Abstract
To overcome drug resistance and reduce the side effects of cisplatin, a widely used antineoplastic agent, major efforts have been made to develop next generation platinum-based anticancer drugs. Because cisplatin-DNA adducts block RNA polymerase II unless removed by transcription-coupled excision repair, compounds that react similarly but elude repair are desirable. The monofunctional platinum agent pyriplatin displays antitumor activity in mice, a cytotoxicity profile in cell cultures distinct from that of cisplatin, and a unique in vitro transcription inhibition mechanism. In the present study, we incorporated pyriplatin globally or site-specifically into luciferase reporter vectors to examine its transcription inhibition profiles in live mammalian cells. Monofunctional pyriplatin reacted with plasmid DNA as efficiently as bifunctional cisplatin and inhibited transcription as strongly as cisplatin in various mammalian cells. Using repair-defective NER-, MMR-, and SSBR-deficient cells, we demonstrate that NER is mainly responsible for removal of pyriplatin-DNA adducts. These findings reveal that the mechanism by which pyriplatin generates its antitumor activity is very similar to that of cisplatin, despite the chemically different nature of their DNA adducts, further supporting a role for monofunctional platinum anticancer agents in human cancer therapy. This information also provides support for the validity of the proposed mechanism of action of cisplatin and provides a rational basis for the design of more potent platinum anticancer drug candidates using a monofunctional DNA-damaging strategy.
Introduction
cis-Diamminedichloroplatinum(II) (cDDP, cisplatin) and its close analogues carboplatin and oxaliplatin are used to treat about half of all patients receiving chemotherapy for cancer (Figure 1) (1). Broader application of platinum-based anticancer drugs, however, is limited by intrinsic or acquired drug resistance and side effects including emetogenesis, nephrotoxicity, and neurotoxicity. Extensive efforts have been made to synthesize and test new platinum-based anticancer agents, with the promise that compounds with improved antitumor activity and fewer toxic side effects will be discovered.
Figure 1.
Chemical structures of platinum(II) anticancer agents cisplatin, oxaliplatin, and pyriplatin.
The mechanism of action of platinum(II) anticancer agents has been extensively explored, and many details of the cellular response to these compounds are now understood. Cisplatin attacks nuclear DNA to form Pt-DNA cross-links. The major adducts are intrastrand cis-{Pt(NH3)2}2+ cross-links including 1,2-d(GpG) cross-links, which efficiently block RNA polymerase II until removed by DNA damage repair pathways (2, 3). Besides the conventional bifunctional platinum-based anticancer drugs that contain two reactive sites for DNA-binding, another class of compounds, monofunctional platinum anticancer agents with only one DNA reactive site, have been developed and tested. Monofunctional platinum(II) complexes such as chlorodiethylene-triamineplatinum(II) chloride {[PtCl(dien)]Cl} and [PtCl(NH3)3]Cl do not inhibit DNA-dependent polymerases and are ineffective against cancer cells (4, 5). Another monofunctional compound, the aminophosphine-containing platinum(II) complex, cis-[PtCl(C6H11NH(CH2)2PPh2-N,P)(C6H11NH(CH2)2PPh2-P)], binds rapidly to DNA forming monodentate adducts at guanine residues. Although this compound has demonstrated anticancer activity, it only slightly inhibits DNA synthesis and has little influence on DNA conformation (6). Some monofunctional platinum(II)-polyamide complexes, designed for recognition of specific DNA sequences, are capable of forming covalent bonds with DNA, but failed to evoke better cytotoxicity against cancer cells than cisplatin (7, 8). A series of cationic monofunctional platinum(II) complexes, cis-[Pt(NH3)2(N-donor)Cl]+, has been synthesized and analyzed, and some of the compounds demonstrated moderate biological activity against in vivo murine tumor models (9). Like cisplatin these complexes inhibit DNA polymerase despite their very different DNA-binding modality (10). The spectrum of activity of one of these compounds, cis-diammine(pyridine)chloroplatinum(II) [cDPCP, or “pyriplatin” (Figure 1)], against a panel of human cancer cell lines differs significantly from those of cisplatin or oxaliplatin, rendering pyriplatin a lead compound for generating new platinum anticancer drug candidates (11).
When attached to the N7 position of a guanine residue in duplex DNA, pyriplatin generates no significant structural distortion (12). An in vitro study revealed that pyriplatin is a substrate for nucleotide excision repair (NER), but it eludes this repair pathway much more readily than cisplatin. Studies of RNA polymerase II activity on a DNA duplex containing a single cis-{Pt(NH3)2(py)}2+-dG adduct revealed a transcription inhibition mechanism distinct from that of bifunctional platinum compounds like cisplatin (13). An X-ray crystal structure analysis indicated that pyriplatin bound to the N-7 position of a guanosine residue can be accommodated in the Pol II active site, where it forms a standard Watson-Crick base pair with cytosine of the growing RNA strand. Blockage of subsequent pol II translocation from the damaged site leads to inhibition of the pol II transcribing complex. In contrast, for the cis-{Pt(NH3)2}2+ 1,2-d(GpG) cross-link, delivery of the damaged nucleosides to the active site is inhibited by a translocation barrier (14). Details of transcription inhibition by pyriplatin in live mammalian cells, especially in comparison to other bifunctional platinum-based anticancer drugs, and the repair pathways that are responsible for removal of pyriplatin-DNA adducts, remain to be elucidated, however.
To address these deficiencies, we have in the present study investigated the transcription inhibition profiles of pyriplatin and compared them to those of cisplatin and oxaliplatin. We examined the ability of cisplatin, oxaliplatin, and pyriplatin to inhibit the transcription of a Gaussia luciferase reporter gene, utilizing globally platinated expression vectors in live mammalian cells. Different repair-deficient cell lines, including NER-, mismatch repair (MMR)-, and single strand break repair (SSBR)-deficient cells, were utilized to reveal repair pathways that might be involved in removal of pyriplatin-DNA adducts. In addition, a site-specific pyriplatin-dG adduct was incorporated into the Gaussia luciferase expression vector. The transcription inhibition effects from this single pyriplatin-dG adduct in a 3,986-bp plasmid, as well as the mechanisms by which the repair-deficient cells process the site-specific lesion, were investigated. Our results shed light on the transcription inhibition effects and repair mechanisms of pyriplatin-DNA adducts. Moreover, they provide details about the mechanisms by which this monofunctional platinum compound generates its antitumor activity and suggest how this activity can be improved in the design of novel anticancer drug candidates based on monofunctional platinum complexes.
