Abstract
Genomic copy number alterations are a feature of many human diseases including cancer. We have evaluated the effectiveness of an oligonucleotide array, originally designed to detect single-nucleotide polymorphisms, to assess DNA copy number. We first showed that fluorescent signal from the oligonucleotide array varies in proportion to both decreases and increases in copy number. Subsequently we applied the system to a series of 20 cancer cell lines. All of the putative homozygous deletions (10) and high-level amplifications (12; putative copy number >4) tested were confirmed by PCR (either qPCR or normal PCR) analysis. Low-level copy number changes for two of the lines under analysis were compared with BAC array CGH; 77% (n = 44) of the autosomal chromosomes used in the comparison showed consistent patterns of LOH (loss of heterozygosity) and low-level amplification. Of the remaining 10 comparisons that were discordant, eight were caused by low SNP densities and failed in both lines. The studies demonstrate that combining the genotype and copy number analyses gives greater insight into the underlying genetic alterations in cancer cells with identification of complex events including loss and reduplication of loci.
Comparative genomic hybridization (CGH; Kallioniemi et al. 1992) has been used extensively to document gains and losses of genomic DNA in diseases such as cancer (Albertson et al. 2000; Jain et al. 2001) and mental retardation (Ghaffari et al. 1998; Veltman et al. 2002). The recent development of CGH using arrays of either genomic (Pinkel et al. 1998) or cDNA clones (Pollack et al. 1999) has improved the resolution of these analyses, allowing better detection and mapping of localized changes such as gene amplification or homozygous deletions.
CGH by these methods only catalogs the number of copies of a DNA sequence. It cannot, for example, distinguish one copy of each parental chromosome from two copies of one parental chromosome, both of which will generate a signal equivalent to two copies. However, in cancer and other human diseases, the provenance of the chromosome or genomic region undergoing copy number alteration is often important, for example, in uniparental disomy disorders (Nicholls et al. 1989). Therefore, a platform that provides information pertaining to both copy number and the status of each parental allele would be beneficial.
Kennedy et al. (2003) have devised a generic sample preparation method that uses a small number of oligonucleotide primers, coupled to allele discrimination on synthetic DNA microarrays. The method (whole-genome sampling assay, or WGSA) uses a simple restriction enzyme digestion, followed by linker-ligation of a common adaptor sequence to every fragment, allowing multiple loci to be amplified using a single primer complementary to this adaptor. PCR then converts the genomic DNA into a predictable sample of reduced complexity that is hybridized to the arrays. Completion of the human genome sequence has made it possible to conduct in silico digests of total genomic DNA and predict which fragments will amplify using this methodology. SNPs that reside on these fragments are then identified, and oligonucleotides corresponding to these SNPs are synthesized onto high-density microarrays. Matching the SNP content on the chip to that produced in the target allows one to maximize the information gained from each array.
In this study, we have explored the effectiveness of WGSA and high-density oligonucleotide arrays, originally designed to detect single-nucleotide polymorphisms, in generating both genotype and copy number data in the same experiment.
RESULTS
Validation of SNP Genotyping Data
The Affymetrix p501 array was designed as a prototype array for the WGSA and contained oligonucleotides representing 8473 SNPs predicted to be present on XbaI fragments that were 400–800 bp in length. Further experimentation identified a set of 6587 SNPs that met the following selection criteria: displayed three genotype clusters when data for 133 ethnically diverse individuals were analyzed, demonstrated appropriate Mendelian inheritance across 33 families, displayed high (>99.9%) reproducibility in 12 replicates, displayed call rates of >90% across more than 300 experiments, and had genotype distributions that were in Hardy-Weinberg equilibrium and mapped to unique positions within the genome. This set of 6587 SNPs had a 99.5% concordancy rate with genotype calls generated by single-base extension methodology, average heterozygosity of 35.2% (±11%) in 133 ethnically diverse individuals with a median spacing of 260 kb through euchromatic regions of the genome (Kennedy et al. 2003; H. Matsuzaki, pers. comm.).
The call rate using the WGSA p501 array was estimated at 82% (SD 6.9%) across 86 experiments (data not shown). The reproducibility was tested by pairwise analysis across 18 aliquots of the same DNA (NCI-BL2126), giving an average concordance of 99.65%. We compared the genotyping data from the p501 array to data from the ABI LMS-MD10 microsatellite marker set in a subset of six(COLO829, HCC38, NCI-H209, NCI-H2171, NCI-H2126, and NCI-H1395) of the 20 lines under investigation. For both platforms, loss of heterozygosity was identified by comparing the genotype data for the tumor line to a lymphoblastoid cell line from the same individual. Of the 400 microsatellite markers in the LMS-MD 10 set, we were able to map 369 onto the NCBI-33 build of the human genome sequence, and these were positioned in relation to the SNP data from the array. A total of 1558 (70.4%) of the markers from the LMS-MD 10 set gave informative results from the sixcell lines, of which 1477 (94.8%) reported results consistent with one or both of the flanking SNPs. Of the 81 microsatellite genotype calls that did not agree with either flanking SNP, 34 were consistent with one of the flanking microsatellite genotyping calls and could therefore represent small-scale mapping errors in the human genome sequence. Therefore, of the 1558 informative genotypes using the micro-satellite marker set, only 47 (3.0 %) were clearly different from the data from the p501 SNP array. These differences could result from retention of small intervals spanning the microsatellite marker but not extending to the flanking SNPs, errors in the microsatellite genotyping data, or potentially errors in the SNP genotyping data.
