Abstract
Genome-wide physical maps are crucial to many aspects of advanced genome research. We report a genome-wide, bacterial artificial chromosome (BAC) and plant-transformation-competent binary large-insert plasmid clone (hereafter BIBAC)-based physical map of the soybean genome. The map was constructed from 78,001 clones from five soybean BAC and BIBAC libraries representing 9.6 haploid genomes and three cultivars, and consisted of 2905 BAC/BIBAC contigs, estimated to span 1408 Mb in physical length. We evaluated the reliability of the map contigs using different contig assembly strategies, independent contig building methods, DNA marker hybridization, and different fingerprinting methods, and the results showed that the contigs were assembled properly. Furthermore, we tested the feasibility of integrating the physical map with the existing soybean composite genetic map using 388 DNA markers. The results further confirmed the nature of the ancient tetraploid origin of soybean and indicated that it is feasible to integrate the physical map with the linkage map even though greater efforts are needed. This map represents the first genome-wide, BAC/BIBAC-based physical map of the soybean genome and would provide a platform for advanced genome research of soybean and other legume species. The inclusion of BIBACs in the map would streamline the utility of the map for positional cloning of genes and QTLs, and functional analysis of soybean genomic sequences.
Soybean, Glycine max (L.) Merr., is the world's top legume crop and foremost source of edible plant oil and proteins. To develop tools essential for continued genetic improvement of the crop, DNA-marker-based genetic linkage maps have been developed (e.g., Lark et al. 1993; Shoemaker and Specht 1995; Keim et al. 1997; Cregan et al. 1999a; Iqbal et al. 2001; http://soybase.agron.iastate.edu), 93 genes and >900 quantitative trait loci (QTLs) of agronomic importance have been mapped with the genetic maps (http://soybase.agron.iastate.edu), several large-insert bacterial artificial chromosome (BAC) and plant-transformation-competent binary plasmid clone (hereafter BIBAC) libraries have been constructed (Marek and Shoemaker 1997; Danesh et al. 1998; Salimath and Bhattacharyya 1999; Meksem et al. 2000), and a large collection of expressed sequence tags (ESTs) has been generated (Shoemaker et al. 2002; http://soybean.ccgb.umn.edu). However, further advances, such as development of DNA markers for a genomic region of interest for fine mapping of genes and QTLs, isolation of clones containing a gene and/or QTL of interest for positional cloning, mapping of the developed ESTs (Wu et al. 2002), and large-scale genome sequencing, are limited because of the shortage of essential and powerful infrastructure.
Genome-wide physical maps have provided powerful tools and infrastructure for advanced genomics research of human and several model species. They are not only crucial for large-scale genome sequencing (Hodgkin et al. 1995; Adams et al. 2000; The Arabidopsis Genome Initiative 2000; The International Human Genome Sequencing Consortium 2001), but also provide powerful platforms required for many other aspects of genome research, including targeted marker development, efficient positional cloning, and high-throughput EST mapping (Zhang and Wu 2001). Whole-genome physical maps have been constructed for Caenorhabditis elegans (Coulson et al. 1986; Hodgkin et al. 1995), Arabidopsis thaliana (Marra et al. 1999; Mozo et al. 1999; Chang et al. 2001), Drosophila melanogaster (Hoskins et al. 2000), human (The International Human Genome Mapping Consortium 2001), rice (Oryza sativa; Tao et al. 2001; Chen et al. 2002), and mouse (Mus musculus; Gregory et al. 2002). However, no genome-wide physical map has been reported for soybean and other legume species.
Several approaches have been developed to construct whole-genome physical maps with large-insert BAC and BIBAC clones (Gregory et al. 1997; Marra et al. 1997; Zhang and Wing 1997; Tao and Zhang 1998; Ding et al. 1999; Zhang and Wu 2001). We helped pioneer the strategies and technologies of whole-genome physical mapping from BAC and BIBAC clones by restriction fingerprint analysis on DNA sequencing gels (Zhang and Wing 1997; Tao and Zhang 1998). The DNA sequencing gel-based fingerprinting method (Coulson et al. 1986; Gregory et al. 1997; Zhang and Wing 1997; Tao and Zhang 1998; Ding et al. 1999; Zhang and Wu 2001) not only has a significantly higher resolution (≤1 nt) than that of the agarose gel-based method (10–500 bp; Marra et al. 1997; Zhang and Wu 2001; Z. Xu, S. Sun, and H.-B. Zhang, unpubl.), but is also economical and highly amenable to analysis by automated DNA sequencers (Gregory et al. 1997; Ding et al. 1999; Z. Xu, Y.-L. Chang, K. Ding, and H.-B. Zhang, unpubl.) and to high-throughput technologies (Zhang and Wu 2001; Z. Xu, Y.-L. Chang, K. Ding, and H.-B. Zhang, unpubl.). Using these techniques and strategies, we previously developed a BAC/BIBAC-based integrated physical and genetic map of Arabidopsis (Chang et al. 2001), and the whole-genome BAC-based physical maps of O. sativa ssp. indica (Tao et al. 2001) and chicken (Ren et al. 2003).
Soybean has a genome size of 1115 Mb/1C (Arumuganathan and Earle 1991), and ∼40%–60% of its genome is repetitive sequence and heterochromatic (Goldberg 1978; Gurley et al. 1979; Singh and Hymowitz 1988). Although the genome of soybean is smaller in size than the genomes of human and mouse, for which BAC-based physical maps have been developed (The International Human Genome Mapping Consortium 2001; Gregory et al. 2002), development of a genome-wide physical map of the soybean genome is more difficult. This is because soybean is a recently diploidized tetraploid (last duplication only 8 million years ago [Mya]) and has an average of 2.55 duplicated segments with as many as six copies per gene (Shoemaker et al. 1996).
Efforts were made to develop a regional, BAC-based physical map of the soybean genome using the soybean cv. Williams 82 and cv. Faribault BAC libraries. However, the map only covered ∼20% of the soybean genome (Marek et al. 2001). Here we report a genome-wide, BAC- and BIBAC-based physical map of the soybean genome. We also tested and discussed the feasibility and strategies of integrating the physical map with the existing soybean genetic linkage maps (http://soybase.agron.iastate.edu; Lark et al. 1993; Shoemaker and Specht 1995; Keim et al. 1997; Cregan et al. 1999a; Iqbal et al. 2001).
