SUMMARY
Polyglutamine (polyQ) stretches exceeding a threshold length confer a toxic function on proteins that contain them and cause at least nine neurological disorders. The basis for this toxicity threshold is unclear. Although polyQ expansions render proteins prone to aggregate into inclusion bodies (IBs), IB formation may be a neuronal coping response to more toxic forms of polyQ. The exact structure of these more toxic forms is unknown. Here we show that monoclonal antibody (mAb) 3B5H10 recognizes a species of polyQ protein in situ that strongly predicts neuronal death. The epitope selectively appears among some of the many low-molecular weight conformational states expanded polyQ assumes and disappears in higher molecular-weight aggregated forms, such as IBs. These results suggest that protein monomers and possibly small oligomers containing expanded polyQ stretches can adopt a conformation that is recognized by 3B5H10 and is toxic or closely related to a toxic species.
INTRODUCTION
Misfolding and self-aggregation of specific proteins are a common feature of most common age-related neurodegenerative diseases, including Huntington’s disease (HD), Alzheimer’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis. In HD, an abnormal expansion in the polyglutamine (polyQ) stretch of the huntingtin protein (htt) results in protein misfolding and neurodegeneration, especially in the striatum1. Eight proteins containing polyQ tracts, but otherwise unrelated to htt, also result in protein misfolding and neurodegeneration upon polyQ expansion2. For each of these “proteinopathies,” an open question is which of the many putative misfolded conformations and/or aggregated states of the culprit protein is responsible for neurodegeneration.
To determine the species of misfolded proteins that are critical for disease pathogenesis, tools for detecting species that form naturally in live neurons are needed. Unfortunately, with the exception of some recently developed antibodies that recognize specific secondary and tertiary protein structures3–7, tools are generally lacking to quantify and distinguish among simultaneously existing protein species in situ.
Even with antibody “tools” to identify multiple misfolded species in situ, it is difficult to determine the pathologic significance of any single species within the context of other simultaneously present species. In clinical research, survival-based statistical techniques, such as Cox analysis, are routinely employed to determine how each of multiple, simultaneously present, putative risk factors relates to an outcome of interest8,9. Analogously, when multiple species of misfolded protein are simultaneously present in a neuron, each species can be related to neuronal death by survival-based statistical techniques, such as Cox analysis. To employ survival analysis, however, the levels of multiple protein species in each of thousands of neurons must be measured, and the survival time for each neuron must be determined.
In HD, formation of the end-stage, very large aggregated species of mutant htt (mHtt), termed an inclusion body (IB), can be a coping response10,11. IBs may sequester toxic species of mHtt, which are distributed diffusely throughout the neuron. However, the species of diffuse mHtt that leads to neurodegeneration remains elusive.
Here, we developed novel methods that allowed us to identify and characterize pathogenic species within the diffuse fraction of mHtt in neurons. First, we used conformation-specific antibodies to distinguish among species of htt that exist in situ. Next, we assessed how well each of these species predicted survival in thousands of neurons by individually tracking the neurons over long periods of time with an automated microscope10,12. Finally, we applied a modified form of Cox analysis to this dataset to understand which species best predicted survival and then biochemically and structurally characterized this species. Our results show that, from the antibodies tested, monoclonal antibody 3B5H10 recognizes a species of mHtt in situ that best predicts neuronal death. This epitope is exposed in certain conformations of monomeric and possibly small oligomeric polyQ species but disappears in higher-molecular weight aggregated forms, such as IBs. Therefore, protein monomers and possibly small oligomers containing disease-associated polyQ can adopt a conformation recognized by 3B5H10 that is pathogenic or closely related to a pathogenic species.
RESULTS
Developing novel α-polyglutamine monoclonal antibodies
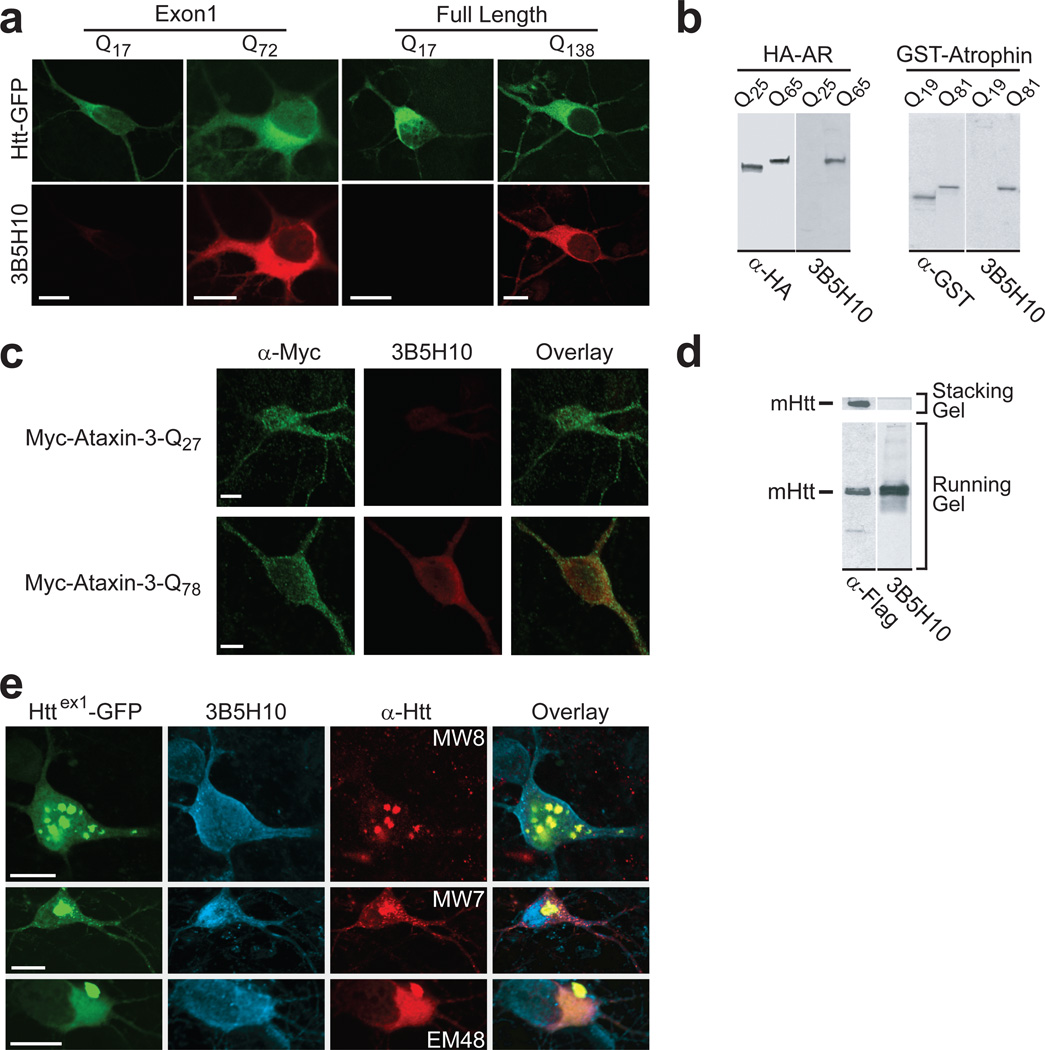
We reasoned that antibodies might be useful probes to distinguish species of diffuse htt in situ and possibly to identify the species most tightly linked to neurodegeneration. We immunized six mice against a natively prepared GST-N-terminal fragment of htt containing the first 171 amino acids and a disease-associated polyQ (Q66) expansion. Among 480 hybridomas, six produced monoclonal antibodies (mAbs) that preferentially bound mHtt (Supplementary Fig. 1 online). One, 3B5H10, was further characterized. By immunocytochemistry, we observed that 3B5H10 preferentially labeled neurons transiently expressing disease-associated polyQ expansions in full-length13 or the exon1 fragment of htt (httex1)10 (Fig. 1a). 3B5H10 specifically recognizes the polyQ expansion in htt, as the antibody binds a synthetic polyQ (K2Q39K2) peptide as seen by SELDI-TOF-MS (Supplementary Fig. 2 online) and recognizes disease-associated polyQ expansions in other neurodegeneration-causing proteins that otherwise share no homology with each other or with htt2 (e.g., androgen receptor14, atrophin15, and ataxin-316) (Fig. 1b,c).