Validation of Copy Number Analysis
Application of the WGSA array to determine genomic copy number will only be possible if the fluorescent intensity from each feature shows a dosage response to variations in copy number. This was tested in two ways, by applying samples with varying numbers of the X-chromosome and by spiking a series of identical DNA aliquots with varying concentrations of PCR product to increase the copy number of 42 SNPs from twofold (control sample) to 1000-fold.
In the X-chromosome copy number experiment, we used the average value from males to represent the case of 1X and the average value of females to represent 2X; we also collected data from 3X-, 4X-, and 5X-containing cell lines. Using (I) to indicate chip intensity, the dosage-response assumption can be written Ia ≅ Cab × Ib, where Ia is the intensity for a region with copy number a, Ib is the intensity on the same region with copy number b, and Cab is a constant determined by a and b. S̃ (see Methods) can be viewed as an approximation of log intensity (all references to the log function refer to the natural log e unless otherwise stated). Therefore, if the assumption is true, a log transformation leads to S̃a ≅ S̃b + C̃ab. The results using average female values as a baseline (173 SNPs on the X-chromosome) give estimated intensity ratios of 0.584, 1, 1.484, 1.822, and 2.243, equating to copy numbers of 1.17, 2, 2.97, 3.64, and 4.49, with a correlation of 0.981, 1, 0.943, 0.935, and 0.939 for onefold, twofold, threefold, fourfold, and fivefold actual copy numbers, respectively (baselined on female values with a copy number of two; therefore, the estimated copy number equals 2 × the estimated intensity ratio). When plotting estimated copy number against actual copy number, a straight-line response is seen (R2 = 0.9976). The slope of the relationship, 0.83, differs from the ideal value of 1 but is similar to data from spotted array CGH experiments (Pinkel et al. 1998; Pollack et al. 1999).
The response of the copy number estimation at higher copy number values was simulated by spiking aliquots of the same DNA (NCI-BL2126) with PCR products for 42 SNPs selected because of their high call rates. The copy number was increased from 2 (control) by 2, 5, 10, 25, 50, 75, 100, 250, 500, 750, and 1000 copies. Of the 42 SNPs tested, 40 gave an increase in fluorescence in response to increased copy number up to and including the 1000-fold spike with a correlation between log intensity and log copy number of 0.92 (Fig. 1). Two of the SNPs showed no increase in fluorescence in response to increasing copy number; this subset therefore indicates that only a small fraction of SNPs on the array would fail to report copy number change.
Figure 1.
Plot of log(copy number) against log(intensity) for the spiking experiment in which 18 aliquots of the same DNA were spiked with varying concentrations of 42 SNPs from twofold up to 1000-fold. The black dots indicate the results for the individual SNPs across the 12 spiking concentrations (spiked with an extra 0, 2, 5, 10, 25, 50, 75, 100, 250, 500, 750, and 1000 copies); the green dots and line show the mean for all 42 SNPs. Two SNPs did not report increased fluorescence with increased copy number, and these are highlighted in red.
Copy Number Changes in Cancer Cell Lines
Having confirmed the utility of the oligonucleotide array for detecting increases in copy number, we analyzed a set of 20 cancer cell lines. The analysis of the fluorescence data from the tumor samples identified a total of 14 putative high-level amplifications in which a minimum of three consecutive SNPs reported ratios >2.5 (equivalent to a copy number of 5, also visible in the unprocessed fluorescence data). Of these, 12 loci were tested, and each was shown to have a copy number in excess of 5 by qPCR (Table 1). An example of genomic amplification of the c-MYC locus in COR-L96-CAR can be seen in Figure 2A; the profile of this amplification was also obtained by qPCR (SYBR Green) using amplicons designed to SNPs from the array; the comparison with the data from the p501 array is shown in Figure 3. A total of 10 putative homozygous deletions were also identified (again reported by three consecutive SNPs and visible in the unprocessed fluorescence data), all of which were assessed and confirmed by conventional PCR (Table 1). Figure 2B shows an example of a homozygous deletion of the p16/INK4 locus in LB1047-RCC.