Materials and Methods
Preparation of Globally Platinated Transcription Probes
For global platination experiments, 125 μg/ml (45.4 nM) of pGLuc, prepared as described in Supplementary Information, was treated with 0, 0.25, 0.51, 1.02, 2.04, 4.07 μM cisplatin, 0, 0.23, 0.45, 0.91, 1.81, 3.63 μM oxaliplatin, or 0, 0.42, 0.84, 1.68, 3.36, 6.71 μM pyriplatin in 25 mM Na-HEPES, 10 mM NaCl, pH 7.4 buffer for 16 h at 37 °C in the dark. A control plasmid without platinum was treated similarly. The reaction mixtures were then dialyzed against water and subsequently against TE buffer (10 mM Tris-HCl, 2 mM EDTA, pH 8.0) to remove unbound plati-num. Quantification of Pt content for these globally platinated plasmids was obtained by flameless atomic absorption spectroscopy on a Perkin-Elmer AAnalyst 600 system. DNA concentrations were measured by UV-vis absorption spectroscopy at 260 nm on a HP 8453 UV-visible spectrometer. The number of platinum complexes bound per nucleotide, rb, was computed from this information.
Preparation of a Pyriplatin Modified Insertion Strand
A 16-mer oligonucleotide containing a site-specific cis-{Pt(NH3)2(py)}2+ -dG adduct was prepared in the following manner. A 25.7 mM aqueous solution of pyriplatin was activated by addition of 0.98 equiv of AgNO3 followed by agitation for 8 h in the dark at room temperature. The suspension was centrifuged. To a 0.2 mM solution of 5′-CCTCCTCG*TCTCTTCC (Integrated DNA Technologies), where the asterisk denotes the base to be platinated, in 10 mM NaH2PO4, pH 6.3, was added 1.2 equiv of activated pyriplatin. The reaction mixture was incubated overnight in the dark at 37 °C. The reaction was stopped by freezing the solution. The pyriplatin-modified insertion strand was purified by ion exchange HPLC [Agilent 1200 HPLC system, Dionex DNAPac PA-100, linear gradient, 0.34 to 0.45 M NaCl in 25 mM Tris-HCl (pH 7.4) over 11 min]. After purification, the platinated DNA solution was dialyzed against H2O and lyophilized. The platination level was confirmed by UV-vis and atomic absorption spectroscopy, which yielded a Pt/DNA ratio of 1.03±0.01. The insertion strand was further analyzed for nucleotide composition by enzyme digestion to confirm the validity of the platination site following a published previously protocol (data not shown) (15).
Preparation of Site-Specifically Platinated Plasmids
Site-specifically platinated pGLuc8temG plasmid containing a cis-{Pt(NH3)2(py)}2+ -dG adduct between the CMV promoter and the luciferase expression gene was prepared following the strat-egy published previously (16). The gapped plasmid was obtained with the use of Nt.BbvCI and Nt.BspQI nicking restriction enzymes (New England Biolabs), followed by annealing with excess 24-mer complementary strand to remove the nicked strand. A 300 μg quantity of pGLuc8temG plasmid was digested with 15 U of Nt.BbvCI at 37 °C for 1 h. The reaction mixture was heated at 80 °C for 20 min to deactivate the enzyme and then extracted with phenol/chloroform/isoamyl alcohol to remove the enzyme. The resulting aqueous phase was dialyzed against H2O overnight at 4 °C. The plasmid was further digested with 15 U Nt.BspQI at 50 °C for 1 h, and the enzyme was heat-deactivated and removed by a phenol/chloroform/isoamyl alcohol extraction. The nicked plasmid was mixed with 1000 equiv of complementary DNA strand 5′-TTTTGGAAGAGACGAGGAGGTTTT in a buffer of 10 mM Tris-HCl, 2 mM MgCl2, 0.4 M NaCl, pH 7.4, heated at 80 °C for 5 min, and subsequently cooled at 4 °C for 5 min for 10 cycles. The gapped plasmid was purified by isopycnic centrifugation using a CsCl gradient at 58,000 rpm, 20 °C for 24 h, and quantitated by UV-vis spectroscopy.
The 16-mer insertion strands, either containing a site-specific cis-{Pt(NH3)2(py)}2+ -dG adduct or no platinum, were phosphorylated with T4 polynucleotide kinase at 37 °C for 3 h. The enzyme was removed by a phenol/chloroform/isoamyl alcohol extraction. The phosphorylated strands were ethanol-precipitated and stored at -80 °C at a concentration of 100 pmol/μL. A 120 μg quantity of the gapped plasmid was annealed with 100 equiv of the insertion strand in a buffer of 10 mM Tris-HCl, 2 mM MgCl2, 0.4 M NaCl, pH 7.4 from 90 °C to 4 °C at -1 °C/min in a thermocycler, followed by a ligation with 240 U T4 DNA ligase at 16 °C for 16 h. The platinated plasmid was dialyzed against H2O at 4 °C overnight and further purified by treatment with 30 U of BsmBI at 55 °C for 1 h. The closed-circular form of plasmid was purified and concentrated by isopycnic centrifugation, followed by n-butanol extraction and ethanol precipitation. The plas-mids were quantitated by a Quant-iT™ PicoGreen® dsDNA Kit (Invitrogen) and stored at −80 °C in TE buffer.
To carry out a restriction analysis on ligated platinated or unplatinated plasmids, a 60 ng quantity of pGLuc8temG plasmid was incubated with 2 U BsmBI at 55 °C for 30 min. The plasmids were analyzed using 0.8% agarose gel electrophoresis containing 0.5 μg/mL ethidium bromide. The gels were documented with a BioRad Fluor-S MultiImager.
Cell Lines and Tissue Culture
XPF (GM08437) cells were obtained from the Coriell Cell Depositories at Coriell Institute. XPFcorr cells were generously offered by Dr. Gan Wang at Wayne State University. U2OS-MOCK and XPF-1128 cells were offered by Dr. Nora Graf in the Department of Chemistry at the Massachusetts Institute of Technology. PARP-1+/+ and PARP-1−/− mouse embryonic fibroblasts (MEFS) were kindly provided by Prof. Paul Chang at the Massachusetts Institute of Technology. HEC59 and HEC59+Chr2 cells were obtained from Dr. Thomas Kunkel from the National Institutes of Health.
All cells were grown in a humidified incubator at 37 °C under 5% CO2. XPF, U2OS-MOCK, and XPF-1128 cells were maintained in DMEM with 10% FBS, and 1% penicillin/streptomycin. XPFcorr cells were maintained in DMEM with 10% FBS, 0.5 mg/mL G418 sulfate, and 1% penicillin/streptomycin. HEC59 cells were grown in DMEM/F12 supplemented with 10% FBS and 1% penicillin/streptomycin. HEC59+Chr2 cells were grown in DMEM/F12 supplemented with 10% FBS, 1% penicillin/streptomycin, and 0.4 mg/mL G418 sulfate. PARP-1+/+ and PARP-1−/− MEFS were maintained in DMEM medium with 10% FBS and 1% penicillin/streptomycin.
Transient Transfection of Cells and GLuc Reporter Transcription Assays
Transfection of the platinated plasmids into mammalian cells was carried out as reported pre-viously (17). The details are reported in the Supplementary Information section.