RESULTS
Source BAC and BIBAC Fingerprinting
We fingerprinted a total of 84,946 BACs and BIBACs from the five BAC and BIBAC libraries (Table 1) on 1332 autoradiographs using the DNA sequencing gel-based restriction fingerprinting method (Zhang and Wing 1997; Chang et al. 2001; Tao et al. 2001; Ren et al. 2003). The autoradiographs of the BAC and BIBAC fingerprints were scanned into image files and edited with the Image program (Sulston et al. 1988). Of the clones, 6945 (8.18%) were deleted during fingerprint editing because they either failed in fingerprinting or had no inserts. Therefore, a total of 78,001 clones were successfully fingerprinted and integrated into the FPC database. The 78,001 clones represented 9.580× genome equivalents of soybean, of which the clones equivalent to 3.052×, 2.262×, 4.030×, 0.121×, and 0.115× haploid genomes were from the Forrest HindIII BIBAC library, the Forrest BamHI BIBAC library, the Forrest EcoRI BAC library, the Faribault EcoRI BAC library, and the Williams 82 HindIII BAC library, respectively (Table 1). To minimize the influence of the lower (>1 base) resolution of the higher-molecular-weight fingerprint bands on the accuracy of physical map contig assembly, we used only the bands between 58 and 773 bases for the physical map assembly. Consequently, an average band number of 27.22 for the Forrest HindIII BIBACs, 23.43 for the Forrest BamHI BIBACs, 39.12 for the Forrest EcoRI BACs, 28.46 for the Faribault EcoRI BACs, and 32.03 for the Williams 82 HindIII BACs (Table 1) was used in the physical map assembly of the soybean genome.
Table 1.
BACs and BIBACs Fingerprinted and Used for the Soybean Physical Map
| Libraries | Cloning site | No. of clones fingerprinted | Average insert size (kb) | No. of clones used in mapping | Average no. of bands/clone | Redundancy genome equivalent |
|---|---|---|---|---|---|---|
| Forrest | ||||||
| BIBAC-H | HindIII | 30,720 | 125 | 27,221 | 27.63 | 3.052× |
| BIBAC-B | BamHI | 21,504 | 125 | 20,181 | 23.43 | 2.262× |
| BAC-E | EcoRI | 30,720 | 157 | 28,620 | 39.12 | 4.030× |
| Faribault | ||||||
| BAC | EcoRI | 1142a | 120 | 1125 | 28.46 | 0.121× |
| Williams 82 | ||||||
| BAC | HindIII | 860a | 150 | 854 | 32.03 | 0.115× |
| Combined libraries | 84,946 | 78,001 | 9.580× |
The BACs were identified with 267 SSR markers and 105 RFLP markers by the laboratories of R. Shoemaker, Iowa State University and N. Young, University of Minnesota (Marek et al. 2001) from the soybean cv. Williams 82 (Marek and Shoemaker 1997) and cv. Faribault (Danesh et al. 1998) BAC libraries. The distribution of the markers in the soybean genetic map (Cregan et al. 1999a) were from Marek et al. (2001)
Physical Map Contig Assembly and Manual Editing
The FPC database of 78,001 BAC and BIBAC fingerprints was subjected to overlap analysis using the computer program FPC 4.7 (Soderlund et al. 2000). The FPC program assembled 4792 overlapping BAC/BIBAC contigs using the cutoffs ranging from 1e - 30 to 1e - 10 and a fixed tolerance of 2, whereas 4933 clones remained as singletons (Table 2). The physical length of the automated contigs was estimated to be 1481.5 Mb, based on 364,908 unique bands, with each being equivalent to 4.06 kb (Table 2).
Table 2.
Status of the Soybean Physical Map Before and After Manual Editing
| Automatic contig assembly | After manual editing | |
|---|---|---|
| Date | Jan. 2002 | Aug. 2002 |
| Number of clones in FPC database | 78,001 | 78,001 |
| Number of singletons | 4933 | 4954 |
| Number of contigs | 4792 | 2905 |
| Contigs containing: | ||
| >200 clones | 0 | 6 |
| 101–200 clones | 0 | 88 |
| 51–100 clones | 21 | 321 |
| 26–50 clones | 1176 | 504 |
| 10–25 clones | 1606 | 920 |
| 3–9 clones | 1777 | 850 |
| 2 clones | 212 | 216 |
| Unique bands of the contigs | 364,570 | 346,884 |
| Physical length of the contigs in megabase pairs | 1480a | 1408a |
| DNA markers in the contigs | 0 | 388 |
Each fingerprint band was estimated to represent an average of 4.06 kb. It was estimated by the average insert size of the BAC and BIBAC clones divided by the average number of bands per clone used for the map contig assembly
To verify and extend the contigs, we manually edited each of them using two methods. First, we manually checked every contig and disassembled potential chimeric contigs that were apparently not overlapped according to the clone fingerprint patterns, and/or that apparently conflicted with either DNA marker data or the existing soybean BAC contig data (Marek et al. 2001). Then all questionable contigs were split or killed. Second, to identify potential junctions between contigs, we searched the entire FPC fingerprint database for matches to the terminal clone fingerprints of every contig using the End Extension function of the FPC program with the cutoffs ranging from 1e - 28 to 1e - 10. We merged the contig pairs if their terminal clones shared 10 or more bands and their overall fingerprint patterns supported the junction. We also coalesced the contig pairs if they hybridized with two or more neighboring DNA markers and could be merged into a single contig using the cutoff values between 1e - 15 and 1e - 10. As a result, the total number of contigs of the physical map was reduced to 2905, with 4954 clones (6.35%) remaining as singletons (Table 2). The 2905 contigs consisted of 346,884 unique bands, collectively spanning 1408 Mb in physical length. The longest contig (ctg127) contained 319 clones, encompassing 1345 unique bands and spanning 5.5 Mb in physical length. The fingerprint database of all 78,001 BACs and BIBACs and all contigs of the soybean physical map are posted at http://hbz.tamu.edu and made available to the public. Figure 1 shows an example of the contigs of the physical map and the distribution of the BACs and BIBACs from the five soybean libraries within the contig.
Figure 1.