Figure 1.
mAb 3B5H10 binds low molecular weight disease-associated polyQ expansions. (a) 3B5H10 preferentially labeled striatal neurons transiently expressing disease-associated polyQ expansions in an exon1 fragment or full-length htt. Striatal neurons transfected with Httex1-eGFP (Q17, Q72) or GFP-Full-Length-Htt (Q17, Q138) (top row; green) were labeled with mAb 3B5H10 (bottom row; red) and imaged by confocal microscopy. Scale bar=10 µm. (b, c) 3B5H10 recognizes disease-associated polyQ expansions in other neurodegeneration-causing proteins. (b) HEK293 extracts containing HA epitope–tagged androgen receptor (AR) (wt=Q25, mutant=Q65)14 or GST-tagged atrophin fragments (wt=Q19, mutant=Q81)15 were blotted with 3B5H10 and α-HA or α-GST antibodies, respectively. 3B5H10 preferentially recognized versions with disease-associated polyQ expansions. (c) Striatal neurons transfected with Myc-Ataxin-3 (wt=Q27, mutant=Q78)16 were labeled with α-Myc polyclonal and 3B5H10 mAb and imaged by confocal microscopy. α-Myc (green) recognizes both wt and mutant ataxin-3, whereas 3B5H10 (red) preferentially labeled mutant ataxin-3. Scale bar=5 µm. (d–e) 3B5H10 recognizes visibly non-aggregated, diffuse forms of mHtt. (d) Protein extracts from HEK293 cells expressing FLAG epitope-tagged mHtt (171-Q68-FLAG) were blotted with α-FLAG or 3B5H10. Aggregated forms of mHtt that are retained in the stacking portion of the gel (see α-FLAG lane) selectively lose 3B5H10 immunoreactivity. (e) Striatal neurons transfected with Httex1-(Q46, Q72, or Q97)-eGFP were labeled with Alexa 647-conjugated 3B5H10 and MW8, MW7, or EM48 α-htt antibodies. Fluorescence from GFP (green), Alexa 647 (blue), and Cy3-conjugated secondary antibodies (red) to detect MW8, MW7, or EM48 was collected with confocal microscopy. Scale bar=10 µm.
In western blots of cell lysates transfected with fragments of mHtt, 3B5H10 did not recognize aggregated species that remained in the stack (Fig. 1d). Immunocytochemistry and immunogold electron microscopy with striatal neurons transfected with mutant httex1 revealed that 3B5H10 recognized diffuse mHtt but not IBs (Fig. 1e, Supplementary Fig. 3 online). In contrast, several other α-htt antibodies recognized both diffuse mHtt and IBs (MW7, which recognizes the polyproline region of htt located immediately C-terminal to the polyQ stretch4, and EM48, which was raised against the first 256 amino acids of htt without a polyQ stretch17) or just IBs (MW8, which recognizes the AEEPLHRPK epitope near the polyproline region of htt4)(Fig. 1e).
In brain tissue sections from HD mouse models, we confirmed that 3B5H10 recognizes diffuse mHtt over IBs (Supplementary Fig. 4 online). In 12-month-old BACHD mice, an HD model expressing full-length mHtt (Q97)18, aggregates are relatively abundant, but 3B5H10 primarily stained diffuse mHtt. When aggressive antigen retrieval using formic acid was employed, 3B5H10 staining of BACHD tissue revealed more prominent staining of neuropil aggregates (data not shown). In 15-week-old R6/2 mice, an HD model expressing an N-terminal fragment of mHtt (~Q150)19, essentially all mHtt is aggregated into IBs, and 3B5H10 staining is poor. However, after R6/2 brain tissue is treated with 90% formic acid, 3B5H10 staining (primarily of IBs) increases. 3B5H10 staining after these aggressive antigen retrievals is likely attributable to the unmasking of polyQ epitopes not normally exposed in vivo.
We also confirmed that 3B5H10 recognizes full-length mutant ataxin-3 in another animal model of polyQ neurodegeneration, the YAC-SCA3 mouse20 (Supplementary Fig. 4 online). Together, our results suggest that select antibodies can distinguish among different species of polyQ protein in situ7.
Decoding pathogenic significance of htt species in situ
We next sought to understand the pathogenic significance of epitopes recognized by 3B5H10 and other α-htt antibodies. To do so, we turned to multivariate Cox analysis. Cox analysis is one of the most widely used survival models in clinical research for discovering and measuring factors that predict an outcome of interest8,9. The technique allows one to quantitatively rank how important numerous risk factors present at the same time are for an outcome of interest.
Applying multivariate survival analysis to the question of which mHtt epitope (simultaneously-present risk factors) best predicts neurodegeneration requires an estimation of the amount of each epitope in individual live neurons and a determination of how long each of those neurons lives. However, to estimate the amount of each epitope in individual live neurons, we were faced with the conundrum that antibody staining requires fixation. Thus, while we could measure epitope levels in a given neuron, we couldn’t then directly analyze the survival time of that neuron.
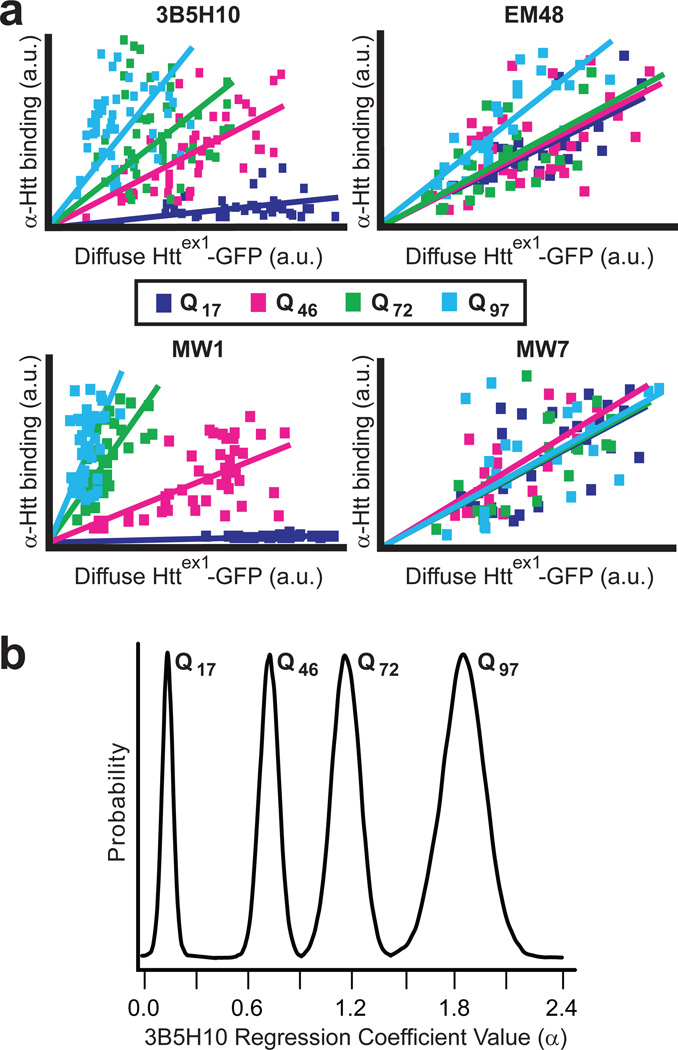
To overcome this issue, we first measured how the abundance of different htt epitopes in striatal neurons recognized by these antibodies varied with incremental changes in htt expression and polyQ length in situ. We transfected striatal neurons with httex1 fused to enhanced GFP (eGFP) and containing Q17, Q46, Q72, or Q97 (Httex1-Qn-eGFP), fixed the neurons, and then compared mAbs 3B5H10, MW7, EM48, and MW14 by immunostaining experiments. Similar to 3B5H10, MW1 preferentially binds diffuse mHtt; however, it evidently binds to an extended unfolded polyQ stretch21, whereas 3B5H10 binds to a compact structure of polyQ (unpublished observations). We found that the four antibodies differed significantly from each other in their quantitative binding to diffuse htt in situ (Fig. 2a). Particularly interesting was the fact that 3B5H10 and MW1, despite both preferentially recognizing expanded polyQ stretches, exhibit distinct quantitative binding profiles, suggesting that they are distinguishing different conformational epitopes of polyQ in situ. More importantly, the binding of each one of the antibodies tested was predictable and estimable from the length of the polyQ expansion and the level of diffuse htt. Therefore, to estimate the amount of each epitope in individual live neurons, we can measure a live neuron’s diffuse htt-eGFP levels (via eGFP fluorescence in live fluorescence microscopy) and then mathematically infer the amount of each epitope in that neuron with data from Figure 2a and regression analysis. Using regression analysis, however, means that the amount of each epitope estimated for a given neuron carries an inherent estimation error. To account for this uncertainty in estimation, we applied Bayesian regression analysis, a technique whose output is a probability plot of regression coefficient values (Fig. 2b, Supplementary Fig. 5 online).
Figure 2.
Quantitative binding of α-htt antibodies 3B5H10, EM48, MW1, or MW7 to httex1 is distinguishable and predictable. (a) 3B5H10, EM48, MW1, and MW7 differ significantly in their quantitative binding to diffuse htt in situ, suggesting each recognizes a unique htt species. Quantitative binding for each of the antibodies to httex1 can be estimated by regression analysis when the fluorescence of the eGFP tag fused to httex1 and httex1’s polyQ length are known. Striatal neurons transfected with Httex1-(Q17,Q46,Q72, or Q97)-eGFP were fixed at 24 hours and subjected to immunocytochemistry with one of the four α-htt antibodies. Fluorescence was measured by confocal microscopy (17–48 neurons per condition). For this analysis, only neurons without IBs were measured. (b) The significant data scatter around the linear regression lines in Figure 2a suggested that predicting the amount of antibody binding to a given mHtt-transfected neuron carries significant estimation error. To account for this error, we reanalyzed the data in Figure 2a with Bayesian statistics. The output of Bayesian regression analysis is a probability plot demonstrating how likely the actual regression coefficient (α) is a particular value. Bayesian regression plots for 3B5H10 are presented in Figure 2b, while α for the other antibodies are presented in Supplementary Figure 5 online.