Table 1.
Cell Lines Containing Larger-Scale Genomic Alterations, Homozygous Deletions, and Genomic Amplification, Together With Their Chromosomal Locations, Flanking SNPs, and Size of Region
|
Copy number
|
|||||||
|---|---|---|---|---|---|---|---|
| Cell line | Genomic alteration | Chromosomal position | p501 | PCR | Flanking SNPs | Size (Mb) | Status |
| NCI-H1395 | Amplification | 1q21.3 | 14 | 11.6 | TSC0602316-TSC0902438 | 4.7 | Known |
| HCC38 | Homozygous deletion | 3p12.2 | 0 | 0 | TSC0041186-TSC0261189 | 2.7 | Known |
| COR-L96-CAR | Amplification | 5p13.1 | 19 | — | TSC0260201-TSC0066115 | 2.5 | Known |
| NCI-H209 | Homozygous deletion | 5q14.3 | 0 | 0 | TSC0052315-TSC0061600 | 1.0 | Novel |
| HCC1395 | Homozygous deletion | 6q16.3 | 0 | 0 | TSC0553269-TSC0152381 | 2.1 | Novel |
| HCC1395 | Homozygous deletion | 6q16.3 | 0 | 0 | TSC0833631-TSC0050825 | 3.7 | Novel |
| NCI-H2171 | Amplification | 8q12.2 | 11 | 85.8 | TSC0272325-TSC0681497 | 3.2 | Known |
| HCC1395 | Homozygous deletion | 8q11.21 | 0 | 0 | TSC0048903-TSC0065447 | 1.6 | Novel |
| Cor-L96-CAR | Amplification | 8q24.21 | 27 | 74 | TSC0719292-TSC0741747 | 1.9 | Known |
| NCI-H2171 | Amplification | 8q24.21 | 15 | 31 | TSC0719292-TSC0741747 | 1.9 | Known |
| BB132-MEL | Homozygous deletion | 9p23 | 0 | 0 | TSC0823256-TSC0048714 | 2.2 | Known |
| HCC38 | Homozygous deletion | 9p21.3 | 0 | 0 | TSC0827951-TSC0544304 | 10.2 | Known |
| LB1047-RCC | Homozygous deletion | 9p21.3 | 0 | 0 | TSC0055892-TSC0049516 | 2.3 | Known |
| NCI-H2126 | Homozygous deletion | 9p21.3 | 0 | 0 | TSC0056694-TSC0602274 | 2.2 | Known |
| HCC1395 | Homozygous deletion | 11p13 | 0 | 0 | TSC0741958-TSC0345031 | 0.6 | Known |
| NCI-H2171 | Amplification | 11p13 | 5 | 8.6 | TSC0055572-TSC0059555 | 1.0 | Known |
| NCI-H2171 | Amplification | 11q14.1 | 7 | 27.6 | TSC0050602-TSC1007318 | 1.2 | Known |
| 1542T-P41A | Amplification | 11q22.3 | 9 | 20 | TSC0050600-TSC0308740 | 16.1 | Known |
| 1156-Q-E | Amplification | 12p | 9 | — | TSC0046300-TSC0585919 | 36.6 | Known |
| NCI-H2171 | Amplification | 12p11.23 | 9 | 7.6 | TSC0081620-TSC0055751 | 5.5 | Known |
| 833-KE | Amplification | 12p13.31 | 6 | 6.6 | TSC0052512-TSC0083456 | 25.5 | Known |
| NCI-H2171 | Amplification | 12p13.31 | 10 | 9.2 | TSC0556975-TSC0056780 | 2.7 | Known |
| NCI-H2171 | Amplification | 14q11.2 | 7 | 22.6 | TSC1031933-TSC0549368 | 1.6 | Novel |
| I82 | Amplification | 20q13.13 | 7 | 7 | TSC0615769-TSC0543744 | 7 | Known |
The copy number estimation from both the p501 array and confirmation data is included (— indicates no data); homozygous deletions were confirmed by PCR of SNPs from the affected region, whereas amplifications were confirmed by qPCR using TaqMan dual-labelled probes. The status of the amplifications and homozygous deletions is (Novel) not previously reported in the literature or (Known) previously identified.
Figure 2.