Example of the BAC/BIBAC contigs of the soybean physical map. This contig (ctg16) was anchored to the molecular linkage group MLG F of the soybean genetic map by the SSRmarker Satt343f (Cregan et al. 1999a; http://soybase.agron.iastate.edu/). The highlighted clones were the positive clones of the SSRmarker. The contig contains 120 BAC and BIBAC clones and 651 unique bands, estimated to span ∼2643 kb in physical length. The clones prefixed with “IS” or “UM” were from the Williams 82 and Faribault BAC libraries, respectively; and the clones prefixed with “E,” “B,” or “H” were from the Forrest EcoRI BAC, BamHI, and HindIII BIBAC libraries, respectively.
Integration of the Physical Map With the Existing Soybean Genetic Maps
Soybean is an ancient tetraploid species, which presents a significant challenge to develop a robust integrated physical and genetic map. To test the feasibility of anchoring the physical map contigs to the existing soybean composite genetic map (Cregan et al. 1999a; http://soybase.agron.iastate.edu/), we screened the Forrest EcoRI BAC or HindIII BIBAC libraries by colony filter hybridization with seven RFLP markers and 15 SSR markers. From one to 10 positive clones for each probe were identified (data not shown). The results obtained from SSR markers were further confirmed by PCR-based BAC library screening (http://www.siu.edu/~pbgc/DataBase/datap1.htm). All of the seven RFLP markers were shown to be multiple-copy in the soybean haploid genome by Southern analysis (also see http://soybase.agron.iastate.edu/), and the positive clones identified with each of these DNA markers were located to multiple contigs. In the case of the SSR markers, the positive BACs identified by eight of the 15 SSR markers were observed in single contigs, whereas the positive clones of the remaining seven SSR markers (47%) were observed in two or more contigs.
We also integrated the regional physical map data of soybean (Marek et al. 2001) into the whole-genome physical map constructed in this study. Using the method described above, we fingerprinted the 2002 positive clones from the Williams 82 and Faribault BAC libraries identified using 267 SSR and 105 RFLP markers (Marek et al. 2001). After editing, fingerprint data were successfully obtained from 1851 of the 2002 BACs, which contained 264 SSRs and 102 RFLPs. We then integrated the BAC fingerprint database with our whole-genome BAC fingerprint database and used the combined data for whole-genome physical map contig assembly.
The screening of the Forrest BAC and BIBAC libraries and integration of the positive BACs of the Williams 82 and Faribault libraries together identified 781 marker-containing contigs of the physical map. Of the 388 markers (115 RFLPs and 273 SSRs) used, the positive BACs of each of all 115 RFLP markers, except for one (A469), were located to two or more contigs because they have multiple loci in the soybean genome (Marek et al. 2001), whereas the positive clones of each of 82 of the 273 SSR markers (30.0%) were located to a single contig, indicating a single locus in the soybean genome if a contig is assumed to represent one locus (see Fig. 2 and Supplemental Fig. S1 available online at www.genome.org). Therefore, the 83 contigs of the single-locus markers (1 RFLP and 82 SSRs) were unambiguously anchored to the soybean genetic map. In addition, 16 contigs were hybridized with two or more neighboring markers and thus were also unambiguously anchored to the soybean genetic map. Further efforts will be needed to definitively anchor the remaining contigs to the linkage map.
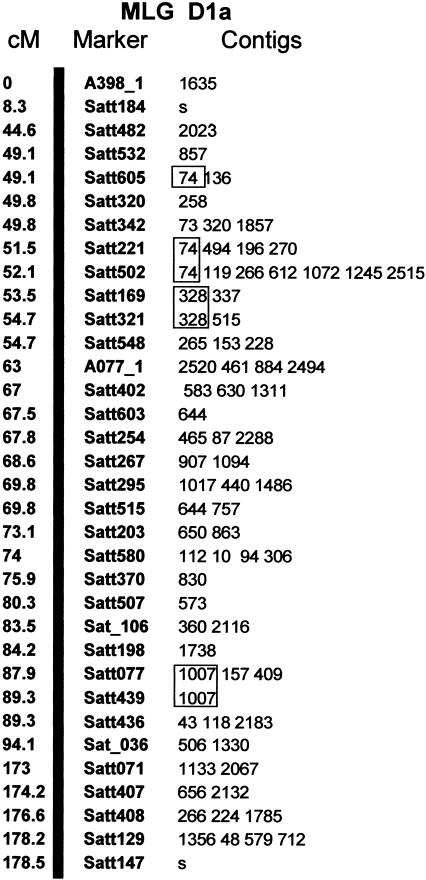
Figure 2.
BAC/BIBAC contigs of the soybean physical map containing DNA markers selected from the MLG D1a of the existing soybean composite genetic map (http://soybase.agron.iastate.edu/; Cregan et al. 1999a). The soybean genetic map consists of 20 molecular linkage groups (MLGs). The DNA markers used are listed in the middle column, positions (centimorgans) of the markers in the linkage group in the left column, and the contig(s) containing the marker in the right column of the linkage group. The letter “s” indicates singleton. Note that the contigs shown in rectangles were hybridized with two or more neighboring DNA markers, seven SSRmarkers each were hybridized to only one contig, and two SSRmarkers each were anchored to a singleton. A total of 10 contigs were unambiguously anchored to the genetic map, whereas the exact positions of the remaining contigs remain to be further refined. The names of the DNA markers were after Cregan et al. (1999a) and http://soybase.agron.iastate.edu/, the markers prefixed with “Satt,” “Sat,” or “Sct” representing SSRmarkers and the remaining markers being RFLP markers. The markers suffixed with _1, _2, or _3 indicate that they reside at two or more loci in the soybean genome.
