Abstract
The detection of tumor suppressor gene promoter methylation in sputum-derived exfoliated cells predicts early lung cancer. Here we identified genetic determinants for this epigenetic process and examined their biological effects on gene regulation. A two-stage approach involving discovery and replication was employed to assess the association between promoter hypermethylation of a 12-gene panel and common variation in 40 genes involved in carcinogen metabolism, regulation of methylation, and DNA damage response in members of the Lovelace Smokers Cohort (n=1434). Molecular validation of three identified variants was conducted using primary bronchial epithelial cells. Association of study-wide significance (P<8.2×10−5) was identified for rs1641511, rs3730859, and rs1883264 in TP53, LIG1, and BIK, respectively. These SNPs were significantly associated with altered expression of the corresponding genes in primary bronchial epithelial cells. In addition, rs3730859 in LIG1 was also moderately associated with increased risk for lung cancer among Caucasian smokers. Together, our findings suggest that genetic variation in DNA replication and apoptosis pathways impacts the propensity for gene promoter hypermethylation in the aerodigestive tract of smokers. The incorporation of genetic biomarkers for gene promoter hypermethylation with clinical and somatic markers may improve risk assessment models for lung cancer.
Keywords: DNA damage response, promoter hypermethylation, single nucleotide polymorphism, sputum, smoker
Introduction
Lung cancer is the leading cause of cancer-related mortality in both men and women in the United States and occurs largely from chronic exposure to tobacco carcinogens (1). The development of this disease over 30 to 40 years involves field cancerization, characterized as the acquisition of genetic and epigenetic changes in oncogenes and tumor suppressor genes (TSG) throughout the airway epithelium (2–4). The silencing of TSGs through promoter methylation is now recognized as a major and causal epigenetic event that occurs during lung cancer initiation and progression (4). Genes involved in all aspects of normal cell function that include cell cycle regulation, differentiation, adhesion, and death, are silenced by promoter methylation in lung tumors (4). Based on the silencing of key TSGs in the lungs of smokers, we hypothesized that the detection of promoter methylation of TSGs in exfoliated cells in sputum would provide an assessment of the extent of field cancerization that in turn may predict early lung cancer. This hypothesis has been validated in several studies (5–7), suggesting that gene methylation in sputum could be used as a molecular marker for identifying people at high risk for cancer incidence.
The fact that promoter methylation of TSGs is a promising marker for early detection of lung cancer makes understanding factors that influence the individual susceptibility for this epigenetic process throughout the respiratory epithelium a high priority. The precise mechanism by which carcinogens disrupt the capacity of cells to maintain the epigenetic code during DNA replication and repair is largely unknown. Carcinogens within tobacco induce single- and double-strand DNA breaks which, if left unrepaired or if mis-repaired, lead to mutagenic and/or carcinogenic changes in cells (8). Accumulating evidence from our group and others suggests that extensive DNA damage could also be responsible for acquisition of promoter methylation of TSGs during lung carcinogenesis. Support for this supposition was provided through several recent studies from our group (9–12). A highly significant association was observed between DNA repair capacity (DRC) for double strand breaks measured in lymphocytes and the propensity for gene methylation detected in sputum from cancer-free smokers from the Lovelace Smokers Cohort (LSC) (9). A subsequent study in the same cohort identified the dietary factors including folate, leafy green vegetables, and multivitamin use as protective against the acquisition of gene methylation possibly through the modulation of DRC and/or the reduction of DNA damage induced by tobacco-derived carcinogens due to their anti-oxidative effect (10). Chronic exposure of human bronchial epithelial cells to non-cytotoxic levels of tobacco-derived carcinogens induced de novo methylation of TSGs, epithelial-to-mesenchymal transition, and eventually transformation (11). Cuozzo et al. (12) provides a mechanistic link between DNA damage and methylation by demonstrating activation of homologous recombination following introduction of a double strand break and subsequent methylation of the recombinant gene. Together, these studies suggest that chronic DNA damage and reduced DRC could be important determinants for inducing gene methylation.