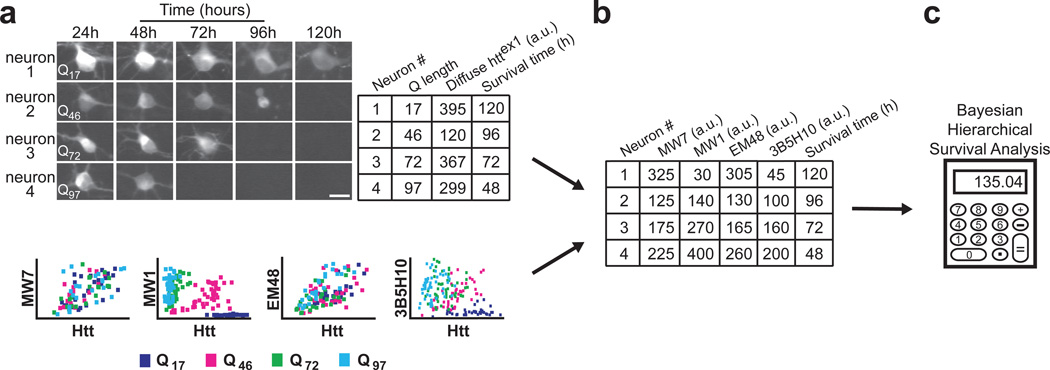
With the ability to estimate how much of each mHtt epitope exists in a given live neuron, we next turned to determining how long that neuron lives. Such data allow us to relate multiple, simultaneously present risk factors (mHtt epitopes) to an outcome of interest (neuronal death) by multivariate Cox analysis. We again transfected striatal neurons with httex1 fused to eGFP and containing Q17, Q46, Q72, or Q97, and we performed longitudinal survival analysis. Beginning approximately 24 hours after transfection, thousands of those neurons were tracked individually and daily for 7–10 days with an automated microscope. Diffuse levels of htt in individual neurons that did not form IBs during the total experimental time were quantified by measuring eGFP intensity10. Simultaneously, we determined how long each neuron lived. Thus, using the Bayesian regression analysis in Figure 2b, we were able to assemble a dataset that contained the htt epitope levels and survival times for thousands of individual neurons (Fig. 3).
Figure 3.
Novel methodology distinguishes which of several simultaneously existing in situ epitopes of diffuse httex1 best predicts neurotoxicity. (a) The survival of individual neurons and the levels of diffuse Httex1-(Q17,Q46,Q72, or Q97)-eGFP they contained were determined by automated microscopy and recorded in a spreadsheet. Next, we used the regression coefficients from Figure 2 to relate diffuse Httex1-Qn-eGFP levels to antibody binding values (bottom; copied from Figure 2a). To account for the inaccuracy inherent in estimating antibody binding values from the graphs at bottom, we technically calculated regression coefficients using Bayesian methods (Fig. 2b, Supplementary Fig. 5 online). Scale bar=25 µm. (b) Using the regression coefficients from the plots in Figure 2, we can estimate how much antibody staining would have occurred in each neuron. These antibody staining values are then noted in the spreadsheet. (c) Finally, using each neuron’s estimated amount of antibody staining and its survival time, we compared the epitopes with each other using Bayesian hierarchical survival (Cox) analysis, which determines the antibody that best predicts degeneration. Hierarchical Bayesian methods ensure that “estimation errors” from each step of the analysis propagate through to the final readout. A more detailed schematic of our approach is illustrated in Supplementary Figure 6 online and detailed in Supplementary Methods online.
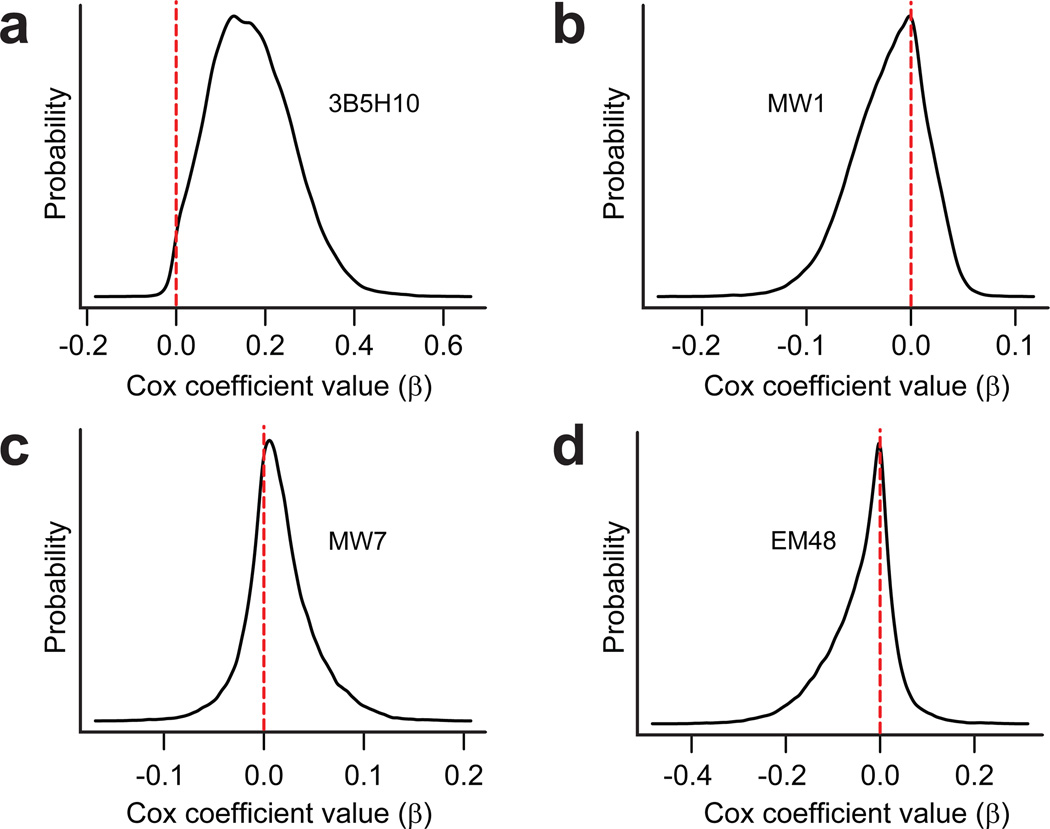
However, to employ multivariate Cox analysis on this dataset, we had to account for the inherent uncertainty in epitope values, an uncertainty which derived from having to estimate these values by Bayesian regression. We addressed this estimation error issue with a well-established method called hierarchical Bayesian analysis22,23. Details of this hierarchical model are presented in Supplementary Methods online and depicted in Figure 3 and Supplementary Figure 6 online. When comparing the prognostic values of epitopes recognized by 3B5H10, MW1, EM48, and MW7, we discovered that only the 3B5H10 epitope significantly predicts if and when neurodegeneration will occur (Fig. 4, Table 1).
Figure 4.
A species of htt recognized by 3B5H10 best predicts striatal neurodegeneration. The significance to neurodegeneration of htt species formed in situ and distinguished by 3B5H10, EM48, MW1, or MW7 was assessed by Cox analysis with a hierarchical Bayesian statistical approach (Fig. 3). Each graph plots the Cox coefficient (β) value for a particular antibody on the x axis and the probability of that coefficient value on the y axis. A positive coefficient signifies that antibody staining is associated with decreased survival. A negative coefficient signifies improved survival. (a) Cox coefficient (β) distribution for 3B5H10. (b) Cox coefficient (β) distribution for MW1. (c) Cox coefficient (β) distribution for MW7. (d) Cox coefficient (β) distribution for EM48.
TABLE 1.
Mean of the probability distributions for each antibody’s Cox coefficient (β) from Figure 4. Of the four antibodies, 3B5H10 bound a species of httex1 that predicted degeneration the best (mean probability (P) of 3B5H10’s Cox coefficient [Coef] is positive (~99% of the area under the curve is above 0)). Cox coefficients associated with the other antibodies tested had probability distributions centered around zero.
| Cox analysis results | ||
|---|---|---|
| Antibody | Mean(Coef)# | P(Coef<0) |
| 3B5H10 | 0.169 | 0.0109 |
| MW1 | −0.0216 | 0.704 |
| MW7 | 0.0157 | 0.302 |
| EM48 | −0.0404 | 0.713 |
Coef = Cox coefficient (β)
Positive Coef predicts decreased survival
Negative Coef predicts increased survival
We were surprised that our analysis demonstrated that 3B5H10 is a better predictor of neurodegeneration than MW1, given that the binding of both antibodies is grossly correlated to the length of the polyQ expansion. However, closer inspection of each antibody’s increased binding with longer polyQ stretches reveals differences in the rate of increase between 3B5H10 and MW1 (Fig. 2a). Picking up on these differences, survival analysis concludes that the pattern of increase in the case of 3B5H10 more closely predicts the corresponding changes in risk of death than MW1. To investigate further the ability of the system to resolve subtle differences in the prognostic power of antibodies that recognize polyQ expansions, we evaluated the commonly used mAb 1C2, which was raised against the polyQ-containing transcription factor TBP. We found that the relationship of 1C2 binding to htt expression was intermediate between MW1 and 3B5H10 (Supplementary Fig. 7 online). However, the differences in the pattern of binding between 3B5H10 and 1C2 were smaller than those between 3B5H10 and MW1; small enough, in fact, to exceed the sensitivity limits of survival analysis. In general, when differences between two risk factors in a survival model fall below a certain threshold, the survival analysis loses reliable biological meaning.
