptional unit. Mol. Cell. Biol. 9: 1128-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner, B., Williams, G., Campbell, R.D., and Sanderson, C.M. 2002. Antisense transcripts in the human genome. Trends Genet. 18: 63-65. [DOI] [PubMed] [Google Scholar]
- Lipman, D.J. 1997. Making (anti)sense of non-coding sequence conservation. Nucleic Acids Res. 25: 3580-3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makalowski, W. and Boguski, M.S. 1998. Evolutionary parameters of the transcribed mammalian genome: An analysis of 2,820 orthologous rodent and human sequences. Proc. Natl. Acad. Sci. 95: 9407-9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misener, S.R. and Walker, V.K. 2000. Extraordinarily high density of unrelated genes showing overlapping and intraintronic transcription units. Biochim. Biophys. Acta 1492: 269-270. [DOI] [PubMed] [Google Scholar]
- Miyajima, N., Horiuchi, R., Shibuya, Y., Fukushige, S., Matsubara, K., Toyoshima, K., and Yamamoto, T. 1989. Two erbA homologs encoding proteins with different T3 binding capacities are transcribed from opposite DNA strands of the same genetic locus. Cell 57: 31-39. [DOI] [PubMed] [Google Scholar]
- Miyata, T. and Yasunaga, T. 1978. Evolution of overlapping genes. Nature 272: 532-535. [DOI] [PubMed] [Google Scholar]
- Morelli, C., Magnanini, C., Mungall, A.J., Negrini, M., and Barbanti-Brodano, G. 2000. Cloning and characterization of two overlapping genes in a subregion at 6q21 involved in replicative senescence and schizophrenia. Gene 252: 217-225. [DOI] [PubMed] [Google Scholar]
- Normark, S., Bergstrom, S., Edlund, T., Grundstrom, T., Jaurin, B., Lindberg, F.P., and Olsson, O. 1983. Overlapping genes. Annu. Rev. Genet. 17: 499-525. [DOI] [PubMed] [Google Scholar]
- Okazaki, Y., Furuno, M., Kasukawa, T., Adachi, J., Bono, H., Kondo, S., Nikaido, I., Osato, N., Saito, R., Suzuki, H., et al. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420: 563-573. [DOI] [PubMed] [Google Scholar]
- Shendure, J. and Church, G.M. 2002. Computational discovery of sense-antisense transcription in the human and mouse genomes. Genome Biol. 3: RESEARCH0044. [DOI] [PMC free article] [PubMed]
- Shintani, S., O'hUigin, C., Toyosawa, S., Michalova, V., and Klein, J. 1999. Origin of gene overlap: The case of TCP1 and ACAT2. Genetics 152: 743-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavov, D., Hattori, M., Sakaki, Y., Rosenthal, A., Shimizu, N., Minoshima, S., Kudoh, J., Yaspo, M.L., Ramser, J., Reinhardt, R., et al. 2000. Criteria for gene identification and features of genome organization: Analysis of 6.5 Mb of DNA sequence from human chromosome 21. Gene 247: 215-232. [DOI] [PubMed] [Google Scholar]
- Svaren, J., Apel, E.D., Simburger, K.S., Jenkins, N.A., Gilbert, D.J., Copeland, N.A., and Milbrandt, J. 1997. The Nab2 and Stat6 genes share a common transcription termination region. Genomics 41: 33-39. [DOI] [PubMed] [Google Scholar]
- Williams, T. and Fried, M. 1986. A mouse locus at which transcription from both DNA strands produces mRNAs complementary at their 3′ ends. Nature 322: 275-279. [DOI] [PubMed] [Google Scholar]
- Wolfsberg, T.G. and Landsman, D. 1997. A comparison of expressed sequence tags (ESTs)to human genomic sequences. Nucleic Acids 25: 1626-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelin, R., Dahary, D., Sorek, R., Levanon, E.Y., Goldstein, O., Shoshan, A., Diber, A., Biton, S., Tamir, Y., Khosravi, R., et al. 2003. Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol. 21: 379-386. [DOI] [PubMed] [Google Scholar]
- Zhuo, D., Zhao, W.D., Wright, F.A., Yang, H.Y., Wang, J.P., Sears, R., Baer, T., Kwon, D.H., Gordon, D., Gibbs, S., et al. 2001. Assembly, annotation, and integration of UNIGENE clusters into the human genome draft. Genome Res. 11: 904-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://posnania.cbio.psu.edu/research/overlapping_genes.html; Overlapping Genes Project.