Examples of amplification and deletions together with genotyping data generated by the p501 array compared with BAC CGH data. (Top panel) The fluorescence ratio plot from the p501 array showing the smoothed fluorescent intensity data of the sample divided by the figure from the reference sample. (Second panel) The p-value plot for the individual SNPs calculated by comparison to the mean and standard deviation of 29 normal DNAs with deletions represented by red lines and amplifications in green. (Third and fourth panels) The genotypes for the tumor and matched normal samples, respectively; homozygous SNPs are represented by a red line below the center point whereas heterozygous markers are represented by a green line above the center point. (Bottom panel in C and D only) The results for BAC-array-based CGH (where available). (A) Genomic amplification at the C-MYC locus (arrow) on Chromosome 8 in the prostate line COR-L96-CAR. (B) A homozygous deletion (arrow) at the p16/INK4 locus on Chromosome 9 in the renal cell carcinoma cell line LB1047. (C) Chromosome 18 from the breast cancer cell line HCC1937. (i) This chromosome has a copy number of 2 (intensity ratio of 1) from the pter until 18q12.1. (ii) The copy number drops to 1 (intensity ratio of 0.5) from 18q12.1 until the 18qter, although the genotyping data indicate that this line is heterozygous across the full length of Chromosome 18. (D) Chromosome 5 from the small cell lung cancer line NCI-H209. This chromosome shows a complex pattern with partial amplification of the p-arm (i), followed by a drop in fluorescent intensity to 0.5 with corresponding LOH determined from the genotyping data (ii), until 5q23.2 (iii), where the intensity ratio recovers to 1 (copy number of 2); however, this region still represents LOH as determined by the genotyping data and therefore represents duplication of a single parental chromosome. There is also a homozygous deletion at 5q14.3 (arrow). This was not detected with the BAC array, as the array does not have a clone that covers this region.
Figure 3.
Comparison of copy number estimation for the amplification of the c-MYC proto-oncogene in COR-L96-CAR using the p501 array (A; where copy number equals two times the intensity ratio) and qPCR (B).
The p501 array also detected regions with more subtle copy number changes, namely, amplification events to 3 copies or a reduction in copy number from 2 to 1. For two of the lines (HCC1937 and NCI-H209), we were able to compare the p501 array results with data from BAC-array-based CGH. Of the 44 autosomal chromosomes from these two lines, 34 showed consistent copy number patterns in the two analysis protocols when compared for low copy number changes over extended regions. Chromosomes 17, 19, 20, and 22 gave poor resolution in both samples using the p501 array, thereby accounting for 80% of the inconsistencies. These chromosomes had the lowest SNP density with 1 in 0.71, 1.25, 0.77, and 1.25 Mb, respectively, compared with the average density for the rest of the genome of 1 in 0.44 Mb. For the remaining 18 autosomal chromosomes, the data from the p501 array tended to show a greater variability; however, the underlying pattern was discernable and consistent with that of BAC-array-based CGH (Fig. 2C,D).
Copy Number Changes Combined With Genotyping Data
A comparison was made between the genotyping data from the array and the copy number estimation for the 20 cancer cell lines. This analysis highlighted complexpatterns of chromosomal gains and losses that would not be detectable by either CGH or microsatellite analysis alone.
Copy Number Reduction Without Loss of Heterozygosity (LOH)
Six of the cell lines under analysis contained at least one chromosomal region (∼10 Mb or greater) showing a 50% reduction in fluorescence intensity (to 0.5) observed in the copy number analysis, which corresponded to a region of heterozygosity defined by a minimum of three informative SNPs (eight regions in total). One interpretation of this result is that the cell line has a karyotype with an average ploidy of four. If only two copies of certain chromosomes are present (either these chromosomes are not duplicated with the rest of the genome or two copies, one from each parent, are subsequently lost), then a 50% drop in fluorescence intensity would be observed, and each chromosome would be derived from a different parent. Sky karyotypes (data not shown) were available for two of the lines showing this pattern, allowing average chromosomal number to be estimated; these lines contained 68 (COLO829) and 88 (HCC1937) chromosomes, respectively. Figure 2C shows Chromosome 18 in HCC1937, where the q-arm shows this pattern of copy number reduction without LOH.
LOH Without Copy Number Reduction
All but one of the lines under analysis contained at least one region of ∼10 Mb or greater in which LOH did not correspond with a decrease in copy number. At least 94 such regions were identified in the lines under analysis, averaging 4.7 per line. In this case, the LOH may have arisen through mitotic recombination or two separate genomic events may have occurred, loss of one of the parental regions and subsequently duplication of the other parental copy. This pattern of LOH without reduction in copy number can be seen on Chromosome 5qter of NCI-H209 (Fig. 2D, region iii) and provides a general illustration of the complexity of genetic changes in cancer. In addition to the region of LOH without copy number change, there is also an area of loss accompanied by copy number reduction (Fig. 2D, region ii), a confirmed homozygous deletion (arrowed), and a region of slight copy number increase (Fig. 2D, region i).