3B5H10 binds a very small species of htt
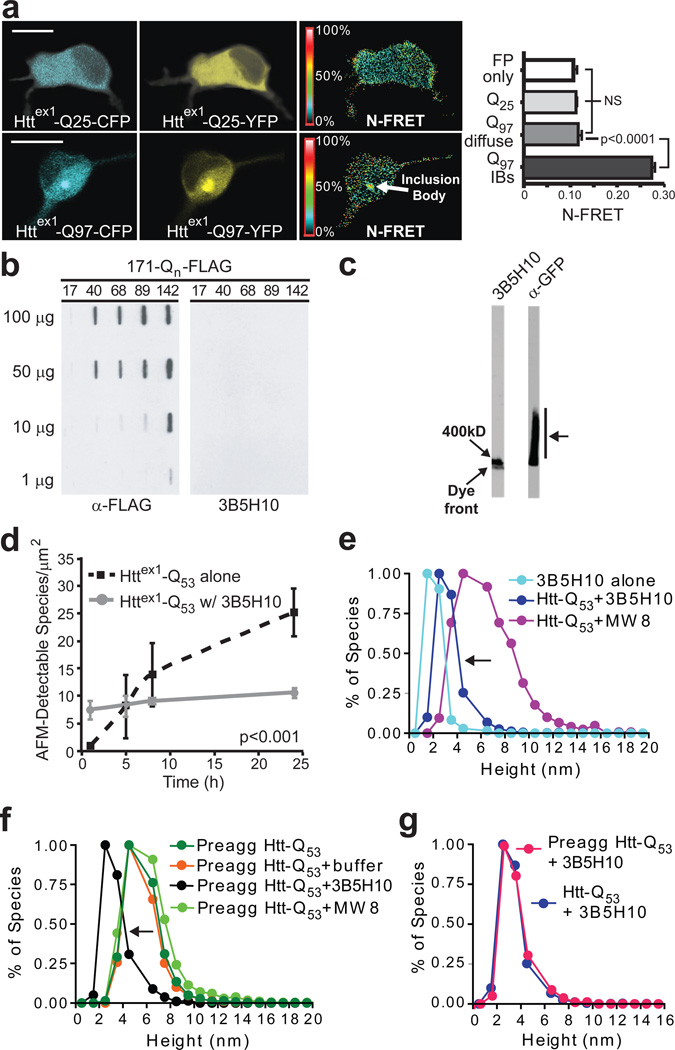
Given the prognostic value of the 3B5H10 epitope, we sought to characterize this epitope further. While 3B5H10 does not recognize IBs (Fig. 1), we considered whether 3B5H10 might bind oligomers or other aggregation intermediates in the diffuse htt fraction of neurons since these species have been proposed to be toxic in HD, PD, and AD24,25. To search for oligomers in live neurons in the diffuse fraction of mHtt, we measured concentration-corrected Förster resonance energy transfer (N-FRET)26 between htt fragments. Primary cortical neurons transfected with httex1 fragments containing wild-type (wt) (Q25) or expanded (Q97) polyQ stretches and tagged with cyan fluorescent protein (CFP) or yellow fluorescent protein (YFP) (Httex1-Q25-CFP, Httex1-Q25-YFP, Httex1-Q97-CFP, Httex1-Q97-YFP) were imaged27. Although we detected significant FRET from mHtt within IBs (Fig. 5a), FRET levels in regions of neurons containing diffuse htt were low and similar whether that neuron contained mutant or wt-htt.
Figure 5.
3B5H10 does not recognize large oligomers of mHtt. (a) N-FRET (mean±s.d.) from cortical neurons was high in IBs formed from Httex1-Q97-CFP and Httex1-Q97-YFP (Q97 IBs) but low in regions of neurons with diffuse mHtt (Q97 diffuse) and not significantly different than in neurons with Httex1-Q25-CFP and Httex1-Q25-YFP (Q25) or CFP and YFP (FP only). Scale bar=10 µm. (b) HEK293 extracts containing Htt-171-(Q17, Q40, Q68, Q89, or Q142)-FLAG were loaded on a 0.20-µm membrane slot-blot and probed with α-FLAG or 3B5H10. α-FLAG revealed submicroscopic aggregates of Htt-171-(Qn)-FLAG that were unrecognizable by 3B5H10. (c) Agarose gel electrophoresis of extracts from PC12 cells stably expressing truncated (no polyproline) Httex1-Q103-GFP were blotted with α-GFP or 3B5H10. While oligomeric Httex1-Q103-GFP was present (arrow), 3B5H10 only stained the dye front, which represents monomer and possibly small oligomers. (d) 3B5H10 prevents mHtt aggregation detected by AFM. Httex1-Q53 aggregation was triggered with or without 3B5H10. (e) AFM-detectable species observed with 3B5H10 alone or with 3B5H10 or MW8 added to monomeric Httex1-Q53. 3B5H10 addition to htt stabilizes a 2–3 nm globular species, consistent with the size of a monomeric htt:antibody complex. (f) AFM-detectable species observed with 3B5H10, MW8, buffer, or nothing added to pre-aggregated oligomers of Httex1-Q53. 3B5H10 had similar effects when added to pre-formed fibrils (Supplementary Fig. 10 online). (g) The final size of AFM-detectable species when 3B5H10 is added to monomeric Httex1-Q53 (Fig. 5e) or pre-aggregated oligomers (Fig. 5f) is statistically indistinguishable (Spearman’s rank correlation coefficient). This size is most consistent with a monomeric htt:antibody complex.
Since a FRET signal from oligomeric association of mHtt in the SH-SY5Y cell line has been previously reported with monomeric versions of CFP and YFP (mCFP/mYFP) attached to httex1 25, we transfected striatal neurons with these previously reported constructs (Httex1-Q17-mCFP, Httex1-Q17-mYFP, Httex1-Q58-mCFP, Httex1-Q58-mYFP) but again found that FRET levels in regions of neurons containing diffuse mHtt were low and not significantly different from wt-htt (Supplementary Fig. 8a online). In a final attempt to increase the sensitivity of our FRET assay, we transfected striatal neurons with wt (Q17) and mutant (Q46) httex1 constructs C-terminally tagged with CyPet or YPet, a pair of fluorophores evolutionarily optimized to improve sensitivity over traditional FRET pairs like CFP and YFP28. Once again, the FRET signals from the diffuse fraction of mutant and wt-htt transfected neurons were indistinguishable (Supplementary Fig. 8b online).
Since we were unable to obtain a FRET signal in the diffuse fraction of mHtt transfected live neurons, we attempted to detect oligomeric intermediates in the diffuse fraction through immunocytochemical staining with an α-oligomer antibody that recognizes polyQ protofibrillar oligomers3. While in vitro aggregation of Httex1-Q5329 led to α-oligomer antibody staining on a dot-blot, the antibody failed to label diffuse mHtt in striatal neurons by immunocytochemistry despite labeling and fixation conditions that were highly analogous to our dot-blot procedure (Supplementary Methods online; data not shown). Thus, we were unable to experimentally demonstrate the presence of aggregated species in the diffuse mHtt fraction of neurons in situ.
Despite our failure to detect oligomeric species in the diffuse mHtt fraction of neurons, 3B5H10 might still be more sensitive at detecting htt aggregation intermediates than FRET or the α-oligomer antibody. To test this possibility in vitro, we performed a filter retardation assay with extracts from HEK293 cells transfected with N-terminal 171–amino acid fragments of htt containing different polyQ stretches (Q17, Q40, Q68, Q89, Q142) and a C-terminal FLAG epitope (Htt-171-Qn-FLAG)30. FLAG blotting revealed that aggregation intermediates were formed and retained on the membrane (> 0.20 µm); however, 3B5H10 failed to detect these aggregates (Fig. 5b).
We were concerned that the SDS concentrations used in our filter retardation assay may disrupt the stability of some htt oligomers, so we turned to agarose gel electrophoresis, which utilizes a reduced SDS concentration, no reducing agent, and no heating of the sample before gel loading. Using extracts from a PC12 cell line stably expressing a fragment of mHtt tagged to GFP (Q103)31, we detected a range of oligomers with an α-GFP antibody. However, 3B5H10 failed to recognize these oligomers, binding only to the leading edge of the agarose gel (Fig. 5c). Thus, 3B5H10 appears to bind only a monomeric or small oligomeric species of mHtt, even in the presence of larger oligomeric species.
To investigate whether 3B5H10 binds to smaller oligomeric structures, we used atomic force microscopy (AFM) and GST-Httex1-Q5329. We reasoned that having 3B5H10 present upon GST cleavage and subsequent Httex1-Q53 aggregation might stabilize the species that it binds and arrest the production of more aggregated species. 3B5H10 addition almost completely prevented the expected time-dependent increase in oligomer and fibril formation by Httex1-Q53 (Fig. 5d). Confirming our AFM studies, dynamic light scattering (DLS) performed on a solution of purified httex1 with an N-terminal thioredoxin tag and a C-terminal hexahistidine tag (Thio-Httex1-Q39-His6) showed that 3B5H10 addition inhibited aggregation and kept particle size small and stable for up to a month (Supplementary Fig. 9 online). AFM measurements of the height (nm) of the mHtt species formed in the presence of 3B5H10 revealed that these globular structures were most consistent with complexes of a single antibody and single mHtt molecule. The structures were larger than samples of 3B5H10 alone but significantly smaller than structures that emerged when Httex1-Q53 was incubated with MW8, an antibody that recognizes aggregated forms of mHtt (Fig. 5e)7.