Several sequence patterns within gene promoters that contain CpG islands and embryonic targets of polycomb-repressive complex 2 are predictive for gene predisposition for methylation in cancer, but cannot discriminate the inter-individual susceptibility for gene silencing (13–17). Sequence variants in promoters associated with reduced gene transcription lead to allele-specific methylation (ASM) and silencing in glutathione S-transferase pi (GSTP1) and O6-methylguanine-DNA methyltransferase (MGMT) in tumors and premalignant tissues (18,19). Mechanisms independent of effects on gene transcription were also identified for ASM of the reversion-induced LIM gene (20). Several studies conducting chromosome-wide or genome-wide surveys on non-imprinted, autosomal regions in human lymphocytes suggest that the majority of TSGs are not silenced by sequence variant dependent ASM (21,22). Based on the likelihood that DNA damage induced by tobacco carcinogens is an important step in the acquisition of de novo methylation and that reduced carcinogen detoxification and DRC have been associated with lung cancer (9–12,23), we tested the hypothesis that genetic variation in some genes involved in these pathways are associated with susceptibility for smokers to acquire gene-specific promoter methylation detected in sputum that contains exfoliated lung cells. A two-stage approach involving discovery and replication was employed to assess the association between promoter methylation of a 12-gene panel in members of the LSC and common variation in 40 genes involved in carcinogen metabolism, regulation of methylation, and DNA damage response, the latter including DNA damage repair, cell cycle regulation, and apoptosis. Molecular validation of significant variants was conducted using primary bronchial epithelial cell cultures.
Materials and Methods
Study Cohort and Sample Collection
The LSC was established in 2001 to conduct longitudinal studies on molecular markers of respiratory carcinogenesis in biological fluids such as sputum from people at risk for lung cancer (9). The enrollment initially focused on female smokers and was expanded to include male smokers in 2004. Enrollment was restricted to current and former smokers age 40 to 74 y with a minimum of 20 pack-years of smoking. Detailed information regarding sample collection was described in Supplementary Materials and Methods. All participants signed a consent form, and the Western Institutional Review Board approved this project.
Methylation of a 12-gene panel was successfully assessed in cytological adequate sputum samples from 1434 cohort members who are either Caucasian or Hispanic and for whom the genotyping call rate was ≥ 75%. Members with other ethnicities were not included in this study because of their low representation in the LSC (overall < 6%). Cohort members were split into two populations for the discovery (n=713) and replication (n=721) based on their methylation index and several non-genetic risk factors for gene methylation including gender, ethnicity, current smoking status, and age at enrollment (10). The demographic characteristics for the cohort members in the discovery and replication stages are shown in Table 1.
Table 1.
Summary of LSC members in the discovery and replication stages *
| Variables | Discovery (n=713) | Replication (n=721) | P values |
|---|---|---|---|
| Age (years, mean ± SD) | 56.5 ± 9.5 | 56.4 ± 9.6 | 0.74 † |
| Gender (male %) | 20.9 | 21.4 | 0.83 ‡ |
| Ethnicity (Hispanics %) | 19.1 | 19.4 | 0.87 ‡ |
| Current smokers (%) | 58.2 | 58.1 | 0.97 ‡ |
| Packyears (mean ± SD) | 40.4 ± 20.7 | 38.7 ± 19.5 | 0.14 § |
| 5–28 ‖ | 30.6 | 33.0 | 0.35 ‡ |
| 28–40 | 30.3 | 31.5 | |
| 40–166 | 39.1 | 35.5 | |
| Methylation index (median, Min-Max) | 2 (0 – 10) | 2 (0 – 10) | 0.51 § |
The overall cohort was split into the discovery and replication groups by a method that kept the two groups balanced with respect to all of the variables included in this table.
Two-sided two-sample t test between discovery and replication.
χ2 test for differences between discovery and replication.
Two-sided Wilcoxon rank sum test between discovery and replication.
Moderate smokers (n=108) with packyears < 20 were included in the data analysis. The average packyears for the 108 moderate smokers are 16.6 ± 3.3.