Results
Transcription Inhibition Strategy
To investigate the extent to which pyriplatin-DNA adducts inhibit transcription, and to gauge possible mechanisms for repairing its monofunctional adducts, transcription assays were carried out utilizing platinated mammalian expression vectors in live mammalian cells of different origin (Table S1). A Gaussia luciferase expression vector, pGLuc, which encodes a secretable form of the enzyme under control of a CMV promoter, was employed. Pyriplatin was incorporated into pGLuc either globally or site-specifically between the CMV promoter and the luciferase gene. Platinated and unplatinated control plasmids were transfected into cells using cationic liposomes. Subsequently, the cell media containing the secreted luciferase were collected at various time intervals. An advantage of the secreted luciferase system is that a time-dependent cellular response to the platinated plasmids can be monitored without lysing the cells, as is necessary using other internal reporter enzyme systems (18, 19). The transcription inhibition activity of pyriplatin, and of cisplatin and oxaliplatin as controls, was determined by quantification of expressed luciferase using coelenterazine as substrate. NER-, MMR-, and SSBR-deficient cells were employed both to monitor transcription inhibition activity of pyriplatin and to identify potential repair mechanisms of pyriplatin-DNA adducts in live cells.
Construction of Globally Platinated Plasmids
pGLuc vectors were globally platinated with different platinum anticancer agents by allowing the plasmids to react with varying concentrations of the compounds in buffer. Platination levels were determined by atomic absorption and UV-vis spectroscopy (12). In Figure 2, the formal ra-tio of platinum to nucleotide in the reaction (rf) is plotted against the amount of platinum bound per nucleotide (rb) for cisplatin, oxaliplatin, and pyriplatin. The slope of the rb vs. rf plot for pyriplatin is identical to that of cisplatin, but much larger than that for oxaliplatin. In other words, pyriplatin reacts with DNA as efficiently as cisplatin, and both compounds react more efficiently than oxaliplatin.
Figure 2.
Plots of rb vs. rf determined for pyriplatin, cisplatin, and oxaliplatin using pGLuc plasmid DNA.
Construction of a Plasmid Containing a Site-Specific Pyriplatin Monofunctional Adduct
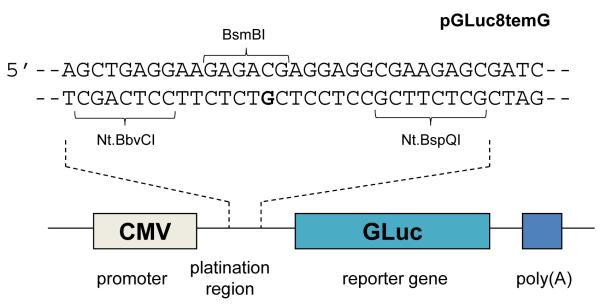
A GLuc vector containing a site-specific cis-{Pt(NH3)2(py)}2+-dG adduct was constructed following the “gapping” strategy reported previously (16). pGLuc was modified to include two unique nicking restriction sites, Nt.BbvCI and Nt.BspQI, for incorporation of 16-mer insertion strands containing either a site-specific cis-{Pt(NH3)2(py)}2+-dG adduct or no platinum (Figure 3). This new plasmid, pGLuc8temG, was then digested with the nicking restriction enzymes Nt.BbvCI and Nt.BspQI to obtain the desired gapped plasmid. The 16-mer insertion strands, containing either a site-specific pyriplatin adduct or no platinum, were annealed and ligated into the gapped plasmids to form the final constructs. The site-specifically platinated and unplatinated control plasmids were designated as pGLuc8temG+IS-PtPy and pGLuc8temG+IS, respectively. Restriction analysis with BsmBI was carried out to confirm the presence of the monofunctional pyriplatin-DNA adduct, and the result is presented in Figure S1. The site-specific pyriplatin adduct in the BsmBI restriction site efficiently inhibits the restriction digestion.
Figure 3.
DNA sequence for building a site-specifically platinated Gaussia luciferase reporter containing a pyriplatin-dG adduct; the platination site is highlighted in bold
Transcription Inhibition Profiles of Pyriplatin in NER-Deficient Cells
Transcription inhibition profiles were obtained for globally and site-specifically platinated probes in NER-deficient cells. Plasmids were transfected into XPF cells, which lack the XPF gene, and into XPFcorr cells, in which the XPF function was restored by introduction of the cDNA for XPF (20). pGLuc globally platinated with pyriplatin was examined first, and expres-sion levels were read after 8, 16, 24, 32, and 44 h of incubation. GLuc expression levels normalized to that of unplatinated plasmid were plotted as a function of Pt/DNA ratios at various time points (Figure 4), or as a function of time at specific Pt/DNA ratios (Figure S2). After 44 h, the transcription levels were substantially restored in both cell lines, indicating repair of pyriplatin-DNA adducts. There was stronger transcription inhibition by pyriplatin in XPF cells. For example, at a Pt/DNA ratio of 23.2, the transcription level recovered from 19.8% at 8 h to 43.4% at 44 h in XPF cells, whereas in XPFcorr cells, transcription recovered from 27.0% at 8 h to 56.3% at 44 h. In addition, D0 values, where D0 is the number of Pt adducts per plasmid required to reduce transcription levels to 37% of control (21), were obtained to quantitate transcription inhibition differences between the two cell lines (Table 1). The increase in D0 values at different time points indicates restoration of transcription, and the higher D0 values in XPFcorr cells suggest a role for NER in repairing pyriplatin-DNA damage.
Figure 4.
Transcription profiles of globally platinated probes with pyriplatin in (A) XPF and (B) XPFcorr cells at 8 h (◆), 16 h (∎), 24 h (▵), 32 h (◻), and 44 h (●). (C) Transcription inhibition by pyriplatin at average loadings of 12.5, 23.2, and 44.1 platinum atoms per plasmid at 44 h in XPF and XPFcorr cells; *, P < 0.05; **, P < 0.01.
Table 1.
D0 valuesa of globally platinated probes with pyriplatin assayed at different time intervals after transfection for XPF and XPFcorr cells.
| Time after Transfection (h) |
XPF (Pt/plasmid) |
XPFcorr (Pt/plasmid) |
|---|---|---|
| 8 | 14.12 ± 2.82 | 17.45 ± 1.35 |
| 16 | 17.30 ± 2.61 | 19.93 ± 0.86 |
| 24 | 19.60 ± 1.99 | 21.86 ± 1.21 |
| 32 | 21.22 ± 1.91 | 23.85 ± 1.23 |
| 44 | 22.34 ± 2.05 | 26.16 ± 1.28 |
D0 value is defined as the number of Pt lesions per plasmid required to reduce transcription levels to 37% of the control.