Remarkably, we also found that the addition of 3B5H10 to pre-formed oligomeric structures led to their disaggregation, as monitored by AFM (Fig. 5f). In contrast, pre-formed oligomers persisted after the addition of buffer or MW8. When the 3B5H10 dissolution process was allowed to proceed to completion, 3B5H10 dissolved pre-formed oligomers into AFM-detectable species that had the dimensions of a monomer of mHtt complexed to 3B5H10 (Fig. 5g). Thus, even when oligomers are present, 3B5H10 appears incapable of stably binding these structures. Importantly, no SDS was used in these experiments, so oligomeric species that would have been unstable in the minimal SDS of agarose gel electrophoresis experiments (Fig. 5c) were presumably available for 3B5H10 binding. We also discovered that 3B5H10 dissolves not just oligomers, but also pre-formed fibrils in a dose- and time-dependent manner (Supplementary Fig. 10 online). In contrast, 3B5H10 had no effect on fibrils formed by a different amyloidgenic protein, α-synuclein. Nor did MW8 disrupt mHtt fibrils (data not shown). Notably, the final size of pre-formed fibrils dissolved by 3B5H10 is the same as structures formed when 3B5H10 is added to monomeric mHtt, a size most consistent with a complex of monomeric mHtt and antibody7.
3B5H10 preferentially recognizes monomers of mHtt
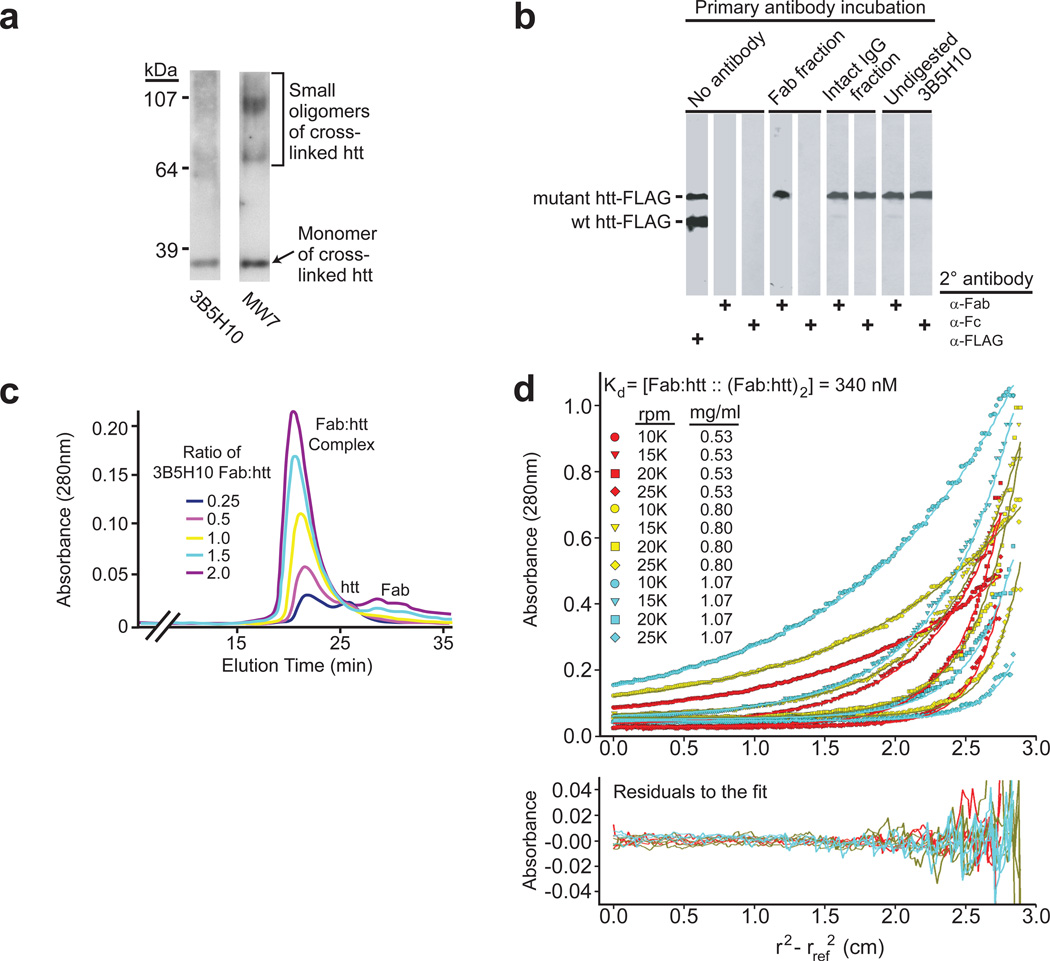
While our results suggested that 3B5H10 preferentially recognizes a very small species of mHtt, we sought more definitive experimental validation by cross-linking experiments, size-exclusion chromatography, and sedimentation equilibrium analytical ultracentrifugation. Because 3B5H10’s ability to slowly dissociate htt oligomers could confound our analysis of the species it binds, we tested the antibody’s ability to bind monomers and small oligomers of htt pre-stabilized via chemical cross-linking. Blotting of cross-linked htt by MW7 revealed the presence of monomers and small oligomeric species; however, the epitope that 3B5H10 recognizes is preferentially present in monomers, although a lesser amount of it may also be present in small oligomers (Fig. 6a).
Figure 6.
Epitope recognized by 3B5H10 is preferentially present in low molecular weight htt species. (a) A solution of Httex1-Q53 was chemically cross-linked and probed with MW7 or 3B5H10. MW7 staining confirms the presence of monomers and small oligomers. The epitope 3B5H10 recognizes is preferentially present in monomers over small oligomers (Full blot: Supplementary Figure 11 online). (b) Monovalent 3B5H10 Fab retains specificity for mHtt over wt-htt, similar to intact bivalent 3B5H10. 3B5H10 Fab was cleaved from the intact IgG by papain proteolysis and purified by ion-exchange and size-exclusion chromatography. HEK293 extracts containing Htt-171-(Q17 or Q68)-FLAG were combined and blotted with i) vehicle, ii) chromatographic fractions corresponding to purified Fab or intact 3B5H10 IgG, or iii) undigested 3B5H10. This was followed by secondary antibody incubation with α-FLAG, α-Fc (which recognizes only intact antibodies), or α-Fab (which recognizes Fabs or intact antibodies). (c) Purified 3B5H10 Fab and Thio-Httex1-Q39-His6 were combined at different molar ratios (Fab:htt = 0.25, 0.5, 1, 1.5, 2) and analyzed by size-exclusion chromatography. A single peak consistent with a 1:1 Fab:htt complex was observed: peaks of pure 3B5H10 Fab or Thio-Httex1-Q39-His6 appeared if either was in molar excess of the other. (d) Purified 3B5H10 Fab and Thio-Httex1-Q39-His6 were combined and analyzed by equilibrium sedimentation analytical ultracentrifugation. The data best fit a model in which 3B5H10 Fab binds httex1 in a 1:1 ratio. At high concentrations, the complex dimerizes. Predictions for this model are overlaid on the raw data curves. The even distribution of residuals suggests no bias in the fit.
To further characterize the species 3B5H10 binds to, we turned to size-exclusion chromatography. Because 3B5H10 IgG is bivalent and could potentially bind two epitopes or antigen molecules simultaneously on size-exclusion chromatography32, we first purified monovalent Fabs (mass=47.65 kD) from 3B5H1013. Importantly, western blot analysis on HEK293 cell extracts containing Htt-171-(Q17 or Q68)-FLAG proteins showed that, like the intact antibody, 3B5H10 Fab retained a strong binding preference for mHtt (Fig. 6b). We then mixed 3B5H10 Fab and pure Thio-Httex1-Q39-His6 (27.10 kD)33 at different molar ratios (Fab:mHtt — 0.25, 0.5, 1, 1.5, 2) and separated the constituents by size-exclusion chromatography. The 1:1 mixtures contained a peak whose elution time was consistent with the expected molecular mass of a complex between Fab and mHtt (74.75 kD). Higher or lower ratios led to the appearance of an additional peak corresponding to unbound Fab or mHtt (Fig. 6c), suggesting that 3B5H10 Fab preferentially binds monomeric mHtt in a complex with a 1:1 stoichiometry.
To confirm these results, we analyzed the mass and stoichiometry of the 3B5H10 Fab:mHtt complex by sedimentation equilibrium analytical ultracentrifugation, a method that determines molecular mass independently of shape. The best fit for data acquired at three Fab:mHtt concentrations and four rotor speeds was a model in which 3B5H10 Fab and Thio-Httex1-Q39-His6 form a stable monomeric 1:1 complex that dimerizes at higher concentrations (>0.05 mg/ml) (Fig. 6d). Monte Carlo analysis revealed a dimerization Kd of 340 nM (95% confidence interval, 240–490 nM). The molecular mass predicted for the 1:1 complex was 73.4 kD, which is 98.1% of its calculated mass (74.75 kD, not shown). Importantly, a model in which the Fab binds a dimer of mHtt was tested and explicitly rejected (Supplementary Fig. 12 online). Supporting these results, small-angle X-ray scattering also indicates that 3B5H10 Fab preferentially binds mHtt in a 1:1 ratio (unpublished data). We therefore conclude that 3B5H10, which binds an epitope of mHtt that is an especially strong predictor of neuronal death, preferentially recognizes monomers (and possibly very small oligomers) of mHtt and that the preferred stoichiometry for the binding of the 3B5H10 Fab to mHtt is 1:1.