Sputum Processing and Gene Promoter Methylation
A detailed procedure for sputum collection and processing has been described (10). Briefly, sputum samples were stored in Saccomanno’s fixative. Sputum adequacy, defined as the presence of deep lung macrophages or Curschmann’s spiral (7), was assessed by a pathologist. Twelve genes were selected for analysis of methylation in sputum based on our previous studies establishing their association with risk for lung cancer and their specificity to methylation in epithelial cells (5–7). These twelve genes include p16, MGMT, DAPK, RASSF1A, PAX5α, PAX5β, GATA4, GATA5, SULF2, PCDH20, DAL1, and JPH3. Our ongoing study to identify the best gene panel for early lung cancer detection has screened over 40 genes and suggests that a panel of approximately 12 genes (inclusive of those described above) may provide the best sensitivity and specificity with additional genes not conferring additional risk (Leng unpublished). Genomic DNA isolated from the sputum samples was bisulfite-modified. Given the low percentage (<3%) of lung epithelial cells in sputum samples that also varied significantly between individuals, a two-stage nested MSP was used to detect methylated alleles (5–7). Our assay can reproducibly detect one methylated allele in a background of 20,000 unmethylated alleles (4). Genomic DNA isolated from cell lines that were obtained from and authenticated by the American Type Culture Collection (Manasssas, VA) with methylated or unmethylated genes from the 12-gene panel was included in each batch of the MSP assays for quality control. Experiments were conducted in cell lines passes for a maximum of 6-months post resuscitation.
Candidate Gene and tag SNP Selection
Genes (n = 40) were selected based on the literature derived citations establishing the relevance of genes to the regulation of gene methylation, lung cancer, and pulmonary function (Supplementary Table 1, available online). Tag SNPs (n = 718) were selected using pairwise r2 algorithm based on the phase 2 HapMap CEU database, linkage disequilibrium (LD) database for DNA repair and carcinogen metabolism genes from the University of Southern California (24; Gilliland unpublished), or the LD database for methylation regulatory and apoptotic genes from LRRI (Leng unpublished) for Caucasian and Hispanic populations. Fifty ancestry informative markers (AIMs) were chosen to assess ancestry admixture (25). The description of the two LD databases, strategy for tag SNP selection, efficiency of selected SNPs for tagging unmeasured SNPs, and the selection of AIMs were described in detail in Supplementary Materials and Methods.
SNP Genotyping and Quality Control
A 768-plex oligo pool assay was designed to genotype lymphocyte DNA from the LSC using the Illumina GoldenGate technology. Detailed information about genotyping and quality control was provided in Supplementary Materials and Methods.
Real Time PCR for Measuring Gene Expression in Cell Lines
Normal human bronchial epithelial cells (NHBECs) that were obtained by bronchoscopy from current or former smokers (n = 58) were used in this study. Expression of genes was assessed in cells at passage 1 or 2. Cells were harvested in TRI reagent (Sigma-Aldrich, St. Louis, MO) at approximately 80% confluence and mRNA was isolated following TRI reagent instructions. TaqMan real time PCR was conducted to measure the expression of candidate genes in cDNA using the ΔCT method with both PCNA and β-actin as the endogenous controls. PCNA and β-actin were selected as the endogenous controls in NHBECs because they show minimal variation in expression across the NHBEC samples and are highly correlated (Pearson correlation coefficient = 0.83, P<0.0001).
Ascertainment of Population Admixture
Population admixture was analyzed using the model-based, Bayesian Markov Monte Carlo algorithm implemented in STRUCTURE 2.3.3 (26). A detailed description of ascertaining population admixture in the LSC members was described in the Supplementary Materials and Methods and in reference 27.
Statistical Analysis
Genetic association analysis was first conducted for all 600 SNPs that successfully passed the quality check in the 713 members in the discovery stage (Supplementary Table 2, available online). Only SNPs associated with methylation index with P values ≤ 0.10 in the discovery stage were then analyzed in the replication stage (n=721). SNPs had to pass the replication stage with P < 0.05. The SNPs that satisfied the criteria for both the discovery and replication stages also were analyzed with the combined data set. A Bonferroni adjustment over all SNPs included in the analysis (n=600) was used (P=8.2×10−5).
The methylation index based on a 12- gene panel is a discrete count variable defined as the number of genes in the panel that are methylated and ranges from 0 to 10 in the LSC (Figure 1). Poisson regression was conducted to examine the association between the independent variables, the SNPs/haplotypes, and the outcome variable, the methylation index, with adjustment for covariates selected a priori that include age, gender, ethnicity, current smoking status, and packyears. The additive model was tested for each SNP; and common homozygotes, heterozygotes, and rare homozygotes were coded as 0, 1, and 2, respectively. The Poisson regression model is a generalized linear model that can be used to estimate the mean frequency ratio (FR) and its 95% confidence interval. The FR was calculated from the regression coefficient for an explanatory variable in the Poisson regression model, β, through the equation FR=eβ. A FR value > 1 suggests that a larger value of a covariate is associated with a higher methylation index, while a FR value < 1 suggests that a larger value of a covariate is associated with a lower methylation index.