Transcription profiles of site-specifically platinated pGLuc in XPF and XPFcorr cells were also determined. The transcription levels from a plasmid carrying a single, site-specific pyriplatin-DNA adduct were 37%, 49% and 54% in XPF cells, and 80%, 115% and 123% in XPFcorr cells at 24, 48, and 72 h, respectively. We also studied site-specific pGLuc plasmids containing a cis-{Pt(NH3)2}2+ intrastrand 1,2-d(GpG) cross-link, the major type of cisplatin-DNA adduct, as a control. Transcription levels in the presence of the cis-{Pt(NH3)2}2+ 1,2-d(GpG) cross-link were 53%, 59% and 62% in XPF cells, and 102%, 128% and 132% in XPFcorr cells at 24, 48, and 72 h, respectively (Figure 5). Transcription levels greater than 100% after 48 h in XPFcorr cells for both cisplatin and pyriplatin suggest, perhaps, the stimulation of transcription from newly repaired plasmids. Restoration of XPF function significantly decreased the transcription inhibition induced by a site-specific pyriplatin-dG adduct, further confirming that NER can remove pyrip-latin adducts on DNA. The results from a cis-{Pt(NH3)2}2+ 1,2-d(GpG) cross-linked plasmid are in agreement with our previously published data (17), reinforcing the pivotal role of NER in the repair of cisplatin-DNA cross-links. It is noteworthy that a cis-{Pt(NH3)2(py)}2+ -dG monofunctional adduct inhibits transcription better than a cis-{Pt(NH3)2}2+ 1,2-d(GpG) cross-link in XPF cells.
Figure 5.
Transcription profiles of site-specifically platinated probes containing a cis-{Pt(NH3)2(py)}2+ -dG adduct or a cis-{Pt(NH3)2}2+ 1,2-d(GpG) intrastrand cross-link in XPF and XPFcorr cells.
Osteosarcoma cancer cells (U2OS) with downregulated XPF gene expression (22) were also examined. Transcription profiles of plasmids globally platinated with pyriplatin or oxaliplatin in XPF-normal (U2OS-MOCK) and XPF-knockdown (XPF-1128) cells are shown in Figures 6, S3, S4, and S5. For oxaliplatin, recovery of transcription occurred in both cell lines in a similar manner over a 44 h time period (Figure S4), and there was stronger transcription inhibition in XPF-1128 cells (Figure S5). For pyriplatin, there was little evidence of transcription recovery in U2OS-MOCK cells, and almost none in XPF-1128 cells (Figure S3). Stronger transcription inhibition was also observed in XPF-1128 cells (Figure 6). Calculated D0 values for both oxaliplatin and pyriplatin are tabulated in Table S2. At most time points, pyriplatin has only slightly smaller D0 values than oxaliplatin in U2OS-MOCK cells and XPF-1128 cells.
Figure 6.
Transcription inhibition by pyriplatin at average loadings of 12.5 (upper left), 23.2 (upper right), 44.1 (lower left), and 86.8 (lower right) platinum atoms per plasmid in U2OS-MOCK and XPF-1128 cells; ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001
Transcription Inhibition Profiles of Pyriplatin in MMR-Deficient Cells
In addition to cells deficient in NER, MMR-deficient (HEC59) and -proficient (HEC59+Chr2) cells were examined in transient transfection assays with globally platinated plasmids in order to investigate the potential role of MMR in removing Pt-DNA adducts, including those from cisplatin, oxaliplatin, and pyriplatin. For cisplatin, there was slightly more transcription inhibition in MMR-proficient compared to -deficient cells over 44 h (Figure S6). Transcription levels increased over 44 h in both HEC59 and HEC59+Chr2 cells. At an adduct level of 19.4 Pt/DNA, transcription increased from 19.2% to 35.2% of control in HEC59 cells and from 10.1% to 31.6% in HEC59+Chr2 cells (Figure S6). There was identical transcription inhibition of oxaliplatin in MMR-deficient and -proficient cells, and little recovery over time in both cell lines (Figure S7). As for pyriplatin-modified transcription probes, there were identical transcription profiles in both MMR-deficient and -proficient cells over 44 h, and slightly greater transcription inhibition occurred in HEC59+Chr2 cells (Figure 7). D0 values for cisplatin, oxaliplatin, and pyriplatin in both cell lines are shown in Table S3. Pyriplatin and cisplatin have identical D0 values in HEC59 and HEC59+Chr2 cells, and the values are greater than those of oxaliplatin. There was a small increase in D0 from 8 h to 44 h in both cell lines for cisplatin, oxaliplatin, and pyriplatin. D0 values were slightly smaller in MMR-proficient cells, especially for cisplatin and pyriplatin.
Figure 7.
Transcription inhibition by pyriplatin in MMR-deficient (HEC59) and proficient (HEC59+Chr2) cells. (A) Transcription profiles of globally platinated probes as a function of Pt adducts/DNA at 8 h (◆), 16 h (∎), 24 h (▵), 32 h (◻), and 44 h (●). (B) Transcription inhibition of pyriplatin at 12.5 and 23.2 Pt adducts/DNA as a function of time.
Transcription assays with site-specifically platinated plasmids showed identical results to those obtained for globally platinated transcription probes. Site-specific pGLuc plasmids containing a cis-{Pt(NH3)2}2+ 1,2-d(GpG) cross-link or a cis-{Pt(NH3)2(py)}2+-dG monofunctional adduct were examined. Transcription levels were determined after 24, 48, and 72 h (Figure S8). The transcription inhibition effects of the cis-{Pt(NH3)2(py)}2+-dG adduct were identical to those of a cis-{Pt(NH3)2}2+ 1,2-d(GpG) cross-link in both HEC59 and HEC59+Chr2 cells. There was a small increase in transcription inhibition by pyriplatin in HEC59+Chr2 cells. The transcription levels in the presence of a cis-{Pt(NH3)2}2+ 1,2-d(GpG) cross-link were 66%, 76%, and 90% in HEC59 cells, and 65%, 76%, and 89% in HEC59+Chr2 cells at 24, 48, and 72 h, respectively (Figure S8).
Transcription Inhibition Profiles of Pyriplatin in PARP-Knockdown Cells
PARP-1 knockdown mouse embryonic fibroblasts (MEFS) were studied to reveal the transcription inhibition profiles of cisplatin, oxaliplatin, and pyriplatin. A small recovery in transcription levels was observed for all three platinum compounds tested, especially in PARP-1+/+ cells (Figures S9, S10). Pyriplatin had D0 values identical to those of cisplatin and oxaliplatin in PARP-1+/+ and PARP-1−/− cells (Table S4), further indicating that pyriplatin inhibits transcription as strongly as cisplatin and oxaliplatin. A slight increase of D0 values occurred from 8 h and 44 h in both cell lines for cisplatin, oxaliplatin, and pyriplatin, and the D0 values were slightly smaller in PARP-1−/− cells.