DISCUSSION
In this study, we developed novel methods to understand which of several simultaneously existing in situ epitopes of diffuse mHtt best predicts neurotoxicity. We used a series of antibodies to distinguish species of htt in situ and then automated microscopy with survival analysis to determine whether any of them predicted toxicity in a primary striatal neuron model of HD. Among four antibodies we compared (3B5H10, MW1, MW7 and EM48), the newly developed mAb 3B5H10 bound a species of htt that best predicted neurodegeneration.
The epitope recognized by 3B5H10 is contained within the polyQ stretch of mHtt but disappears as mHtt aggregates into IBs. The epitope exists in both full-length and an exon1 fragment of mHtt but is negligibly present in wt-htt. Further, the epitope also exists in mutant forms of other polyQ-containing proteins that cause neurodegeneration, including the androgen receptor, atrophin, and ataxin-3. Finally, we determined that the epitope exists in certain conformational folds of polyQ associated with low molecular weight htt species (monomers and possibly very small oligomers).
We considered whether 3B5H10 is capable of binding oligomers of mHtt even though it preferentially binds a monomeric species. While 3B5H10 strongly stains the diffuse mHtt fraction of neurons, we did not detect oligomers in this fraction by FRET or an α-oligomer antibody. Slot blot, agarose gel electrophoresis, AFM, and DLS (Fig. 5, Supplementary Figs. 9–10) confirmed that the epitope recognized by 3B5H10 is present in a very low molecular weight species of htt (likely monomer and possibly small oligomer). Chemical cross-linking experiments (Fig. 6a) confirmed the epitope 3B5H10 recognizes is present in monomers and, to a lesser degree, in very small oligomers of htt. 3B5H10’s tendency to bind monomers over oligomers was further supported by results from size-exclusion chromatography (Fig. 6c), sedimentation equilibrium analytical ultracentrifugation (Fig. 6d), and small-angle X-ray scattering (unpublished observations).
A recent study on the formation of different conformations of mHtt found that 3B5H10 may bind a conformation of amyloid formed in vitro at 4°C.34 This structure, unlike amyloid formed in vitro at 37°C, has a loop/turn organization that exposes loose hairpins of polyQs. Additionally, certain biochemical purification techniques may facilitate de novo exposure of the 3B5H10 epitope on oligomers35, analogous to unmasking of the 3B5H10 epitope upon aggressive antigen retrieval of HD animal model brains (Supplementary Fig. 4 online). Combined with our data showing very faint 3B5H10 staining of cross-linked small htt oligomers (Fig. 6a), we conclude that under certain in vitro conditions or under certain biochemical and immunohistochemical methods, the conformation of polyQ recognized by 3B5H10 may appear in aggregated species of htt.
The ability of 3B5H10 to dissociate pre-formed oligomers and fibrils into monomeric htt (Fig. 5f,g, Supplementary Fig. 10 online) was surprising. We are aware of only one other α-htt antibody that demonstrates this property, MW77. However, MW7 appears to dissolve pre-formed fibrils into AFM-detectable species with dimensions that are consistent with an oligomer rather than a monomer. We speculate that 3B5H10 may promote oligomer and fibril dissociation by sequestering monomers that could be dynamically associating and dissociating with oligomers or fibril ends36,37.
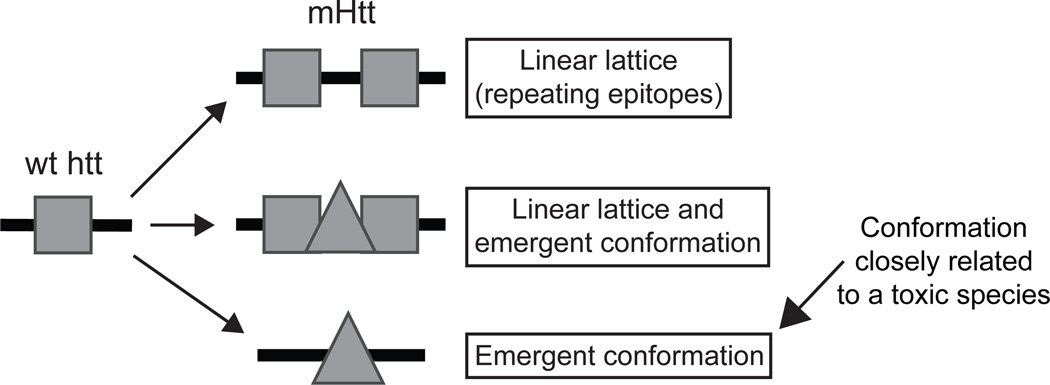
Since 3B5H10 and MW1 both bind polyQ expansions in a length-dependent manner and preferentially recognize diffuse mHtt in situ4,33, it is surprising that only 3B5H10 binding predicts neurodegeneration by multivariate Cox analysis (see legend for Supplementary Fig. 5 online). One explanation is that MW1 and 3B5H10 bind mostly distinct conformers of mHtt, only one of which may have prognostic value. Previous studies demonstrated that MW1 recognizes expanded polyQ as a “linear lattice”21,33. In such a model, the antibody binds weakly to a relatively unstructured epitope of wt polyQ. As the length of the polyQ stretch approaches that associated with disease, the unstructured epitope repeats. Since antibodies are bivalent, the presence of two epitopes in tandem results in dramatically increased binding by MW1, due to increased avidity (Supplementary Fig. 13 online). In contrast, our results and our unpublished structural data demonstrate that 3B5H10 recognizes a compact, structured epitope of polyQ that is minimally present in wt-htt and is exposed or created in mHtt. Thus, rather than preferentially binding expanded polyQ via increased avidity, 3B5H10 demonstrates a strong affinity for mutant polyQ. Supporting these conclusions, MW1 Fab forms a 3:1 complex33 with the exact version of htt (Thio-Httex1-Q39-His6) that we found forms a 1:1 complex with 3B5H10 Fab (Fig. 6c–d, Supplementary Fig. 13 online). Thus, the “linear lattice” epitope that MW1 recognizes is no larger than 13 glutamines, consistent with crystallographic studies of MW1 complexed with a polyQ peptide21. Also supporting these conclusions, the Fab of MW1, which is monovalent and therefore can not bind its target through an avidity mechanism, loses most of its preference for mutant polyQ over wt polyQ33. In contrast, the Fab of 3B5H10 retains a preference for mutant polyQ (Fig. 6b, Supplementary Fig. 13 online). These observations, combined with our structural studies (unpublished data), suggest that low molecular weight species of mHtt may exist in more than one conformation and that the one recognized by 3B5H10 might be toxic or closely related to a toxic species (Fig. 7)38–40.
Figure 7.
The “linear lattice” versus “emergent conformation” hypotheses for expanded polyQ conformation. The linear lattice model posits that a relatively unstructured epitope of polyQ in wt-htt repeats itself as the polyQ stretch expands. The emergent conformation model posits that expansion of the polyQ stretch induces a conformational change, such that wild-type and mutant polyQ are in distinct conformations. As shown in the middle panel, the linear lattice and emergent conformation hypotheses are not mutually exclusive; both models may be simultaneously true for mHtt. Alternatively, some mHtt molecules may exhibit linear lattice epitope repeats, while others may display an emergent conformation, possibly in the same neuron. Assessment of prognostic value for both epitopes by Cox analysis reveals that the emergent conformation, recognized by 3B5H10, is more toxic or more closely related to a toxic species than the linear lattice epitope, recognized by MW1.
A burgeoning body of work in polyQ disease research suggests that culprit proteins may exert their toxic gain-of-function by enhanced native activity (especially via native protein-protein interactions41,42). Since our exon1 model of HD, by definition, does not encompass all native function of full-length htt, we may miss pathogenic events that rely on native activity of htt outside exon1. While technical barriers prohibited us from designing a full-length primary culture model of HD for these experiments, we have previously extensively validated our exon1 model as a faithful model of HD43. Further, we demonstrate in this paper the ability of 3B5H10 to recognize full-length mHtt, ataxin-3, and androgen receptor, thereby establishing the presence of the predictive epitope in full-length protein. Finally, we note that the majority of known htt interactors bind to the N-terminus of htt44–46 and, in some cases, this has been defined even more narrowly to be the exon1 region46. Thus, we favor the hypothesis that the 3B5H10 conformer of httex1 still mediates toxicity at least partially through native activity, including enhanced interactions with some exon1-interactors and diminished interactions with other exon1-interactors.
We previously found that IB formation is associated with improved neuronal survival10, and in our current studies, we discovered that IB formation leads to a substantial loss of intraneuronal 3B5H10 binding. In contrast, IB formation does not lead to a loss of EM48 or MW7 binding (Fig. 1e). Since 3B5H10 binds a species of mHtt that strongly predicts death (better than the epitopes recognized by EM48 or MW7, for example), IB formation might be protective by preferentially reducing, masking, or refolding the 3B5H10 epitope (Supplementary Fig. 14 online)47.