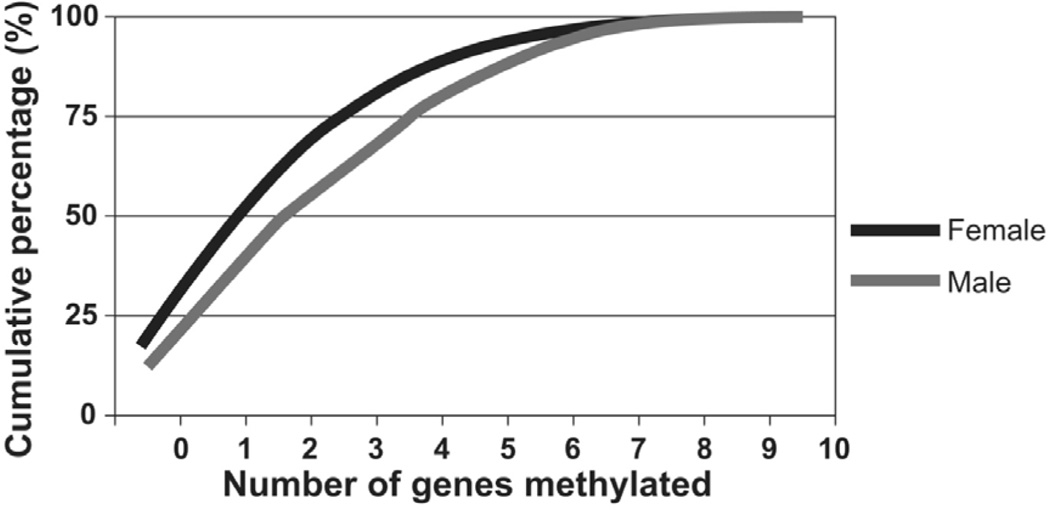
Figure 1.
The distribution of methylation index by gender in the LSC. The median of methylation index in males was higher than that seen in females (3 versus 2 genes methylated, P=8.1×10−12).
The PHASE program was used to reconstruct the haplotypes and to calculate their estimated probabilities from the htSNPs data in a block in Caucasians and Hispanics separately (28,29). The probabilities of the common haplotypes for each individual were used as explanatory variables in the Poisson regression model with adjustment for non-genetic factors to assess the association between the haplotypes and methylation index. Haplotypes with frequency <5% were combined into one group.
The genotype – expression correlation in NHBECs was examined for the SNPs associated with the methylation index. The expression data was presented by setting the level of gene expression with wild homozygote genotypes at 100%. The expression level with heterozygote and variant homozygote was calculated as the ratio relative to that of the wild homozygote. The genotype expression correlation was then analyzed using logarithm-transformed values of the relative quantification of gene expression because this transformation satisfies the normality and homoscedasticity assumption. Logarithm transformed gene expression with high MAF was compared across genotypes using the one way ANOVA. For SNPS with low MAF, a dominant model that combines variant homozygotes and heterozygotes was applied and the difference between the two genotype groups was compared using the student t test. The statistical analyses were conducted using SAS software, version 9.2 (SAS Institute, Cary, NC).
Results
Association between individual SNPs and methylation index
Individual SNP association analysis identified 109 SNPs associated with the methylation index with P value < 0.10 in the discovery stage (Table 2; not shown). These 109 SNPs were then analyzed in the replication stage. Eleven SNPs in LIG1, TP53, BIK, BMF, and BAK1 were associated with methylation index with P ≤ 0.05 in the replication stage (Table 2). Seven of these 11 SNPs were tag SNPs in LIG1. Combined analysis showed that the overall P value for three SNPs including rs1641511, rs3730859, and rs1883264 reached study-wide significance (Bonferroni corrected P = 8.2×10−5). All other SNPs genotyped in the replication stage failed to replicate the associations observed in the discovery stage at the significance level of 0.05. Relaxing the P value from 0.10 to 0.15 for identifying SNPs to be carried over into the replication stage did not identify any additional SNPs reaching study-wide significance.
Table 2.