The transcription profiles of site-specifically platinated pGLuc in PARP-1+/+ and PARP-1−/− cells were also determined. Transcription levels in the presence of a site-specific cis-{Pt(NH3)2(py)}2+-dG adduct were 46%, 56% and 76% in PARP-1+/+ cells, and 61%, 72% and 77% in PARP-1−/− cells at 24, 48, and 72 h, respectively. The transcription levels in the presence of a cis-{Pt(NH3)2}2+ 1,2-d(GpG) cross-link were 52%, 60% and 67% in PARP-1+/+ cells, and 61%, 77% and 84% in PARP-1−/− at 24, 48, and 72 h, respectively (Figure S11). A cis-{Pt(NH3)2(py)}2+-dG adduct inhibited transcription as well as a cis-{Pt(NH3)2}2+ 1,2-d(GpG) cross-link in both PARP-1+/+ and PARP-1−/− cells.
Discussion
Transcription inhibition is one of the major consequences of platinum-DNA damage, and there is a correlation between transcription inhibition by platinum compounds and their efficacy as anticancer agents (23, 24). Numerous reports have presented the transcription inhibition of bifunctional platinum antitumor compounds through reconstituted systems or studies in cell extracts or culture. Different mechanisms of transcription inhibition have been proposed, including hijacking of transcription factors, physical blocking of RNA polymerases, and disruption of chromatin structure (24). Knowledge of transcription inhibition at monofunctional platinum-DNA adducts in the cell is, however, very limited. Monofunctional adducts at guanine residues formed by cis-[PtCl(NH3)2(N7-ACV) [ACV=acyclovir, or 9-(2-hydroxyethoxymethyl)guanine], an active antiviral and antitumor compound, terminate DNA and RNA synthesis in vitro (25, 26). Other monofunctional platinum(II) compounds, such as [PtCl(dien)]Cl and [Pt(NH3)3Cl]Cl, or monofunctional adducts of cisplatin, were unable to inhibit RNA (27-29) or DNA (5) polymerase activity in vitro. Pyriplatin effectively blocked RNA synthesis in vitro, and a detailed mechanism of how this monofunctional platinum antineoplastic compound can inhibit pol II has been put forth (13). The present study significantly extends our knowledge of transcription inhibition by monofunctional platinum anticancer agents to live mammalian cells and provides important information about which repair pathways remove monofunctional Pt-DNA adducts.
Transcription Inhibition of Pyriplatin in NER-Deficient Cells
NER is considered the major pathway for removing cisplatin-DNA 1,2-d(GpG) intrastrand cross-links (3, 30, 31). There are two sub-pathways in NER, namely, transcription-coupled NER (TC-NER) and global genome NER (GG-NER). TC-NER-deficient cells are hypersensitive to cisplatin, indicating its critical role in the platinum-DNA damage response (30). The transcription inhibition profile of a reporter gene containing global cisplatin cross-links, or a site-specific cisplatin cross-link, is dramatically different in XPF and XPFcorr cells (17). Here we studied the transcription inhibition profiles of oxaliplatin and pyriplatin in NER-deficient cells including XPF cells. Transcription of a globally pyriplatin-damaged reporter gene recovers slightly over 44 h, and greater transcription inhibition occurs in XPF cells, indicating a role of NER in the repair of pyriplatin-DNA damage. Compared to the transcription inhibition of cisplatin in XPF and XPFcorr cells that we reported previously (17), however, pyriplatin does not evoke significant transcription recovery with time when globally platinated plasmids were utilized, indicating that pyriplatin-dG adducts are more difficult for the cellular machinery to recognize and repair. We also incorporated pyriplatin site-specifically into the pGLuc plasmid between the CMV promoter and the GLuc expression gene to evaluate whether a pyriplatin-DNA adduct can act as a road-block to the transcribing pol II complex in live cells. Robust transcription inhibition by a single pyriplatin-dG adduct was observed in XPF cells. Restoration of the XPF function significantly restored the transcription levels of luciferase from a plasmid site-specifically modified with pyriplatin. Identical results were obtained from plasmids globally platinated with pyriplatin in XPF-knockdown cells, strongly supporting our conclusion that NER plays a key role in the repair of pyriplatin-DNA damage. Moreover, pyriplatin inhibited transcription as effectively as oxaliplatin in U2OS cells, despite their NER deficient status.
Transcription Inhibition of Platinum Compounds in MMR-Deficient Cells
MMR corrects single base mismatches and looped intermediates generated from DNA polymerase slippage during replication and combination, as well as some forms of DNA damage by endogenous or exogenous toxicants (32). DNA mismatches are recognized by hMutSα, a heterodimer of hMSH2 and hMSH6, and hMutSβ, a heterodimer of hMSH2 and hMSH3. The binding of MMR proteins to different types of platinum-DNA adducts has been studied in vitro. hMSH2 binds with some specificity to DNA globally damaged by cisplatin, and hMutSα recognizes cisplatin 1,2-d(GpG) but not 1,3-d(GpTpG) cross-links (33-35). Cell-based studies revealed that MMR-deficiency is correlated with cisplatin resistance (36, 37). This line of evidence, together with other findings, implies that, in addition to the role of MMR in platinum damage repair, MMR proteins bind to cisplatin-DNA damage and initiate a signal transduction pathway leading to cell cycle arrest and apoptosis (38).
We examined endometrial HEC59 (hMSH2-deficient) and HEC59+Chr2 (hMSH2-proficient) adenocarcinoma cells to investigate the transcription inhibition of pyriplatin, as well as the po-tential role of MMR in the repair or processing cisplatin, oxaliplatin, and pyriplatin lesions. Transcription assays with globally platinated plasmids indicated the inhibitory effects of cisplatin and pyriplatin to be greater in hMSH2-proficient cells. When D0 values are considered, pyriplatin is as active as cisplatin in inhibiting transcription of a reporter gene, and oxaliplatin is the most active compound in these cells. Platinum-DNA adducts from cisplatin and pyriplatin are slightly better transcription inhibitors in hMSH2-proficient cells, and no significant difference was observed between hMSH2-deficient and proficient cells when oxaliplatin plasmids were employed (Table S3). These results further demonstrate that MMR may play a role, albeit a minor one, in mediating the cytotoxicity of cisplatin and pyriplatin. The results are in agreement with a previous model in which MMR proteins bind to cisplatin-DNA cross-links but not those from oxaliplatin (39). When site-specifically platinated plasmids were utilized, there were identical transcription inhibition effects for a cisplatin 1,2-d(GpG) cross-link in hMSH2-deficient and – proficient cells. A cis-{Pt(NH3)2(py)}2+-dG adduct, which is as active as a cisplatin 1,2-d(GpG) cross-link in blocking RNA synthesis, had a moderately stronger transcription inhibition effect in hMSH2-proficient cells. Taken together, these results suggest that MMR, instead of being the major repair pathway for removing pyriplatin-DNA adducts, may play a role in initiating apoptosis signaling pathways in response to pyriplatin-DNA damage.