Physical Map Contig Reliability
We evaluated the reliability of the soybean contig map using several approaches. In our first approach, we assembled automatic contigs from the fingerprints using two different strategies and then compared the resultant contigs. In the first contig assembly strategy, we assembled the contigs using individual stepwise cutoff values between 1e - 30 and 1e - 10. In the second contig assembly strategy, we assembled the contigs using the cutoff values 1e - 25, 1e - 20, 1e - 15, and 1e - 10, respectively, and disassembled and reassembled the contigs that were obviously chimeric using higher-stringency cutoff values. One thousand contigs were randomly selected from the contigs assembled by the two strategies and compared. The result showed that 99.1% of the automated contigs were completely consistent in both clone content and order. In our second approach, we assembled contigs from the clones of the Forrest BamHI, Forrest EcoRI, Forrest HindIII, and Williams 82/Faribault libraries, separately. We randomly selected 100 contigs from the contigs assembled from each of the three Forrest libraries and all 389 contigs of the Williams 82/Faribault libraries, and compared them with their corresponding contigs in the physical map. We found that 93%, 97%, 96%, and 96% of the contigs were shown to be in complete agreement in both clone content and order. For our third approach, we compared 141 RFLP-anchored contigs constructed independently by digesting the marker-positive BACs with a restriction enzyme, followed by Southern hybridization with relevant DNA markers (Marek et al. 2001) against the corresponding contigs of the physical map constructed in this study. By this, 125 of the 141 contigs (88.7%) were shown to be completely consistent in both clone content and order. For our fourth approach, we randomly selected 10 contigs from the physical map, fingerprinted the BACs of the contigs with two enzyme combinations (HindIII/HaeIII and BamHI/HaeIII), respectively, and then reassembled the contigs. As a result, the same contigs as those selected from the physical map were reassembled (data not shown). Finally, we checked the positions of the positive clones of each of the 83 single-locus DNA markers in the physical map. The result showed that the positive clones of every single-locus DNA marker located to the corresponding region of a single contig. Combining the results from all five approaches to the contig verification indicated that the contigs were properly assembled.
DISCUSSION
We have successfully fingerprinted 78,001 clones from five soybean BAC and BIBAC libraries representing a 9.6-fold haploid genome redundancy, created an FPC database for the clone fingerprints, and constructed a genome-wide physical map of the soybean genome. The map consists of 2905 contigs, estimated to span 1408 Mb. The total physical length of the contigs is ∼293 Mb (26.3%) greater than the 1115-Mb genome size of soybean (Arumuganathan and Earle 1991). This indicates that most, if not all, of the contigs overlap adjacent contigs, although the overlaps could not be detected under the conditions used, and/or that the genome size of soybean was underestimated. According to our (Chang et al. 2001; Tao et al. 2001; Ren et al. 2003) and other (Marra et al. 1999; The International Human Genome Mapping Consortium 2001; Chen et al. 2002; Gregory et al. 2002) physical mapping results, the failure to detect overlaps between contigs is likely to be the main cause of the discrepancy between the total physical length of the contigs and the estimated soybean genome size. Therefore, the physical map contigs could be further merged, and the map could be further refined, as additional information such as DNA marker hybridization becomes available (The International Human Genome Mapping Consortium 2001; Gregory et al. 2002). The contigs as well as the clone content and order within the contigs have been confirmed by using different contig assembly strategies, independent contig building methods, different fingerprinting methods, and DNA marker screening results of the source BACs and BIBACs. These results consistently indicated that the contigs of the soybean physical map were properly assembled and, thus, are suitable for advanced genome research of soybean and related species.
This study represents the first report of development of a genome-wide BAC- and BIBAC-based physical map of the soybean genome. The physical map will not only provide a platform for large-scale genome sequencing (Venter et al. 1996; Zhang and Wu 2001), but also facilitate fine mapping of genes and QTLs (Cregan et al. 1999b), positional cloning, comparative analysis of the legume genomes (e.g., Gregory et al. 2002), and many other studies. For clone-by-clone shotgun genome sequencing (Zhang and Wu 2001), the physical map could provide an essential, readily usable platform. Minimally overlapping clone tiling paths needed for clone-by-clone genome sequencing could be directly selected from the constructed contigs, or constructed by electronic chromosome walking using the FPC database of the physical map and the FPC Hitting Tool provided (http://hbz.tamu.edu). A contig is randomly selected and sequenced. The BAC ends of the map contigs are sequenced and used as sequence-tagged connectors (STCs) to extend the sequenced contig by sequence alignment because most of the map contigs overlap adjacent contigs even though the overlaps were not detected under the conditions used. For whole-genome shotgun sequencing (Venter et al. 1996; Zhang and Wu 2001), the physical map could provide a framework for sequence map assembly. The ends of BACs and BIBACs of the physical map are sequenced and used as STCs for anchoring and extending the sequence contigs generated by shotgun sequencing. Furthermore, the BIBACs of the physical map will streamline the positional cloning, genomic sequence functional analysis, and gene/QTL engineering by Agrobacterium-mediated genetic transformation (Clemente et al. 2000; Donaldson and Simmonds 2000). Moreover, the genomes of the two model legumes, Medicago truncatula and Lotus japonicus, are being sequenced. As has been done between the mouse and human genomes (Gregory et al. 2002), the soybean physical map could also be used to study synteny between soybean and the model legumes by contig BAC end sequencing and alignment along the genome sequences of the model legumes. Knowledge of the synteny will greatly facilitate map-based cloning of agronomically important genes and QTLs in the legume species. Finally, soybean is an ancient polyploid. Chromosome doubling and polyploidization is a significant evolutionary process of genomes in higher organisms, including plants, vertebrates, and many other eukaryotes (e.g., Grant 1981; Lundin 1993; Sidow 1996; Leitch and Bennett 1997; Spring 1997; Postlethwait et al. 1998). The genomes of most angiosperms are thought to have incurred one or more polyploidization events during evolution (e.g., Masterson 1994). Therefore, the physical map of the soybean tetraploid genome may also provide a platform for studies of genome duplication, polyploidization, and evolution in polyploid plants.
This study has further confirmed the nature of the ancient tetraploid origin of the soybean genome and provides the first example of developing whole-genome contig maps of polyploid species, which account for ∼70% of the flowering plants. In this study, a total of 781 contigs were identified using 388 DNA markers. Each DNA marker corresponds to an average of 2.0 contigs, with a maximum of 10 contigs per marker. This result is consistent with the average of 2.5 duplicated segments and as many as six copies per gene previously estimated by Shoemaker et al. (1996), thus supporting the hypothesis that the soybean genome is an ancient tetraploid.