The novel methodology we employed may be broadly applicable to the study of diseases associated with protein malfolding. Elucidating toxic species of aggregation prone proteins is difficult because these species may be rare, their existence may depend on endogenous protein interactions that defy biochemical purification, and the tools to study protein conformation in situ are limited. We showed that combining the use of conformation-specific antibodies and automated imaging with longitudinal analysis provides a way to probe protein conformation in situ and elucidate the prognostic significance of one conformer in the context of others. We expect that the better a conformer predicts neurodegeneration, the more tightly it is linked to toxicity and pathogenesis.
METHODS
Antibodies and plasmids
Production of new α-htt monoclonal antibodies and creation of new constructs is described in Supplementary Methods online.
Cell culture, transfection, automated microscopy, and image analysis
Cell culture, transfection, automated microscopy, and image analysis were performed as described10,12,13,48,49 and detailed further in Supplementary Methods online.
Immunocytochemistry, immunohistochemistry, confocal microscopy, and electron microscopy
Immunochemical techniques and subsequent confocal and electron microscopy imaging and quantification are described in Supplementary Methods online.
3B5H10 binding to polyQ peptide
PolyQ (K2Q39K2) or PACAP peptide (basic peptide) was spotted on a PS10 ProteinChip Array (Bio-Rad) and loaded into a ProteinChip SELDI System (Bio-Rad) for data collection. Details on how the how the peptide was spotted are available in Supplementary Methods online.
Hierarchical Bayesian statistical analysis of predictive power for various α-htt antibodies
The statistical model we used to determine how well epitopes recognized by each antibody predict neurotoxicity are outlined in Figure 3 and Supplementary Figure 6 online. The mathematical explanation of our model is available in Supplementary Methods online.
Concentration-corrected Förster resonance energy transfer (N-FRET)
N-FRET was performed as described26 and further detailed in Supplementary Methods online.
Dot blots for α–oligomer staining of htt
Modifications to traditional dot blotting for detection of α-oligomer staining of htt are detailed in Supplementary Methods online.
Filter retardation assay
Lysate was centrifuged at 20,000g at 4°C, and the pellet was resuspended in 20 mM Tris-HCl (pH 8.0), 15 mM MgCl2 and incubated at 37°C for 1 hour. A sample from this insoluble material was diluted in 2% SDS and loaded on a cellulose acetate membrane (Osmonics INC Acetate Plus 0.22 µm 142 mm) previously rinsed three times with 2% SDS, with a slot-blot apparatus (Amersham slot blot manifold (Hoefer PR 648)). After loading the samples, wells were washed again three times with 0.1% SDS and the membrane was blotted. More details are available in Supplementary Methods online.
Agarose gel electrophoresis of PC12 lysates
Conditions for detecting oligomers of mHtt from PC12 lysates in semi-native form are described in Supplementary Methods online.
Atomic force microscopy
AFM experiments were performed using a MFP3D scanning probe microscope (Asylum Research, Santa Barbara, CA). Detailed settings and sample preparation are described in Supplementary Methods online.
Chemical cross-linking
Cleavage and aggregation GST-Httex1-Q53, cross-linking with glutaraldehyde, and immunoblotting with MW7 or 3B5H10 are described in Supplementary Methods online.
Size-exclusion chromatography
Experiments were carried out under a constant htt concentration (0.2 mg/ml), with adjustment of the Fab concentration to achieve Fab:htt ratios of 1:4, 1:2, 1:1, 1.5:1, and 2:1. Total volume for each sample loaded on the size-exclusion column was 500 µl.
Analytical ultracentrifugation
Samples of 3B5H10 Fab and Thio-Httex1-Q39-His6 (1:1 molar ratio) in 10 mM Tris-HCl pH 8.0 and 50 mM NaCl were analyzed with a Beckman/Coulter XL-A Ultima analytical ultracentrifuge using an A Ti60 rotor and cells with 6-channel centerpieces. Experiments were carried out at three different total protein concentrations (0.53, 0.80, and 1.07 mg/ml). Samples were run at four different rotor speeds (10,000, 15,000, 20,000, and 25,000 rpm) at 20°C for 16–31 hours (at each speed) to achieve sedimentation equilibrium, which was confirmed by overlapping scans taken 2 hours apart. Analysis of AU data is described in Supplementary Methods online.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Kazantzev, D. Housman and the Hereditary Disease Foundation (HDF) for pcDNA3.1-Httex1-(Q46, Q97)-GFP plasmids. We also thank R. Truant for eGFP-full-length Htt-β-galactosidase (Q17, Q138) plasmids, M. Diamond for the HA-AR (Q25, Q65) plasmids, J. Burke and CHDI, Inc. for the GST-atrophin-1 (Q19, Q81) plasmids, R. Kopito for Httex1-CFP (Q25, Q97) and Httex1-YFP (Q25, Q97) plasmids, D. Devys for GST-Htt-171 (Q66, Q142) plasmids, R. Pittman for Myc-ataxin-3 (Q27 Q78) plasmids, O. Onodera for Httex1-mCFP (Q17, Q58) and Httex1-mYFP (Q17, Q58) plasmids, P. Bjorkman for the Thio-Httex1-Q39-His6 plasmid, and P. Daugherty for mammalian codon-optimized CyPet and YPet plasmids. We thank P. Patterson for mAbs MW1, MW7, and MW8, and C. Glabe for the α-oligomer polyclonal antibody. We thank members of the Finkbeiner lab for useful discussions Stephen Ordway and Gary Howard for editorial assistance, Kelley Nelson for administrative assistance, and Margaret Sutherland for her interest and support. Primary support for this work was provided by the Lieberman Award of the Hereditary Disease Foundation and the National Institute of Neurological Disease and Stroke (S.F.). Additional support was provided by the National Institute of Aging, the High Q Foundation, the Huntington’s Disease Society of America, the National Center for Research Resources, the Taube-Koret Center for HD Research, the Hellman Family Foundation Program for Alzheimer's Disease Research, and the J. David Gladstone Institutes (S.F.). M.A. and J.M. are supported by the Hillblom Foundation. J.M. and S.M. are supported by the NIH-NIGMS UCSF Medical Scientist Training Program. J.M is supported by a fellowship from the Achievement Rewards for College Scientists (ARCS) Foundation. D.H. is supported by a postdoctoral fellowship from the John Douglas French Alzheimer’s Foundation. J.L. and A.O. are supported by the HDF. The animal care facility was partly supported by an NIH Extramural Research Facilities Improvement Project.
Footnotes
AUTHOR CONTRIBUTIONS
J.M., M.A., and S.F. wrote the manuscript with analytic contributions from S.M. J.M. coordinated data from all authors and performed all immunocytochemistry, FRET, cross-linking, and anti-oligomer dot blot experiments. M.A. optimized 3B5H10 staining conditions and performed all longitudinal survival experiments and the anti-oligomer staining in neurons. J.M. and M.A. cultured neurons for all experiments. J.M., M.A., and M.S. performed initial survival statistics analysis. B.A.S. and J.M. developed final Bayesian survival statistics analysis. E.B., J.C., F.S., and S.F. initiated immunizations and screened hybridomas leading to the identification of 3B5H10. E.B. performed most 3B5H10 Western blots and slot blots. P.K., Y.N., K.W., K.C., J.C., and C.P-L. purified protein, produced Fab, performed size-exclusion experiments, and were responsible for some Western blots. C.P-L. and P.K. performed the dynamic light scattering. J.L. performed all atomic force microscopy. D.H. performed all analytical ultracentrifugation. G.P.L. provided advice and assistance for cross-linking and dot-blot experiments. E. Mitchell performed agarose-gel electrophoresis. A.O. and M.G. performed immunohistochemistry on mouse brains. V.T. performed the synthetic polyQ-peptide binding experiment. E. Masliah performed electron microscopy. X.W.Y. supervised BACHD immunohistochemistry. L.M.T. supervised agarose-gel electrophoresis experiments. P.J.M. supervised atomic force microscopy, cross-linking, and dot blot experiments. K.H.W. supervised aspects of the protein and Fab production as well as size exclusion chromatography and ultracentrifugation experiments. S.F. supervised the entire project.
COMPETING FINANCIAL INTERESTS STATEMENT
No research support for this work was provided by any organization that stands to gain or lose financially through this publication. V.T. was employed by Ciphergen, Inc., prior to its acquisition by Bio-Rad, at the time the research was performed. S.F. has served and is expected to continue to serve as an ad hoc paid consultant in the area of neurodegenerative disease therapeutics. A patent covering a series of antibodies, including some (e.g. 3B5H10 and 4H7H7) used in this study, was issued to the University of California, San Francisco; however, no compensation has been received by any of the authors from the licensing of these antibodies.