Transcription Inhibition of Platinum Compounds in PARP-1-Knockdown Cells
PARP-1 is one of the most abundant nuclear proteins in eukaryotes. The cellular functions of PARP-1 include modulation of chromatin structure and transcription, as well as DNA repair housekeeping (40). When genomic DNA is mildly damaged, PARP-1 is fully activated to recruit repair machinery and signal downstream effectors (41, 42). PARP-1 mainly detects single strand breaks (SSB), obligatory intermediates in base excision repair (BER) and NER. PARP-1 thus participates actively in BER, and there is accumulating evidence for a role in NER (43). The affinity of PARP-1 for cisplatin-damaged DNA has been established by photocross-linking studies, and in vitro binding assays further revealed the binding of PARP-1 to DNA damaged by cisplatin, oxaliplatin, and pyriplatin (44-47). The transcription inhibition effects of pyriplatin in PARP-1 knockdown cells and the question of whether or not PARP-1 has a significant function in the repair of pyriplatin-DNA adducts in live cells have never been explored until now.
To investigate these possibilities, we utilized PARP-1-knockdown MEFS to reveal the transcription inhibition profile of pyriplatin and the possible role of PARP-1 in repairing pyriplatin-DNA adducts. Our results from globally platinated plasmids demonstrate that pyriplatin has identical D0 values compared to cisplatin and oxaliplatin; thus pyriplatin, once it binds to DNA, inhibits transcription as efficiently as these FDA-approved platinum drugs. Cisplatin and oxaliplatin display greater transcription inhibition in PARP-1−/− cells at both high and low numbers of platinum adducts on the plasmids, implying a role for PARP-1 in repair of cisplatin- or oxaliplatin-damaged DNA (Figure S9). When plasmids globally platinated with pyriplatin were examined, transcription inhibition levels were not notably different for PARP-1 normal vs. knockdown cells, especially at 12.5 and 23.2 Pt/DNA (Figure S10), suggesting that SSBR may not be a determinative repair pathway in response to pyriplatin damage. We also used site-specifically platinated plasmids in this reporter assay. Transcription inhibition effects were slightly lower in the PARP-1 knockdown cells for plasmids containing a site-specific cis-{Pt(NH3)2}2+ 1,2-d(GpG) cross-link or a cis-{Pt(NH3)2(py)}2+-dG adduct (Figure S11). Nevertheless, the transcription inhibition effects of the monofunctional platinum dG adduct are as strong as those of the bifunctional platinum 1,2-d(GpG) cross-link. One explanation for the difference between the results from globally and site-specifically platinated plasmids is that PARP-1 is not fully activated by a single plati-num present on the plasmid. The protein may still bind to the adduct, however, possibly shielding the damage from repair. Such an outcome would result in greater transcription inhibition in PARP-1 normal cells. The results from globally and site-specifically platinated plasmids suggest that SSBR is not the major repair pathway to remove pyriplatin-DNA adducts.
Implications for the Mechanism of Action of Cisplatin and Suggestions for Improvement of Monofunctional Platinum Anticancer Agents
The mechanism of cisplatin action is a multistep process including cell entry, activation, DNA binding, and subsequent cellular responses (2, 3). Cisplatin is transported across the plasma membrane by both passive diffusion and carrier-mediated active transport. Once cisplatin enters cells, the relatively low chloride ion concentration of the cytosol favors the formation of activated aquated species. This active form of cisplatin subsequently attacks nucleophilic centers such as DNA bases, which are mainly responsible for the cytotoxicity of the drug. Cisplatin can also bind other molecules such as glutathione or metallothionein, processes that deactivate the compound. Formation of the platinum-DNA cross-links induces a complex series of cellular responses, including transcription inhibition, cell cycle arrest, and apoptosis. Remarkably, the monofunctional compound pyriplatin appears to have a mechanism of action very similar if not identical to that of cisplatin. This commonality of mechanism despite dramatically different structures for the Pt-DNA adducts, provide support for the current consensus mechanism of the platinum anticancer drugs (1).
Previous reports reveal that pyriplatin is an excellent substrate for organic cation transporters (OCTs) (12) but that the potency of pyriplatin is lower than that of cisplatin against ovarian, breast, and a variety of other types of cancer cells (11). From the present results, we conclude that pyriplatin binds to DNA in cancer cells as efficiently as cisplatin. Pyriplatin, once bound to DNA, inhibits transcription as strongly as cisplatin, and pyriplatin-DNA adducts are more difficult to remove by the cellular repair machinery. NER is mainly responsible for removal of pyriplatin-DNA adducts.
Because pyriplatin and cisplatin share similar DNA binding, transcription inhibition, and repair pathways, we propose three possible reasons to account for the lower cytotoxicity of pyriplatin compared to cisplatin. Firstly, the cell may not accumulate pyriplatin as well as cisplatin. Although taken up by OCTs, pyriplatin may not pass as readily into the cytoplasm and nucleus of the cell. Secondly, pyriplatin may be deactivated more readily prior to DNA binding. Thirdly, transcription inhibition by pyriplatin may not correlate with its cytotoxicity, which we believe to be unlikely in mammalian systems. This analysis, combined with previous results on the structures of site-specifically modified DNA, including a transcription complex with Pol II, has guided the rational design of significantly more potent monofunctional platinum anticancer agents (12, 13). In particular, as will be described elsewhere, monofunctional cationic platinum complexes bearing more extended aromatic fused ring heterocyclic ligands than pyridine have recently been obtained that significantly exceed the level of cisplatin cytotoxicity (48). Such compounds are currently being evaluated in pre-clinical studies for possible development as next-generation platinum anticancer drugs.
Summary and Conclusion
In these studies we established transcription inhibition profiles for pyriplatin-DNA adducts in NER-, MMR- and SSBR-deficient cells, as well as in the corresponding repair-proficient cells. Gaussia luciferase reporters were used to measure the effects of pyriplatin modification of DNA, and plasmid-based reporter assays allowed us to examine transcription inhibition by pyriplatin and the DNA repair capacity in live cells. Pyriplatin reacts with plasmid DNA as efficiently as cisplatin and significantly more so than oxaliplatin. It inhibits transcription in live cells to the same extent as cisplatin when globally platinated plasmids were utilized. A single, site-specific cis-{Pt(NH3)2(py)}2+ -dG adduct within the 3,986-bp plasmid dramatically inhibits transcription, at least as strongly as a cis-{Pt(NH3)2}2+ 1,2-d(GpG) cross-link. NER, rather than MMR or SSBR, plays the most important role in removing pyriplatin-DNA adducts. We provide the first evidence that a monofunctional platinum anticancer agent can block RNA synthesis as efficiently as traditional bifunctional platinum anticancer drugs in live mammalian cells, and our data suggest that the repair mechanism of the monofunctional platinum compounds might be similar to that of bifunctional platinum compounds. Our findings support a role for monofunctional platinum anticancer agents in cancer chemotherapy. Because pyriplatin can react with DNA to a level similar to that of cisplatin and, when bound to DNA, inhibit transcription as effectively as cisplatin, monofunctional platinum compounds with improved cellular uptake and reduced ability to bind cellular targets not associated with cytotoxicity may prove to be a promising route to novel platinum-based therapies.