The tetraploid nature of the soybean genome complicated the integration of the physical map contigs with the soybean genetic maps. Positive clones of a single marker were located to two or more contigs, or the markers from different regions of the linkage map were located to the same contigs (Fig. 2; Supplemental Fig. S1). Nevertheless, this result was not surprising in a diploidized ancient polyploid genome that apparently has genome duplication events in addition to the whole-genome duplication (Grant et al. 2000; Wolfe 2001). SSRs may be locus-specific, as defined by single bands on DNA fractionation matrices, when genomic DNA is amplified, but when the primary site is absent such as in a BAC pool, the duplicated site could yield an amplicon even though some mismatches of primer sequence(s) may be present as a result of mutation since the duplication event. It was also observed that single bands revealed on agarose gels, manual sequencing gels, or capillary sequencers did not always imply single fragments, frequently containing multiple fragments from different genomic regions in the polyploid soybean and cotton (data not shown; also see the SSR pattern pictures on manual sequencing gels at the Soybase).
The results obtained here demonstrated that it is feasible to properly integrate the physical map contigs to the existing genetic map and develop a robust integrated physical and genetic map of the soybean genome. First, ∼30% of the SSR markers each were shown to anchor only one contig. Therefore, the contigs containing such SSR markers can be unambiguously anchored to the soybean genetic map. Second, as shown in Figure 2, quite a few of the contigs each contained two or more neighboring DNA markers mapped to the genetic map and thus were also unambiguously anchored to the soybean genetic map even though the DNA markers are multiple-locus in the soybean genome. In this study, we unambiguously anchored 99 of the map contigs using 388 markers. If 3000 or more markers were used, more than 765 of the map contigs would be anchored unambiguously. These contigs then could serve as anchors and starting points to be extended by contig mergence with adjacent contigs although they were identified by multiple-locus markers. Therefore, screening the BACs and BIBACs of the physical map with additional DNA markers, either single-locus or multiple-locus, will result in rapidly and unambiguously anchoring the contigs of the physical map to the genetic map, coalescing the neighboring contigs and drastically reducing the total number of contigs representing the physical map. In this regard, the overlaps between the neighboring contigs, despite being not detected under the conditions used, will provide useful information for contig merger. In return, the coalescence of the contigs will further verify the accuracy of the map contigs. Based on our construction of the integrated physical and genetic map of the 38-Mb rice Chromosome 8 from the rice genome contig map using 59 markers (644 kb/marker on average; Y. Li et al., unpubl.; http://hbz.tamu.edu), at least 3000 markers (372 kb/marker) will be needed to develop the soybean contig map into a robust integrated physical and genetic map. For instance, 1704 (252 kb/marker), 13,695 (241 kb/marker), and 16,992 (194 kb/marker) markers, combined with tens of thousands of BAC end sequences, were used to develop the integrated physical and genetic maps of rice (Chen et al. 2002), human (The International Human Genome Mapping Consortium 2001), and mouse (Gregroy et al. 2002). In addition, other approaches should also be incorporated to develop the contig map into a robust integrated physical and genetic map of the soybean genome. These approaches include BAC end sequencing and alignment, targeted development and mapping of DNA markers from the contigs without markers from the soybean linkage map, comparative sequence analysis of the SSR sites, comparative Southern analysis of RFLP loci, and large-scale EST mapping. The BAC end sequencing and/or EST mapping approaches were previously used in the development of the integrated physical and genetic maps of Arabidopsis (Chang et al. 2001), rice (Chen et al. 2002; Wu et al. 2002), human (The International Human Genome Mapping Consortium 2001) and mouse (Gregory et al. 2002).
The soybean physical map was constructed from the BAC and BIBAC libraries of three genotypes, Forrest, Williams 82, and Faribault. Although >97% of the clones of the physical map were from the Forrest libraries, ∼94% of the clones containing DNA markers were from the Williams 82 and Faribault libraries (Table 1). Fu and Dooner (2002) uncovered the lack of “intraspecific genetic colinearity” in some regions of the maize genome. However, we did not observe in this study sufficient sequence variations between Forrest and Williams 82 or between Forrest and Faribault to cause any significant difficulties for fingerprint analysis and contig assembly. Moreover, we independently constructed, without any problems, a 1-Mb BAC contig by fingerprint analysis using BACs from both the Forrest and Williams 82 libraries for a large disease-resistance gene cluster of the soybean linkage group J including Rmd, Rj2, and Rps2 genes. Such genomic regions were shown to evolve rapidly in the course of genome evolution (C. Wu and H.-B. Zhang, unpubl.). The BACs from Williams 82 and Faribault provided an efficient approach to contig verification and facilitated the physical map construction in this study. Nevertheless, we could not rule out the separate contig assembly of BACs from the genomic regions with sufficient sequence variations between the three soybean genotypes.
METHODS
Source BAC and BIBAC Libraries and DNA Probes
To develop the whole-genome physical map of soybean that is suitable for structural, functional, and comparative genomics research, we used the two BIBAC (Meksem et al. 2000) and one BAC (C. Wu and H.-B. Zhang, unpubl.) libraries of soybean cv. Forrest and the BAC libraries of soybean cv. Williams 82 (Marek and Shoemaker 1997) and cv. Faribault (Danesh et al. 1998; Table 1). These libraries were constructed from the partial digests of nuclear DNA with three restriction enzymes (HindIII, BamHI, and EcoRI) in two different vector systems (bacterial F-factorbased—pBeloBAC 11, pECBAC1, and pECBAC4—and bacterial P1 plasmid-based—pCLD04541). Therefore, they are complementary in genome coverage and would greatly facilitate development of the physical map. The small sizes (about 7.5 kb) of the BAC library vectors, pBeloBAC 11 (Kim et al. 1996), pECBAC1 and pECBAC4 (Frijters et al. 1997), facilitate the utility of the physical map for clone-by-clone-based shotgun genome sequencing. The transformability of the BIBACs in plants facilitates the utility of the physical map in the positional cloning of genes and QTLs important to agriculture, functional analysis of soybean genomic sequences, and gene/QTL engineering by genetic transformation. The Williams 82 and Faribault libraries were previously screened with 389 SSR markers and 223 RFLP markers to construct the regional physical map of the soybean genome (Marek et al. 2001). Inclusion of the positive clones of the markers not only allowed the integration of the regional physical map with the genome-wide physical map developed in this study, but also facilitated the integration of the whole-genome physical map with the existing genetic linkage composite map (Cregan et al. 1999a; http://soybase.agron.iastate.edu/). These libraries are permanently maintained in 384-well microplates and are publicly available at the GENEfinder Genomic Resources (http://hbz.tamu.edu).