REFERENCES
- 1.THDCR G. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 3.Kayed R, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 4.Ko J, Ou S, Patterson PH. New anti-huntingtin monoclonal antibodies: Implications for huntingtin conformation and its binding proteins. Brain Res. Bull. 2001;56:319–329. doi: 10.1016/s0361-9230(01)00599-8. [DOI] [PubMed] [Google Scholar]
- 5.Rakhit R, et al. An immunological epitope selective for pathological monomer-misfolded SOD1 in ALS. Nat. Med. 2007;13:754–759. doi: 10.1038/nm1559. [DOI] [PubMed] [Google Scholar]
- 6.Paramithiotis E, et al. A prion protein epitope selective for the pathologically misfolded conformation. Nat. Med. 2003;9:893–899. doi: 10.1038/nm883. [DOI] [PubMed] [Google Scholar]
- 7.Legleiter J, et al. Monoclonal antibodies recognize distinct conformational epitopes formed by polyglutamine in a mutant huntingtin fragment. J. Biol. Chem. 2009;284:21647–21658. doi: 10.1074/jbc.M109.016923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fleming TR, Lin DY. Survival analysis in clinical trials: Past developments and future directions. Biometrics. 2000;56:971–983. doi: 10.1111/j.0006-341x.2000.0971.x. [DOI] [PubMed] [Google Scholar]
- 9.Roodnat JI, et al. The Cox proportional hazards analysis in words: Examples in the renal transplantation field. Transplantation. 2004;77:483–488. doi: 10.1097/01.tp.0000110424.27977.a1. [DOI] [PubMed] [Google Scholar]
- 10.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 11.Taylor JP, et al. Aggresomes protect cells by enhancing the degradation of toxic polyglutamine-containing protein. Hum. Mol. Genet. 2003;12:749–757. doi: 10.1093/hmg/ddg074. [DOI] [PubMed] [Google Scholar]
- 12.Arrasate M, Finkbeiner S. Automated microscope system for determining factors that predict neuronal fate. Proc. Natl. Acad. Sci. U.S.A. 2005;102:3840–3845. doi: 10.1073/pnas.0409777102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brooks E, Arrasate M, Cheung K, Finkbeiner SM. Using antibodies to analyze polyglutamine stretches. Methods Mol. Biol. 2004;277:103–128. doi: 10.1385/1-59259-804-8:103. [DOI] [PubMed] [Google Scholar]
- 14.Diamond MI, Robinson MR, Yamamoto KR. Regulation of expanded polyglutamine protein aggregation and nuclear localization by the glucocorticoid receptor. Proc. Natl. Acad. Sci. U.S.A. 2000;97:657–661. doi: 10.1073/pnas.97.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Onodera O, et al. Oligomerization of expanded-polyglutamine domain fluorescent fusion proteins in cultured mammalian cells. Biochem. Biophys. Res. Commun. 1997;238:599–605. doi: 10.1006/bbrc.1997.7337. [DOI] [PubMed] [Google Scholar]
- 16.Perez MK, et al. Recruitment and the role of nuclear localization in polyglutamine-mediated aggregation. J. Cell. Biol. 1998;143:1457–1470. doi: 10.1083/jcb.143.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu Z-X, Li S-H, Nguyen H-P, Li X-J. Huntingtin inclusions do not deplete polyglutamine-containing transcription factors in HD mice. Hum. Mol. Genet. 2002;11:905–914. doi: 10.1093/hmg/11.8.905. [DOI] [PubMed] [Google Scholar]
- 18.Gray M, et al. Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J. Neurosci. 2008;28:6182–6195. doi: 10.1523/JNEUROSCI.0857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mangiarini L, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 20.Cemal CK, et al. YAC transgenic mice carrying pathological alleles of the MJD1 locus exhibit a mild and slowly progressive cerebellar deficit. Hum. Mol. Genet. 2002;11:1075–1094. doi: 10.1093/hmg/11.9.1075. [DOI] [PubMed] [Google Scholar]
- 21.Li P, et al. The structure of a polyQ-anti-polyQ complex reveals binding according to a linear lattice model. Nat. Struct. Mol. Biol. 2007;14:381–387. doi: 10.1038/nsmb1234. [DOI] [PubMed] [Google Scholar]
- 22.Bustamante CD, et al. The cost of inbreeding in Aradposis. Nature. 2002;416:531–534. doi: 10.1038/416531a. [DOI] [PubMed] [Google Scholar]
- 23.Gelman A, Carlin JB, Stern HS, Rubin DB. Bayesian data analysis. Boca Raton: Chapman & Hall/CRC; 2004. [Google Scholar]
- 24.Caughey B, Lansbury PT. Protofibrils, pores, fibrils, and neurodegeneration: Separating the responsible protein aggregates from the innocent bystanders. Annu. Rev. Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 25.Takahashi T, et al. Soluble polyglutamine oligomers formed prior to inclusion body formation are cytotoxic. Hum. Mol. Genet. 2008;17:345–356. doi: 10.1093/hmg/ddm311. [DOI] [PubMed] [Google Scholar]
- 26.Xia Z, Liu Y. Reliable and global measurement of fluorescence resonance energy transfer using fluorescence microscopes. Biophys. J. 2001;81:2395–2402. doi: 10.1016/S0006-3495(01)75886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rajan RS, Illing ME, Bence NF, Kopito RR. Specificity in intracellular protein aggregation and inclusion body formation. Proc. Natl. Acad. Sci. U.S.A. 2001;98:13060–13065. doi: 10.1073/pnas.181479798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen AW, Daugherty PS. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat. Biotechnol. 2005;23:355–360. doi: 10.1038/nbt1066. [DOI] [PubMed] [Google Scholar]
- 29.Muchowski PJ, et al. Hsp70 and Hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc. Natl. Acad. Sci. U.S.A. 2000;97:7841–7846. doi: 10.1073/pnas.140202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wanker EE, et al. Membrane filter assay for detection of amyloid-like polyglutamine-containing protein aggregates. Methods Enzymol. 1999;309:375–386. doi: 10.1016/s0076-6879(99)09026-6. [DOI] [PubMed] [Google Scholar]
- 31.Apostol BL, et al. A cell-based assay for aggregation inhibitors as therapeutics of polyglutamine-repeat disease and validation in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 2003;100:5950–5955. doi: 10.1073/pnas.2628045100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klein FAC, et al. Pathogenic and non-pathogenic polyglutamine tracts have similar structural properties: Towards a length-dependent toxicity gradient. J. Mol. Biol. 2007;371:235–244. doi: 10.1016/j.jmb.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 33.Bennett MJ, et al. A linear lattice model for polyglutamine in CAG-expansion diseases. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11634–11639. doi: 10.1073/pnas.182393899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nekooki-Machida Y, et al. Distinct conformations of in vitro and in vivo amyloids of huntingtin-exon1 show different cytotoxicity. Proc. Natl. Acad. Sci. U.S.A. 2009;106:9679–9684. doi: 10.1073/pnas.0812083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sathasivam K, et al. Identical oligomeric and fibrillar structures captured from the brains of R6/2 and knock-in mouse models of Huntington's disease. Hum. Mol. Genet. 2010;19:65–78. doi: 10.1093/hmg/ddp467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collins SR, Douglass A, Vale RD, Weissman JS. Mechanism of prion propagation: Amyloid growth occurs by monomer addition. PLoS Biol. 2004;2:e321. doi: 10.1371/journal.pbio.0020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ellisdon AM, Pearce MC, Bottomly SP. Mechanisms of ataxin-3 misfolding and fibril formation: Kinetic analysis of a disease-assocated polyglutamine protein. J. Mol. Biol. 2007;368:595–605. doi: 10.1016/j.jmb.2007.02.058. [DOI] [PubMed] [Google Scholar]
- 38.Schaffer G, et al. Cellular toxicity of polyglutamine expansion proteins: Mechanism of transcription factor deactivation. Mol. Cell. 2004;15:95–105. doi: 10.1016/j.molcel.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 39.Nagai Y, et al. A toxic monomeric conformer of the polyglutamine protein. Nat. Struct. Mol. Biol. 2007;14:332–340. doi: 10.1038/nsmb1215. [DOI] [PubMed] [Google Scholar]
- 40.Kim MW, Chelliah Y, Kim SW, Otwinowski Z, Bezprozvanny I. Secondary structure of Huntingtin amino-terminal region. Structure. 2009;17:1205–1212. doi: 10.1016/j.str.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duvick L, et al. SCA1-like disease in mice expressing wild type ataxin-1 with a serine to aspartic acid replacement at residue 776. Neuron. 2010:929–935. doi: 10.1016/j.neuron.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nedelsky NB, et al. Native functions of the androgen receptor are essential to pathogenesis in a Drosophila model of spinobulbar muscular atrophy. Neuron. 2010:936–952. doi: 10.1016/j.neuron.2010.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller J, et al. Quantitative relationships between huntingtin levels, polyglutamine length, inclusion body formation, and neuronal death provide novel insight into Huntington‘s disease molecular pathogenesis. J. Neurosci. 2010;30:10541–10550. doi: 10.1523/JNEUROSCI.0146-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harjes P, Wanker EE. The hunt for huntingtin function: Interaction partners tell many different stories. Trends Biochem. Sci. 2003;28:425–433. doi: 10.1016/S0968-0004(03)00168-3. [DOI] [PubMed] [Google Scholar]
- 45.Li S-H, Li X-J. Huntingtin-protein interactions and the pathogenesis of Huntington’s disease. Trends Genet. 2004;20:146–154. doi: 10.1016/j.tig.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 46.Kaltenbach LS, et al. Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet. 2007;3:e82. doi: 10.1371/journal.pgen.0030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fowler DM, et al. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006;4(e6):0100–0107. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saudou F, Finkbeiner S, Devys D, Greenberg ME. Huntingtin acts in the nucleus to induce apoptosis, but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 49.Finkbeiner S, et al. CREB: A major mediator of neuronal neurotrophin responses. Neuron. 1997;19:1031–1047. doi: 10.1016/s0896-6273(00)80395-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.