Supplementary Material
Acknowledgments
The authors are most grateful to Prof. Gan Wang (Wayne State University) for XPFcorr cells, Prof. Paul Chang (Massachusetts Institute of Technology) for PARP-1-knockdown cells, Dr. Thomas Kunkel (National Institutes of Health) for MMR-deficient cells, and Dr. Nora Graf (Massachusetts Institute of Technology) for XPF-knockdown cells. M.M. thanks the Paul Gray Funds for summer research funding. L.S. acknowledges the John Reed Fund for summer research funding.
Grant Support This work was supported by grant CA034992 from the National Cancer Institute to S.J.L.
Footnotes
Disclosure of Potential Conflicts of Interest SJL is a co-founder and serves as head of the Scientific Advisory Board of Blend Therapeutics, a newly formed biopharmaceutical company which is developing nanoparticle drug combinations that may include platinum complexes.
Supporting Information Available Materials and Methods, Tables S1-S4, and Figures S1-S11. This material is available free of charge via the Internet at http://xxx.
References
- 1.Wang D, Lippard SJ. Cellular processing of platinum anticancer drugs. Nat Rev Drug Discov. 2005;4:307–20. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 2.Jamieson ER, Lippard SJ. Structure, recognition, and processing of cisplatin-DNA adducts. Chem Rev. 1999;99:2467–98. doi: 10.1021/cr980421n. [DOI] [PubMed] [Google Scholar]
- 3.Jung Y, Lippard SJ. Direct cellular responses to platinum-induced DNA damage. Chem Rev. 2007;107:1387–407. doi: 10.1021/cr068207j. [DOI] [PubMed] [Google Scholar]
- 4.Brabec V. DNA modifications by antitumor platinum and ruthenium compounds: Their recognition and repair. In: Moldave K, editor. Progress in nucleic acid research and molecular biology. Academic Press; San Diego: 2002. pp. 1–68. [DOI] [PubMed] [Google Scholar]
- 5.Pinto AL, Lippard SJ. Sequence-dependent termination of in vitro DNA synthesis by cis- and trans-diamminedichloroplatinum (II) Proc Natl Acad Sci U S A. 1985;82:4616–9. doi: 10.1073/pnas.82.14.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neplechová K, Kašpárková J, Vrána O, Nováková O, Habtemariam A, Watchman B, et al. DNA interactions of new antitumor aminophosphine platinum(II) complexes. Mol Pharmacol. 1999;56:20–30. doi: 10.1124/mol.56.1.20. [DOI] [PubMed] [Google Scholar]
- 7.Jaramillo D, Wheate NJ, Ralph SF, Howard WA, Tor Y, Aldrich-Wright JR. Polyamide platinum anticancer complexes designed to target specific DNA sequences. Inorg Chem. 2006;45:6004–13. doi: 10.1021/ic060383n. [DOI] [PubMed] [Google Scholar]
- 8.Wheate NJ, Taleb RI, Krause-Heuer AM, Cook RL, Wang S, Higgins VJ, et al. Novel platinum(II)-based anticancer complexes and molecular hosts as their drug delivery vehicles. Dalton Trans. 2007:5055–64. doi: 10.1039/b704973k. [DOI] [PubMed] [Google Scholar]
- 9.Hollis LS, Sundquist WI, Burstyn JN, Heiger-Bernays WJ, Bellon SF, Ahmed KJ, et al. Mechanistic studies of a novel class of trisubstituted platinum(II) antitumor agents. Cancer Res. 1991;51:1866–75. [PubMed] [Google Scholar]
- 10.Hollis LS, Amundsen AR, Stern EW. Chemical and biological properties of a new series of cis-diammineplatinum(II) antitumor agents containing three nitrogen donors: cis-[Pt(NH3)2(N-donor)Cl]+ J Med Chem. 1989;32:128–36. doi: 10.1021/jm00121a024. [DOI] [PubMed] [Google Scholar]
- 11.Lovejoy KS, Serova M, Bieche I, Emami S, D’Incalci M, Broggini M, et al. Spectrum of cellular responses to pyriplatin, a monofunctional cationic antineoplastic platinum(II) compound, in human cancer cells. Mol Cancer Ther. 2011;10:1709–19. doi: 10.1158/1535-7163.MCT-11-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lovejoy KS, Todd RC, Zhang S, McCormick MS, D’Aquino JA, Reardon JT, et al. cis-Diammine(pyridine)chloroplatinum(II), a monofunctional platinum(II) antitumor agent: Uptake, structure, function, and prospects. Proc Natl Acad Sci U S A. 2008;105:8902–7. doi: 10.1073/pnas.0803441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang D, Zhu G, Huang X, Lippard SJ. X-ray structure and mechanism of RNA polyme-rase II stalled at an antineoplastic monofunctional platinum-DNA adduct. Proc Natl Acad Sci U S A. 2010;107:9584–9. doi: 10.1073/pnas.1002565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Damsma GE, Alt A, Brueckner F, Carell T, Cramer P. Mechanism of transcriptional stalling at cisplatin-damaged DNA. Nat Struct Mol Biol. 2007;14:1127–33. doi: 10.1038/nsmb1314. [DOI] [PubMed] [Google Scholar]
- 15.Todd RC, Lovejoy KS, Lippard SJ. Understanding the effect of carbonate ion on cisplatin binding to DNA. J Am Chem Soc. 2007;129:6370–1. doi: 10.1021/ja071143p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ang WH, Brown WW, Lippard SJ. Preparation of mammalian expression vectors incorporating site-specifically platinated-DNA lesions. Bioconjug Chem. 2009;20:1058–63. doi: 10.1021/bc900031a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ang WH, Myint M, Lippard SJ. Transcription inhibition by platinum-DNA cross-links in live mammalian cells. J Am Chem Soc. 2010;132:7429–35. doi: 10.1021/ja101495v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michelini E, Cevenini L, Mezzanotte L, Ablamsky D, Southworth T, Branchini BR, et al. Combining intracellular and secreted bioluminescent reporter proteins for multicolor cell-based assays. Photochem Photobiol Sci. 2008;7:212–7. doi: 10.1039/b714251j. [DOI] [PubMed] [Google Scholar]
- 19.Tannous BA, Kim D-E, Fernandez JL, Weissleder R, Breakefield XO. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol Ther. 2005;11:435–43. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 20.Chen Z, Xu XS, Harrison J, Wang G. Defining the function of xeroderma pigmentosum group F protein in psoralen interstrand cross-link-mediated DNA repair and mutagenesis. Biochem J. 2004;379:71–8. doi: 10.1042/BJ20031143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mello JA, Lippard SJ, Essigmann JM. DNA adducts of cis-diamminedichloroplatinum(II) and its trans isomer inhibit RNA polymerase II differentially in vivo. Biochemistry. 1995;34:14783–91. doi: 10.1021/bi00045a020. [DOI] [PubMed] [Google Scholar]