SNPs with the most significant association with gene methylation based on a 12-gene panel in the LSC *
| Genes | SNPs | Discovery (n=713) | Replication (n=721) | Combined analysis (n=1434) | |||
|---|---|---|---|---|---|---|---|
| FR (95% CI) | P values | FR (95%CI) | P values | FR (95%CI) | P values | ||
| TP53 | rs1641511 | 1.09 (1.01–1.18) | 2.8×10−2 | 1.17 (1.08–1.27) | 1.3×10−4 | 1.13 (1.07–1.19) | 1.8×10−5 |
| LIG1 | rs3730859 | 1.10 (1.02–1.18) | 1.2×10−2 | 1.14 (1.06–1.22) | 4.9×10−4 | 1.11 (1.06–1.17) | 3.0×10−5 |
| BIK | rs1883264 | 0.91 (0.82–1.00) | 5.7×10−2 | 0.82 (0.74–0.91) | 1.3×10−4 | 0.86 (0.80–0.93) | 5.4×10−5 |
| LIG1 | rs7246512 | 0.92 (0.85–1.01) | 7.1×10−2 | 0.86 (0.78–0.94) | 1.3×10−3 | 0.89 (0.84–0.95) | 5.6×10−4 |
| LIG1 | rs436943 | 0.88 (0.80–0.97) | 6.9×10−3 | 0.90 (0.81–0.99) | 2.6×10−2 | 0.89 (0.83–0.95) | 6.4×10−4 |
| LIG1 | rs156641 † | 1.09 (1.02–1.17) | 1.0×10−2 | 1.08 (1.01–1.15) | 3.5×10−2 | 1.08 (1.03–1.14) | 1.3×10−3 |
| BMF | rs3923235 | 1.09 (1.01–1.17) | 2.7×10−2 | 1.08 (1.01–1.17) | 3.2×10−2 | 1.09 (1.03–1.14) | 2.2×10−3 |
| LIG1 | rs156640 | 1.06 (0.99–1.14) | 9.9×10−2 | 1.11 (1.03–1.20) | 6.2×10−3 | 1.08 (1.03–1.14) | 3.5×10−3 |
| LIG1 | rs156645 | 1.08 (1.01–1.16) | 3.0×10−2 | 1.08 (1.01–1.16) | 3.1×10−2 | 1.08 (1.02–1.13) | 3.8×10−3 |
| BAK1 | rs210134 | 1.065 (0.99–1.15) | 9.7×10−2 | 1.09 (1.02–1.17) | 1.6×10−2 | 1.08 (1.02–1.13) | 5.8×10−3 |
| LIG1 | rs4802436 | 0.93 (0.85–1.01) | 9.2×10−2 | 0.91 (0.83–0.99) | 3.3×10−2 | 0.92 (0.86–0.98) | 8.9×10−3 |
From Poisson regression models with adjustment for age, gender, ethnicity, current smoking status, and pack years.
Rs156641 is a redundant tag SNP for rs156645 (r2=1).
Association between haplotype alleles in LIG1 and methylation index
Because multiple tag SNPs were associated with methylation index in LIG1, a haplotype-based analysis was conducted to integrate the genetic variation in each individual genomic locus. LIG1 resides in an 84-kb haplotype block (chr19: 53296285– 53381205, NCBI B36) that contains three genes: PLA2G4C, LIG1, and LOC374920 (Supplementary Figure 1A, available online). Six haplotype tagging SNPs (htSNPs) including rs11564509, rs251692, rs3730859, rs3730895, rs4802436, and rs972800 allowed the construction of the four common haplotype alleles with minimal Rh2 over 0.82 in both Caucasians and Hispanics (24). These six htSNPs accounted for > 84% of the haplotype variation in the LIG1 block in Caucasians and Hispanics in the LSC (Table 3). The likelihood ratio global tests of association indicate that the genetic variation in the haplotype block where LIG1 resides as a locus is strongly associated with methylation index in the entire LSC (P=0.00029; not shown). Hap1 strongly tagged by rs3730859 is the only risk haplotype allele associated with increased methylation index compared to all other haplotype alleles (FR=1.09, P=0.00036, Table 3). Hap3 solely tagged by rs4802436 was associated with a reduction of methylation index compared with all other haplotype alleles (FR=0.93, P=0.023, Table 3). However, when Hap2, Hap4, and others of LIG1 were used as the reference for comparison in a Poisson regression model, only Hap1 was significantly associated with increased methylation index (P=0.0032, Table 3), supporting Hap1 as the most influential haplotype allele defining the global association between this locus and methylation index.
Table 3.