DISCUSSION
This study demonstrates that the Affymetrixp501 oligonucleotide array coupled with the whole-genome sampling assay (WGSA) generates reproducible SNP-based genotyping data that can be used in a range of genomic applications (Schubert et al. 2002; Dumur et al. 2003). We have shown that the SNP array can also be used to detect copy number variations in cancer cell lines. The platform reliably reports high-level amplifications and homozygous deletions extending over regions of <1 Mb to several megabases as demonstrated by the confirmation of 12 amplifications and 10 homozygous deletions each reported by a minimum of three consecutive SNPs. Single SNPs reporting high-level copy number changes with p-values <0.0001 were identified and have been shown to report real events (J. Huang, pers. comm.); however, in this study none has been checked by other methods. In addition, subtler changes resulting in loss or gain of a single copy of larger genomic regions can be detected. The combination of both genotyping and copy number analysis for each data point allows for the identification of genomic alterations that would go undetected in array CGH or genotyping analysis alone. Analysis of the 20 cell lines used in this study identified eight regions of copy number reduction without LOH and 94 regions of LOH without copy number reduction, demonstrating that integration of genotype data and copy number information offers a deeper insight into genomic alterations present within cancer cells than either analysis on its own.
This method differs from spotted-array-based CGH in that the normal and tumor DNAs are hybridized to different arrays in a similar fashion to Affymetrix expression array experiments (Lockhart et al. 1996). This approach has the advantage of being able to build a pool of normal data for the subsequent tumor analysis and reduces the complexity of the hybridization.
To evaluate the reliability of reporting of subtle copy number changes, we compared the results from the p501 array with those from a BAC array. Overall, the data from the p501 array exhibited more variability than BAC-array-based CGH. This may in part be because of the intrinsic variability of the PCR-based approach used in the genome representation/simplification for the WGSA system. In addition, the hybridization kinetics of oligonucleotide probes compared with BAC clones may account for some of the variability, which appears to persist despite the multiple representations (28-fold) of each SNP locus. Finally, although the criteria for the SNPs used in this study would remove those that had common SNPs within their flanking XbaI restriction sites, rare SNPs within the XbaI sites would not be detected and may contribute to variation in PCR product representation that does not reflect genomic copy number. Nevertheless, subtle copy number patterns found in two cancer cell lines analyzed with both the p501 array and BAC-array-based CGH were similar, with 34 out of 44 autosomal chromosomes showing the same patterns; of those 10 chromosomes that gave inconsistent patterns, eight were caused by poor representation on the p501 array owing to low SNP density (Chromosomes 17, 19, 20, and 22). The distribution of SNPs on the array is determined by both the number of publicly available SNPs and the occurrence of “predicted” Xba I sites within the genome, which is a function of both the actual distribution of sites and the level of completion of genome sequence at the time the arrays were designed. Increases in SNP identification/coverage, completion of the genomic sequence, and addition of alternative restriction enzyme fractions will all help in increasing the density of SNPs that can be interrogated by this approach. Furthermore, the selective use of alternative restriction enzyme fractions whose SNP density complements that of XbaI will compensate for the paucity of XbaI sites in certain regions of the genome. It is anticipated that as SNP density increases, resolution and the ability to assess subtle copy number changes will increase.
We and others (J. Huang, pers. comm.) have recently extended our analyses to the Affymetrix GeneChip Mapping 10K assay (Xba_131 array), which contains 11,555 SNPs. This array performs better than the p501, giving average call rates of 93% (n = 30) with an average concordance of 99.5% based on five DNAs analyzed in triplicate (data not shown). The new Xba_131 array can also be used for the copy number analysis. The copy number data, although still variable, is better than that achieved using the p501 array because of the higher SNP density. Chromosomes 17, 19, 20, and 22, which have low SNP densities on the p501 array, now have densities of 1 SNP in 287.7 kb, 666.7 kb, 297.7 kb, and 620.3 kb, respectively; the remaining chromosomes have an average density of 1 in 269.6 kb. Therefore, the SNP densities for the Xba_131 array on Chromosomes 17 and 20 are similar to the mean of the rest of the genome, whereas Chromosomes 19 and 22 still have low SNP densities.
Further improvement in the data from the SNP arrays may be possible using even higher density SNP arrays. It may also be possible to improve the data from SNP arrays by assessing copy number variation for each SNP based on analysis at the individual feature level instead of using the means of the features.
While this article was in preparation, Lucito et al. (2003) published work based on “representational oligonucleotide microarray analysis” (ROMA) using oligonucleotides 70 bases in length with a resolution of 30 kb throughout the genome. Their protocol is similar to other microarray-based technologies with cohybridization of a normal control sample together with the test sample, changes in copy number being reported by changes in the ratios of the Cy3- and Cy5-labeled DNAs. This technology is similar to BAC-array-based CGH with the advantage of very-high-density probes. However, ROMA does not give genotyping data in conjunction with the copy analysis and therefore would not identify regions of LOH without copy number change as described here.