- 22.Graf N, Ang WH, Zhu G, Myint M, Lippard SJ. Role of endonucleases XPF and XPG in nucleotide excision repair of platinated DNA and cisplatin/oxaliplatin cytotoxicity. ChemBioChem. 2011;12:1115–23. doi: 10.1002/cbic.201000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandman KE, Marla SS, Zlokarnik G, Lippard SJ. Rapid fluorescence-based reporter-gene assays to evaluate the cytotoxicity and antitumor drug potential of platinum complexes. Chem Biol. 1999;6:541–51. doi: 10.1016/S1074-5521(99)80086-6. [DOI] [PubMed] [Google Scholar]
- 24.Todd RC, Lippard SJ. Inhibition of transcription by platinum antitumor compounds. Metallomics. 2009;1:280–91. doi: 10.1039/b907567d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balcarová Z, Kašpárková J, Žákovská A, Nováková O, Sivo MF, Natile G, et al. DNA interactions of a novel platinum drug, cis-[PtCl(NH ) + 3 2(N7-Acyclovir)] Mol Pharm. 1998;53:846–55. [PubMed] [Google Scholar]
- 26.Coluccia M, Boccarelli A, Cermelli C, Portolani M, Natile G. Platinum(II)-acyclovir complexes: synthesis, antiviral and antitumour activity. Met Based Drugs. 1995;2:249–56. doi: 10.1155/MBD.1995.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brabec V, Boudný V, Balcarová Z. Monofunctional adducts of platinum(II) produce in DNA a sequence-dependent local denaturation. Biochemistry. 1994;33:1316–22. doi: 10.1021/bi00172a005. [DOI] [PubMed] [Google Scholar]
- 28.Brabec V, Leng M. DNA interstrand cross-links of trans-diamminedichloroplatinum(II) are preferentially formed between guanine and complementary cytosine residues. Proc Natl Acad Sci U S A. 1993;90:5345–9. doi: 10.1073/pnas.90.11.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corda Y, Job C, Anin MF, Leng M, Job D. Spectrum of DNA-platinum adduct recognition by prokaryotic and eukaryotic DNA-dependent RNA polymerases. Biochemistry. 1993;32:8582–8. doi: 10.1021/bi00084a027. [DOI] [PubMed] [Google Scholar]
- 30.Furuta T, Ueda T, Aune G, Sarasin A, Kraemer KH, Pommier Y. Transcription-coupled nucleotide excision repair as a determinant of cisplatin sensitivity of human cells. Cancer Res. 2002;62:4899–902. [PubMed] [Google Scholar]
- 31.Reed E. Nucleotide excision repair and anti-cancer chemotherapy. Cytotechnology. 1998;27:187–201. doi: 10.1023/A:1008016922425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006;106:302–23. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 33.Mello JA, Acharya S, Fishel R, Essigmann JM. The mismatch-repair protein hMSH2 binds selectively to DNA adducts of the anticancer drug cisplatin. Chem Biol. 1996;3:579–89. doi: 10.1016/s1074-5521(96)90149-0. [DOI] [PubMed] [Google Scholar]
- 34.Duckett DR, Drummond JT, Murchie AI, Reardon JT, Sancar A, Lilley DM, et al. Human MutSα recognizes damaged DNA base pairs containing O6-methylguanine, O4-methylthymine, or the cisplatin-d(GpG) adduct. Proc Natl Acad Sci U S A. 1996;93:6443–7. doi: 10.1073/pnas.93.13.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mu D, Tursun M, Duckett DR, Drummond JT, Modrich P, Sancar A. Recognition and repair of compound DNA lesions (base damage and mismatch) by human mismatch repair and excision repair systems. Mol Cell Biol. 1997;17:760–9. doi: 10.1128/mcb.17.2.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Francia G, Green SK, Bocci G, Man S, Emmenegger U, Ebos JML, et al. Down-regulation of DNA mismatch repair proteins in human and murine tumor spheroids: implications for multicellular resistance to alkylating agents. Mol Cancer Ther. 2005;4:1484–94. doi: 10.1158/1535-7163.MCT-04-0214. [DOI] [PubMed] [Google Scholar]
- 37.Lin X, Howell SB. DNA mismatch repair and p53 function are major determinants of the rate of development of cisplatin resistance. Mol Cancer Ther. 2006;5:1239–47. doi: 10.1158/1535-7163.MCT-05-0491. [DOI] [PubMed] [Google Scholar]
- 38.Topping RP, Wilkinson JC, Scarpinato KD. Mismatch repair protein deficiency compromises cisplatin-induced apoptotic signaling. J Biol Chem. 2009;284:14029–39. doi: 10.1074/jbc.M809303200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fink D, Nebel S, Aebi S, Zheng H, Cenni B, Nehmé A, et al. The role of DNA mismatch repair in platinum drug resistance. Cancer Res. 1996;56:4881–6. [PubMed] [Google Scholar]
- 40.Schreiber V, Dantzer F, Amé JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–28. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 41.Malanga M, Althaus FR. The role of poly(ADP-ribose) in the DNA damage signaling network. Biochem Cell Biol. 2005;83:354–64. doi: 10.1139/o05-038. [DOI] [PubMed] [Google Scholar]
- 42.Bouchard VJ, Rouleau M, Poirier GG. PARP-1, a determinant of cell survival in response to DNA damage. Exp Hematol. 2003;31:446–54. doi: 10.1016/s0301-472x(03)00083-3. [DOI] [PubMed] [Google Scholar]
- 43.Ghodgaonkar MM, Zacal N, Kassam S, Rainbow AJ, Shah GM. Depletion of poly(ADP-ribose) polymerase-1 reduces host cell reactivation of a UV-damaged adenovirus-encoded reporter gene in human dermal fibroblasts. DNA Repair (Amst) 2008;7:617–32. doi: 10.1016/j.dnarep.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 44.Zhang CX, Chang PV, Lippard SJ. Identification of nuclear proteins that interact with platinum-modified DNA by photoaffinity labeling. J Am Chem Soc. 2004;126:6536–7. doi: 10.1021/ja049533o. [DOI] [PubMed] [Google Scholar]
- 45.Guggenheim ER, Xu D, Zhang CX, Chang PV, Lippard SJ. Photoaffinity isolation and identification of proteins in cancer cell extracts that bind to platinum-modified DNA. ChemBioChem. 2009;10:141–57. doi: 10.1002/cbic.200800471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu G, Chang P, Lippard SJ. Recognition of platinum-DNA damage by poly(ADP-ribose) polymerase-1. Biochemistry. 2010;49:6177–83. doi: 10.1021/bi100775t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guggenheim ER, Ondrus AE, Movassaghi M, Lippard SJ. Poly(ADP-ribose) polymerase-1 activity facilitates the dissociation of nuclear proteins from platinum-modified DNA. Bioorg Med Chem. 2008;16:10121–8. doi: 10.1016/j.bmc.2008.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park GY, Lippard SJ. unpublished results.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.