The DNA marker probes were selected from the soybean composite genetic map (Cregan et al. 1999a; http://soybase.agron.iastate.edu/). RFLP probes were purchased from Biogenetic Services, Inc.. The inserts of the probe clones were isolated and purified on agarose gels. SSR probes were generated by PCR amplification of the Forrest genomic DNA with the SSR primer pairs obtained from the Soybase (http://soybase.agron.iastate.edu/). The PCR products of the SSR primers were analyzed and purified on agarose gels. In all, 732 RFLP-positive and 1270 SSR-positive BAC clones were selected from the Williams 82 (860 BACs) and Faribault (1142 BACs) BAC libraries (Marek et al. 2001).
BAC and BIBAC Fingerprinting and Contig Assembly
BAC and BIBAC DNA were isolated and fingerprinted according to Chang et al. (2001) and Tao et al. (2001). BAC DNA was double-digested with HindIII and HaeIII, end-labeled with [33P]dATP using reverse transcriptase for 2 h at 37°C, and then subjected to 3.5% (w/v) polyacrylamide DNA sequencing gel electrophoresis at 85 W for 100 min. The gel was dried and autoradiographed.
The fingerprints on the autoradiographs were scanned into image files using a UMAX Mirage D-16L scanner and edited using the Image 4.0 (Soderlund et al. 1997). There was no vector-derived band of the BIBAC clones present in the fingerprint fragment range, and the vector bands derived from the BAC clones were manually removed from the data files. The clones that failed in fingerprinting or had no inserts were deleted during fingerprint editing. Consequently, 78,001 clones, equivalent to 9.580× soybean haploid genomes, were used to assemble the physical map contigs.
To select the tolerance and cutoff values that were suitable for physical map contig assembly of the soybean genome, we first assembled BAC/BIBACs contigs using different tolerance and cutoff values using the FPC program version 4.7 (Soderlund et al. 2000) and then analyzed the contigs containing the RFLP- and SSR-positive clones, especially the positive clones of single-locus SSR markers (single bands on 3.5% Metaphor agarose gels). We assumed that if the contigs were properly assembled, the positive clones of a single-locus DNA marker should be assembled to the same region of a contig. Using this criterion, we conducted a series of tests and finally, tolerance = 2 and cutoff values between 1e - 30 and 1e - 10 were selected for the physical map contig assembly.
The BAC/BIBAC contigs of the soybean genome physical map were assembled as follows. We first assembled automatic contigs under the above criteria and manually edited every automatic contig to ensure that they were accurate. Then we joined automatic contigs into larger contigs using lower stringency cutoff values ranging between 1e - 28 and 1e - 10. The contig pairs were merged if their terminal clones shared 10 or more bands and their overall fingerprint patterns supported joining. We also coalesced the contig pairs if they were anchored to the genetic map (Cregan et al. 1999a) by two or more neighboring DNA markers and formed a single contig under the cutoff values between 1e - 15 and 1e - 10.
BAC and BIBAC Library Screening and Integration of DNA-Marker-Containing BACs
The soybean BAC and BIBAC libraries or the clones of the physical map contigs were double-spotted on Hybond N+ membrane (Amersham) in a 3 × 3 format using the Biomek 2000 robotic workstation (Beckman). The high-density colony filters were prepared according to Zhang et al. (1996). The probes were labeled with [32P]dCTP, and the colony hybridization was performed according to Zhang et al. (1996). The filters were washed three times in 0.1% SDS, 0.5× SSC at 65°C, 30 min each wash.
The positive BAC clones of the Williams 82 and Faribault BAC libraries identified with 264 SSR and 102 RFLP markers (Marek et al. 2001) were fingerprinted, edited, and assembled into contigs along with the Forrest BACs and BIBACs as above. The purposes of this experiment were to integrate the contigs of the Williams 82 and Faribault BAC clones into the genome-wide BAC/BIBAC physical map of the soybean genome under construction and to anchor the BAC/BIBAC contigs that contained the Williams 82 and/or Faribault BACs to the soybean composite genetic map (Cregan et al. 1999a; http://soybase.agron.iastate.edu/).
Acknowledgments
H.B.Z. and C.W. thank Randy Shoemaker, USDA/ARS, Iowa State University, and Nevin D. Young, University of Minnesota for kindly providing the marker-anchored positive clone lists and the soybean Williams 82 and Faribault BAC libraries; and Chantel Scheuring for critically reading the manuscript. This study was supported in part by the National Science Foundation–Plant Genome Program (Award #9872635), the Illinois Soybean Operating Board (Award #ISPOB-24-198-3), and the Texas Agricultural Experiment Station (Acc. #8536-203104).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1405004. Article published online before print in January 2004.
Footnotes
[Supplemental material is available online at www.genome.org and http://hbz.tamu.edu. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: R. Shoemaker, N.D. Young, Z. Xu, and Y.-L. Chang.]