METHODS
Cell Lines
Normal and cancer cell lines were cultured using the suppliers' recommended conditions. DNA was extracted from the cell lines using the QIAGEN “blood and cell culture” DNA Maxi Kit (catalog #13362). The cell lines used in this study were normal lines: 1156-Q-LC, 1542N-P63B, 833-K-LC, BB132-EBV, BB65-EBV, COLO829BL, COR-L96-LCL, HA7-EBV, HCC1954BL, HCC2157BL, HCC2218BL, HCC38BL, J82-EBV, LB1047-EBV, LB2518-EBV, LB373-EBV, LB996-EBV, NA15080, NA17205, NA17217, NA17228, NA17235, NCI-BL1184, NCI-BL1395, NCI-BL1437, NCI-BL2009, NCI-BL209, NCI-BL2126, and NCI-BL2171; tumor lines: 1156-Q-E, 1542T-P41B, 833-KE, BB132-MEL, BB65-RCC, COLO829, COR-L96-CAR, HA7-RCC, HCC38, J82, LB1047-RCC, LB2518-MEL, LB373-MEL, LB996-RCC, NCI-H1395, NCI-H209, NCI-H2126, NCI-H2171, HCC1395, and HCC1937. DNA from cell lines NA04626 (3X), NA01416 (4X), and NA06061 (5X) were obtained from Coriell.
Array Design
The p501 array contains allele-specific hybridization probes complementary to 8473 SNPs predicted to be in the fraction of the genome represented by 400–800-bp fragments of XbaI-digested genomic DNA. Oligonucleotides corresponding to these SNPs were synthesized used a photolithographic methodology, and each SNP is represented by 56 different oligonucleotide probes. The oligonucleotides are 25-mer sequences that interrogate the site of polymorphism on both the sense and antisense strands and contain both a perfect match (PM) and mismatch (MM) sequence to allow signal-to-noise measurements. Additional oligonucleotides that are offset from the site of polymorphism by 1–4 nt are synthesized to allow for data redundancy and to maximize genotype accuracy.
Target Preparation
WGSA relies on genomic representation to reduce the complexity of the genome by ∼98% and thereby improve hybridization kinetics (Lucito et al. 1998; Kennedy et al. 2003). The sample DNA was digested to completion using Xba1 prior to ligation of adaptors and amplification of the ligation products using an adaptor-specific primer. The reagents and protocol used were taken from Kennedy et al. (2003), except 400 ng of sample DNA was used instead of 250 ng. Because of the increased input DNA all reactions volumes involved in the digestion, linker ligation and amplification stages were increased by 60%, thereby maintaining the reaction conditions. After PCR, the samples were concentrated using a QIAGEN minielute PCR purification kit (catalog #28006) and eluted in 30 μL of EB buffer, using one column per PCR, giving a final elution volume of 180 μL. This was reduced to 50 μL with a Microcon YM-30 filter (catalog #42410). The final concentrated DNA was quantified using a Hoeffer DyNA Quant capillary cuvette kit; 20 μg of the PCR product was fragment, reducing the average product size down to 50–150 bp.
Hybridization and Scanning
Hybridization and staining of the probe to the array together with scanning of the chip were carried out as in Kennedy et al. (2003). Basically, the fragmented DNA was labeled with biotin-N6-ddATP using terminal transferase before being mixed with hybridization solution and added to the p501 arrays for hybridization. After hybridization, the array was washed and stained using the Affymetrix Fluidics Station. The staining procedure was designed to amplify the signal from the annealed probe. The sample was first stained with straptavidin followed by treatment with biotinylated anti-streptavidin and finally Streptavidin R0-phycoerythrin conjugate. Scanning was carried out using the Agilent GeneArray Scanner.
Feature Extraction
The p501 array design uses 28 probe pairs for each SNP, 14 for allele A and 14 for allele B. The 14 probe pairs for each allele are equally divided between the sense and antisense strands. A probe pair includes a perfect match cell and a mismatch cell. We use
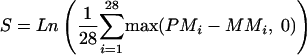 |
as the basic measurement for any given SNP, where PMi is the intensity of the perfect match cell of probe pair i and MMi is the intensity of the mismatch cell of probe pair i. This value measures the average intensity difference between a perfect match and a mismatch on a log scale. The log transformation makes the distribution more Gaussian. After S is calculated for all the SNPs on a given chip, it is scaled to have a mean of zero. In other words:
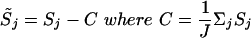 |
j = 1,..., J are all the autosomal SNPs on the chip.
Copy Number Estimation
When estimating copy number, the output from the feature extraction was smoothed, first outliers were removed by taking the median of five SNPs—the test SNP together with the two flanking SNPs for each data point—and the mean of the five SNPs was then taken. This was then converted into copy number by calculating the fluorescence ratio with respect to the mean reading from a series of 29 normal DNAs. Therefore, if an SNP is from a diploid region of the test sample, it will give a reading of 1 when divided by the normal reference sample, representing a copy number of 2.
Significance Calculation
To estimate the significance of the copy number variation in the target cancer cell line, we compare it with a reference set consisting of 29 normal DNA samples. For any given SNP j, we assume that S̃j follows a Gaussian distribution; the mean and variance are estimated using the 29 normal reference samples.
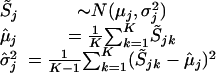 |
where k = 1,..., K represents the normal reference set. Assuming the target cancer cell line has value 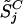 on SNP j, the significance of the difference of
on SNP j, the significance of the difference of 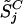 from the normal reference distribution is measured by the p-value:
from the normal reference distribution is measured by the p-value:
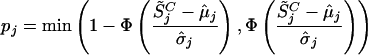 |
This probability indicates how likely the normal population will have values as extreme as the cancer cell line. The smaller it is, the more significant the difference between the cancer and the normal cell lines.
BAC Array CGH
BAC array CGH was carried out using a 1-Mb array containing ∼4100 publicly available BACs. Array construction was based on a modification of the protocol by Hodgson et al. (2001). DNA was extracted from 15-h cultures using the QIAGEN REAL prep kit and amplified using degenerate oligonucleotide primers (5′-CCGACTCGAGNNNNNNATGTGG-3′). Each BAC was amplified under two different annealing temperatures (58°C and 60°C) to improve coverage. PCR products were pooled, purified (QiaQuick kits, QIAGEN), and lyophilized before being resuspended in 50% DMSO/water to a final concentration of ∼300 ng/μL. A minimum of two replicates of each clone was printed on each slide.
DNA was labeled (random prime labeling kit, Invitrogen) and hybridized together with Cot-1 DNA for 48–72 h at 37°C. Slides were washed as described by Hodgson et al. (2001) and scanned using the Affymetrix428 microarray scanner. Analysis was carried out using the Genepixsoftware (Axon) with adjustments made to account for variations in labeling efficiency (print-tip-based data adjustment; Yang et al. 2001).
CA Repeat Genotyping
Genotyping was carried out using the 10-cM microsatellite marker set from ABI (LMS-MD10). The markers were amplified from 12 ng of DNA using ABI True Allele PCR premix(catalog #403061) in a total volume of 10 μL. Cycling was performed on a Kbiosystems “Duncan” thermocycler using a 40-cycle program of 94°C denaturation, 60°C annealing, and 72°C extension each for 30 sec, proceeded by a 10-min soak at 94°C to activate the Amplitaq Gold, with a final 10-min soak at 72°C for complete extension. Samples were prepared and loaded onto either an ABI 3700 or ABI 3100 DNA analyzer as per the manufacturer's instructions. The data were analyzed using the ABI Genescan (V5.1) and Genotyper (V3.6) software. Fragment sizing was standardized between runs by comparison of data from a control DNA (CEPH 1347-02) that was included in every sample set.
Confirmation of Homozygous Deletions
Homozygous deletions were confirmed by PCR. Primers were designed for SNPs reporting the deletion together with flanking SNPs. If possible, primers were designed either side of the SNP; otherwise, primers were placed as near to the SNP as possible. PCRs were carried out in duplicate for the test DNA together with a normal control. PCR products were visualized by gel electrophoresis.
Confirmation of Amplifications
Amplifications were confirmed by qPCR using the ABI 7700 together with TaqMan dual labeled probes. Copy number estimation was calculated using the standard curve method for relative quantitation using separate tubes with relation to two reference loci (APP1 and DCK, Ensembl genes ENSG00000090621 and ENSG00000156136, respectively) and expressed relative to a normal control DNA. Experimental design and calculations were carried out as described by ABI (User Bulletin #2). The profile of the amplification in COR-L96-CAR was confirmed by qPCR of amplicons designed to 18 SNPs from the p501 array reporting the amplification. Copy number was calculated with relation to an unamplified flanking SNP and expressed relative to a normal control DNA. For this experiment, qPCR was carried out using the QuantiTect SYBR Green PCR kit (QIAGEN catalog #204143).
Acknowledgments
The work of the Cancer Genome Project was supported by the Wellcome Trust. Other funding included Cancer Research UK (M.G.) and the Abramson Family Cancer Research Institute (B.L.W.). We thank the staff of the Cancer Genome Project for their contribution to this work.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2012304.
Footnotes
[Supplemental material is available online at www.genome.org and ftp.sanger.pub/p501. The data from all 70 arrays (29 normals, 20 cancer lines, 3 X-copy number, and 18 “spike” DNAs) used in this study will also be made available initially on ftp.sanger.pub/p501, until submission to Array Express is arranged.]