References
- Adams, M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185-2195. [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-815. [DOI] [PubMed] [Google Scholar]
- Arumuganathan, K., and Earle, E.D. 1991. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9: 208-219. [Google Scholar]
- Chang, Y.L., Tao, Q., Scheuring, C., Meksem, K., and Zhang, H.-B. 2001. An integrated map of Arabidopsis thaliana for functional analysis of its genome sequence. Genetics 159: 1231-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M., Presting, G., Barbazuk, W.G., Goicoechea, J.L., Blackmon, B., Fang, G., Kim, H., Frisch, D., Yu, Y., Sun, S., et al. 2002. An integrated physical and genetic map of the rice genome. Plant Cell 14: 537-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente, T.E., LaVallee, B.J., Howe, A.R., Conner-Ward, D., Rozman, R.J., Hunter, P.E., Broyles, D.L., Kasten, D.S., and Hinchee, M.A. 2000. Progeny analysis of glyphosate-selected transgenic soybeans derived from Agrobacterium-mediated transformation. Crop Sci. 40: 797-803. [Google Scholar]
- Coulson, A., Sulston, J., Brenner, S., and Karn, J. 1986. Toward a physical map of the genome of the nematode C. elegans. Proc. Natl. Acad. Sci. 83: 7821-7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregan, P.B., Jarvik, T., Bush, A.L., Shoemaker, R.C., Lark, K.G., Kahler, A.L., Kaya, N., VanToai, T.T., Lohnes, D.J., Chung, J., et al. 1999a. The integrated map of the soybean genome. Crop Sci. 39: 1464-1490. [Google Scholar]
- Cregan, P.B., Mudge, J., Fickus, E.W., Marek, L.F., Danesh, D., Denny, R., Shoemaker, R.C., Matthews, B.F., Jarvik, T., and Young, N.D. 1999b. Targeted isolation of simple sequence repeat markers through the use of bacterial artificial chromosomes. Theor. Appl. Genet. 98: 919-928. [Google Scholar]
- Danesh, D., Penuela, S., Mudge, J., Denny, R.L., Nordstrom, H., Martinez, J.P., and Young, N.D. 1998. A bacterial artificial chromosome library for soybean and identification of clones near a major cyst nematode resistance gene. Theor. Appl. Genet. 96: 196-202. [Google Scholar]
- Ding, Y., Johnson, M.D., Colayco, R., Chen, Y.J., Melnyk, J., Schmitt, H., and Shizuya, H. 1999. Contig assembly of bacterial artificial chromosome clones through multiplexed fluorescence-labeled fingerprinting. Genomics 56: 237-246. [DOI] [PubMed] [Google Scholar]
- Donaldson, P.H. and Simmonds, D.H. 2000. Susceptibility to Agrobacterium tumefaciens and cotyledonary node transformation in short-season soybean. Plant Cell. Rep. 19: 478-484. [DOI] [PubMed] [Google Scholar]
- Frijters, A.C., Zhang, Z., van Damme, M., Wang, G.L., Ronald, P.C., and Michelmore, R.W. 1997. Construction of a bacterial artificial chromosome library containing large EcoRI and HindIII genomic fragments of lettuce. Theor. Appl. Genet. 94: 390-399. [Google Scholar]
- Fu, H. and Dooner, H.K. 2002. Intraspecific violation of genetic colinearity and its implications in maize. Proc. Natl. Acad. Sci. 99: 9573-9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, R.B. 1978. DNA sequence organization in the soybean plant. Biochem. Genet. 16: 45-68. [DOI] [PubMed] [Google Scholar]
- Grant, D., Cregan, P., and Shoemaker, R.C. 2000. Genome organization in dicots: Genome duplication in Arabidopsis and synteny between soybean and Arabidopsis. Proc. Natl. Acad. Sci. 97: 4168-4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, V. 1981. Plant speciation. Columbia University Press, New York.
- Gregory, S.G., Howell, G.R., and Bentley, D.R. 1997. Genome mapping by fluorescent fingerprinting. Genome Res. 7: 1162-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory, S.G., Sekhon, M., Schein, J., Zhao, S., Osoegawa, K., Scott, C.E., Evans, R.S., Burridge, P.W., Cox, T.V., Fox, C.A., et al. 2002. A physical map of the mouse genome. Nature 418: 743-750. [DOI] [PubMed] [Google Scholar]
- Gurley, W.B., Hepburn, A.G., and Key, J.L. 1979. Sequence organization of the soybean genome. Biochem. Biophys. Acta 561: 167-183. [DOI] [PubMed] [Google Scholar]
- Hodgkin, J., Plasterk, R.H.A., and Waterston, R.H. 1995. The nematode Caenorhabditis elegans and its genome. Science 270: 410-414. [DOI] [PubMed] [Google Scholar]
- Hoskins, R.A., Nelson, C.R., Berman, B.P., Laverty, T.R., George, R.A., Ciesiolka, L., Naeemuddin, M., Arenson, A.D., Durbin, J., David, R.G., et al. 2000. A BAC-based physical map of the major autosomes of Drosophila melanogaster. Science 287: 2271-2274. [DOI] [PubMed] [Google Scholar]
- The International Human Genome Mapping Consortium. 2001. A physical map of the human genome. Nature 409: 934-941. [DOI] [PubMed] [Google Scholar]
- The International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Iqbal, M.J., Meksem, K., Njiti, V.N., Kassem, A., and Lightfoot, D.A. 2001. Microsatellite markers identify three additional quantitative trait loci for resistance to soybean sudden-death syndrome (SDS) in Essex × Forrest RILs. Theor. Appl. Genet. 102: 187-192. [Google Scholar]
- Keim, P., Schupp, J.M., Travis, S.E., Clayton, K., Zhu, T., Shi, L., Ferreira, A., and Webb, D.M. 1997. A high density genetic map of soybean based upon AFLP markers. Crop Sci. 37: 537-543. [Google Scholar]
- Kim, U.-J., Birren, B.W., Slepak, T., Mancino, V., Boysen, C., Kang, H.L., Simon, M.I., and Shizuya, H. 1996. Construction and characterization of a human bacterial artificial chromosome library. Genomics 34: 213-218. [DOI] [PubMed] [Google Scholar]
- Lark, K.G., Weisemann, J.M., Matthews, B.F., Palmer, R., Chase, K., and Macalma, T.A. 1993. Genetic map of soybean (Glycine max L.) using an intraspecific cross of two cultivars: `Minsoy' and `Noir 1.' Theor. Appl. Genet. 86: 901-906. [DOI] [PubMed] [Google Scholar]
- Leitch, I.J. and Bennett, M.D. 1997. Polyploidy in angiosperms. Trends Plant Sci. 2: 470-476. [Google Scholar]
- Lundin, L.G. 1993. Evolution of the vertebrate genome as reflected in paralogous chromosome regions in man and the house mouse. Genomics 16: 1-19. [DOI] [PubMed] [Google Scholar]