References
- Albertson, D.G., Ylstra, B., Segraves, R., Collins, C., Dairkee, S.H., Kowbel, D., Kuo, W.L., Gray, J.W., and Pinkel, D. 2000. Quantitative mapping of amplicon structure by array CGH identifies CYP24 as a candidate oncogene. Nat. Genet. 25: 144-146. [DOI] [PubMed] [Google Scholar]
- Dumur, C.I., Dechsukhum, C., Ware, J.L., Cofield, S.S., Best, A.M., Wilkinson, D.S., Garrett, C.T., and Ferreira-Gonzalez, A. 2003. Genome-wide detection of LOH in prostate cancer using human SNP microarray technology. Genomics 81: 260-269. [DOI] [PubMed] [Google Scholar]
- Ghaffari, S.R., Boyd, E., Tolmie, J.L., Crow, Y.J., Trainer, A.H., and Connor, J.M. 1998. A new strategy for cryptic telomeric translocation screening in patients with idiopathic mental retardation. J. Med. Genet. 35: 225-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson, G., Hager, J.H., Volik, S., Hariono, S., Wernick, M., Moore, D., Nowak, N., Albertson, D.G., Pinkel, D., Collins, C., et al. 2001. Genome scanning with array CGH delineates regional alterations in mouse islet carcinomas. Nat. Genet. 29: 459-464. [DOI] [PubMed] [Google Scholar]
- Jain, A.N., Chin, K., Borresen-Dale, A.L., Erikstein, B.K., Eynstein Lonning, P., Kaaresen, R., and Gray, J.W. 2001. Quantitative analysis of chromosomal CGH in human breast tumors associates copy number abnormalities with p53 status and patient survival. Proc. Natl. Acad. Sci. 98: 7952-7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallioniemi, A., Kallioniemi, O.P., Sudar, D., Rutovitz, D., Gray, J.W., Waldman, F., and Pinkel, D. 1992. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 258: 818-821. [DOI] [PubMed] [Google Scholar]
- Kennedy, G.C., Matsuzaki, H., Dong, S., Liu, W., Huang, J., Liu, G., Su, X., Manqiu, C., Chen, W., Zhang, J., et al. 2003. Large-scale genotyping of complex DNA. Nat. Biotechnol. 21: 1233-1237. [DOI] [PubMed] [Google Scholar]
- Lockhart, D.J., Dong, H., Byrne, M.C., Follettie, M.T., Gallo, M.V., Chee, M.S., Mittmann, M., Wang, C., Kobayashi, M., Horton, H., et al. 1996. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14: 1675-1680. [DOI] [PubMed] [Google Scholar]
- Lucito, R., Nakimura, M., West, J.A., Han, Y., Chin, K., Jensen, K., McCombie, R., Gray, J.W., and Wigler, M. 1998. Genetic analysis using genomic representations. Proc. Natl. Acad. Sci. 95: 4487-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucito, R., Healy, J., Alexander, J., Reiner, A., Esposito, D., Chi, M., Rodgers, L., Brady, A., Sebat, J., Troge, J., et al. 2003. Representational oligonucleotide microarray analysis: A high-resolution method to detect genome copy number variation. Genome Res. 13: 2291-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls, R.D., Knoll, J.H., Butler, M.G., Karam, S., and Lalande, M. 1989. Genetic imprinting suggested by maternal heterodisomy in nondeletion Prader-Willi syndrome. Nature 342: 281-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel, D., Segraves, R., Sudar, D., Clark, S., Poole, I., Kowbel, D., Collins, C., Kuo, W.L., Chen, C., Zhai, Y., et al. 1998. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat. Genet. 20: 207-211. [DOI] [PubMed] [Google Scholar]
- Pollack, J.R., Perou, C.M., Alizadeh, A.A., Eisen, M.B., Pergamenschikov, A., Williams, C.F., Jeffrey, S.S., Botstein, D., and Brown, P.O. 1999. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat. Genet. 23: 41-46. [DOI] [PubMed] [Google Scholar]
- Schubert, E.L., Hsu, L., Cousens, L.A., Glogovac, J., Self, S., Reid, B.J., Rabinovitch, P.S., and Porter, P.L. 2002. Single nucleotide polymorphism array analysis of flow-sorted epithelial cells from frozen versus fixed tissues for whole genome analysis of allelic loss in breast cancer. Am. J. Pathol. 160: 73-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman, J.A., Schoenmakers, E.F., Eussen, B.H., Janssen, I., Merkx, G., van Cleef, B., van Ravenswaaij, C.M., Brunner, H.G., Smeets, D., and van Kessel, A.G. 2002. High-throughput analysis of subtelomeric chromosome rearrangements by use of array-based comparative genomic hybridization. Am. J. Hum. Genet. 70: 1269-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L., Tran, D.K., and Wang, X. 2001. BADGE, Beads Array for the Detection of Gene Expression, a high-throughput diagnostic bioassay. Genome Res. 11: 1888-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]