- Marek, L.F. and Shoemaker, R.C. 1997. BAC contig development by fingerprint analysis in soybean. Genome 40: 420-427. [DOI] [PubMed] [Google Scholar]
- Marek, L.F., Mudge, J., Darnielle, L., Grant, D., Hanson, N., Paz, M., Yan, H., Denny, R., Larson, K., Foster-Hartnett, D., et al. 2001. Soybean genomic survey: BAC-end sequences near RFLP and SSR markers. Genome 44: 572-581. [PubMed] [Google Scholar]
- Marra, M.A., Kucaba, T.A., Dietrich, N.L., Green, E.D., Brownstein, B., Wilson, R.K., McDonald, K.M., Hillier, L.W., McPherson, J.D., and Waterston, R.H. 1997. High-throughput fingerprint analysis of large-insert clones. Genome Res. 7: 1072-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra, M., Kucaba, T., Sekhon, M., Hillier, L., Martienssen, R., Chinwalla, A., Chinwalla, A., Crockett, J., Fedele, J., Grover, H., et al. 1999. A map for sequence analysis of the Arabidopsis thaliana genome. Nat. Genet. 22: 265-270. [DOI] [PubMed] [Google Scholar]
- Masterson, J. 1994. Stomatal size in fossil plants: Evidence for polyploid in majority of angiosperms. Science 264: 421-424. [DOI] [PubMed] [Google Scholar]
- Meksem, K., Ruben, E., Zobrist, K., Zhang, H.-B., and Lightfoot, D.A. 2000. Two large insert libraries for soybean: Applications in cyst nematode resistance and genome wide physical mapping. Theor. Appl. Genet. 101: 747-755. [Google Scholar]
- Mozo, T., Dewar, K., Dunn, P., Ecker, J.R., Fischer, S., Kloska, S., Lehrach, H., Marra, M., Martienssen, R., Meier-Ewert, S., et al. 1999. A complete BAC-based physical map of the Arabidopsis thaliana genome, Nat. Genet. 22: 271-275. [DOI] [PubMed] [Google Scholar]
- Postlethwait, J.H., Yan Y.-L., Gates, M.A., Horne, S., Amores, A., Brownlie, A., Donovan, A., Egan, E.S., Force, A., Gong, Z., et al. 1998. Vertebrate genome evolution and the zebrafish genetic map. Nat. Genet. 18: 345-349. [DOI] [PubMed] [Google Scholar]
- Ren, C., Lee, M.-K., Yan, B., Ding, K., Cox, B., Romanov, M.N., Price, J.A., Dodgson, J.B., and Zhang, H.-B. 2003. A BAC-based physical map of the chicken genome. Genome Res. 13: 2754-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimath, S.S. and Bhattacharyya, M.K. 1999. Generation of a soybean BAC library, and identification of DNA sequences tightly linked to the Rps-1k disease resistance gene. Theor. Appl. Genet. 98: 712-720. [Google Scholar]
- Shoemaker, R.C., and Specht, J.E. 1995. Integration of the soybean molecular and classical genetic linkage groups. Crop Sci. 35: 436-446. [Google Scholar]
- Shoemaker, R.C., Polzin, K.C., Labate, J., Specht, J.E., Brummer, E.C., Olson, T., Young, N., Concibido, V., Wilcox, J., Tanmulonis, J.P., et al. 1996. Genome duplication in soybean (Glycine subgenus soja). Genetics 144: 329-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker, R., Keim, P., Vodkin, L., Retzel, E., Clifton, S.W., Waterston, R., Smoller, D., Coryell, V., Khanna, A., Erpelding, J., et al. 2002. A compilation of soybean ESTs: Generation and analysis. Genome 45: 329-338. [DOI] [PubMed] [Google Scholar]
- Sidow, A. 1996. Gen(ome) duplication in the evolution of early vertebrates. Curr. Opin. Genet. Dev. 6: 715-722. [DOI] [PubMed] [Google Scholar]
- Singh, R.J. and Hymowitz, T. 1988. The genomic relationship between Glycine max (L.) Merr. and G. soja Sieb. and Zucc. as revealed by pachytene chromosomal analysis. Theor. Appl. Genet. 76: 705-711. [DOI] [PubMed] [Google Scholar]
- Soderlund, C., Longden, I., and Mott, R. 1997. FPC: A system for building contigs from restriction fingerprinted clones. CABIOS 13: 523-535. [DOI] [PubMed] [Google Scholar]
- Soderlund, C., Humphray, S., Dunham, A., and French, L. 2000. Contigs built with fingerprints, markers, and FPC V4.7. Genome Res. 10: 1772-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spring, J. 1997. Vertebrate evolution by interspecific hybridization: Are we polyploid? FEBS Lett. 400: 2-8. [DOI] [PubMed] [Google Scholar]
- Sulston, J., Mallett, F., Staden, R., Durbin, R., Horsnell, T., and Coulson, A. 1988. Software for genome mapping by fingerprinting techniques. CABIOS 4: 125-132. [DOI] [PubMed] [Google Scholar]
- Tao, Q. and Zhang, H.-B. 1998. Cloning and stable maintenance of DNA fragments over 300 kb in Escherichia coli with conventional plasmid-based vectors. Nucleic Acids Res. 26: 4901-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, Q., Chang, Y.L., Wang, J., Chen, H., Islam-Faridi, M.N., Scheuring, C., Wang, B., Stelly, D.M., and Zhang, H.-B. 2001. BAC-based physical map of the rice genome constructed by restriction fingerprint analysis. Genetics 158: 1711-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter, J.C., Smith, H.O., and Hood, L. 1996. A new strategy for genome sequencing. Nature 381: 364-366. [DOI] [PubMed] [Google Scholar]
- Wolfe, K.H. 2001. Yesterday's polyploids and the mystery of diploidization. Nat. Rev. 2: 333-341. [DOI] [PubMed] [Google Scholar]
- Wu, J., Maehara, T., Shimokawa, T., Yamamoto, S., Harada, C., Takazaki, Y., Ono, N., Mukai, Y., Koike, K., Yazaki, J., et al. 2002. A comprehensive rice transcript map containing 6591 expressed sequence tag sites. Plant Cell 14: 525-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H.-B. and Wing, R.A. 1997. Physical mapping of the rice genome with BACs. Plant Mol. Biol. 35: 115-127. [PubMed] [Google Scholar]
- Zhang, H.-B. and Wu, C. 2001. BAC as tools for genome sequencing. Plant Physiol. Biochem. 39: 195-209. [Google Scholar]
- Zhang, H.-B., Choi, S., Woo, S.-S., Li, Z., and Wing, R.A. 1996. Construction and characterization of two rice bacterial artificial chromosome libraries from the parents of a permanent recombinant inbred mapping population. Mol. Breed 2: 11-24. [Google Scholar]
WEB SITE REFERENCES
- http://hbz.tamu.edu; the GENEfinder Genomic Resources, Texas A&M University.
- http://soybase.agron.iastate.edu/; Soybase.
- http://soybean.ccgb.umn.edu/; The Center for Computational Genomics and Bioinformatics, University of Minnesota.
- http://www.siu.edu/~pbgc/DataBase/datap1.htm; Plant Biotechnology and Genome Core Facility, Southern Illinois University at Carbondale.