Haplotype based association analysis in LIG1 for methylation index in the LSC (n=1434) *
| Haplotype alleles | Frequency | Analysis 1 † | Analysis 2 ‡ | |||
|---|---|---|---|---|---|---|
| CEU | HISP | FR (95%CI) | P values | FR (95%CI) | P values | |
| Hap1 (CGTGAT) | 0.35 | 0.31 | 1.09 (1.04–1.15) | 0.00036 | 1.08 (0.03–1.14) | 0.0032 |
| Hap2 (CACTAT) | 0.24 | 0.30 | 1.03 (0.98–1.09) | 0.27 | Reference | — |
| Hap3 (CACTGT) | 0.17 | 0.13 | 0.93 (0.87–0.99) | 0.023 | 0.96 (0.89–1.03) | 0.26 |
| Hap4 (TGCGAT) | 0.09 | 0.10 | 0.96 (0.88–1.04) | 0.29 | Reference | — |
| Others | 0.15 | 0.16 | 0.90 (0.84–0.97) | 0.0038 | Reference | — |
FR and 95% CI were estimated in a Poisson regression with adjustment for age, gender, ethnicity, current smoking status, and packyears.
All other haplotype alleles were used as reference for comparison in a Poisson regression.
Hap2, Hap4, and others of LIG1 were used as reference for comparison in a Poisson regression to compare the effect of Hap1 and Hap3 in LIG1.
Genotype – expression correlation for LIG1, TP53, and BIK
Characterization of the LD for SNPs located 500kb surrounding each of the top three SNPs using the 1000 Genome Pilot1 CEU population identified 60 SNPs that have high LD (r2 > 0.8) with these three SNPs. Interestingly, all 60 SNPs locate within the expanded regions of the three candidate genes (Supplementary Figure1B – 1D, available online), suggesting that it is unlikely that the association seen for these three SNPs is due to long-range LD with functional SNPs located further away from these candidate regions. The functional potential for these SNPs was assessed by searching the FuncPred: Functional SNP Prediction module of SNPinfo Web Server (30). No known nonsynonymous SNPs from the genes in the four candidate regions were identified to be in high LD with these SNPs. However, 13 of the 60 SNPs were predicted either to affect the binding of transcription factors or microRNAs or to locate in an exonic splicing enhancer or silencer element (Supplementary Table 3, available online). Specifically, seven SNPs surrounding the transcription start site of LIG1 that are in high LD with rs3730859 (r2 > 0.93) were predicted to affect the binding of transcription factors.
Thus, we examined whether the top three SNPs could in cis affect the expression of LIG1, TP53, and BIK in NHBECs. The expression of LIG1, TP53, and BIK was highly correlated in NHBECs with Spearman correlation coefficients from 0.60 to 0.75 (P values <0.0001; not shown). As a control, the expression of PLA2G4C which resides in the same haplotype block and directly upstream of LIG1 had no or weak correlation with the expression of LIG1 and the other genes (Spearman correlation coefficients from 0.16 to 0.42, not shown). Genotype – expression correlation analysis found strong associations between rs3730859 and LIG1, between rs1641511 and TP53, and between rs1883264 and BIK (Table 4). The most striking difference was observed for rs3730859 with the variant homozygotes associated with a 74% reduction of LIG1 expression compared with the wild homozygotes (P=0.002). In addition, as expected, the genotype expression correlation is more significant for LIG1 using PCNA as the endogenous control compared with β-actin because LIG1 expression is cell cycle regulated (31). The genotype expression correlation for TP53 has comparable results between PCNA and β-actin, a gene whose expression during the cell cycle is influenced by the presence of DNA damage. BIK expression is not cell cycle related and as such the significant genotype expression correlation was identified using β-actin as the endogenous control. More importantly, the repressive effect of current smoking status on BIK expression (32) can only be confirmed with β-actin as the endogenous control (36% reduction in current smokers, P=0.048, Leng unpublished), supporting the use of β-actin for the relative quantification of BIK expression. No association between SNPs and PLA2G4C was identified (not shown).These results suggest that SNPs associated with methylation index could in cis affect the expression of LIG1, TP53, and BIK.
Table 4.
Association of genotype and gene expression *
| Gene | SNP | Genotype | n † | Gene expression 1 ‡ | Gene expression 2 § | ||
|---|---|---|---|---|---|---|---|
| Mean ± SD | P values | Mean ± SD | P values | ||||
| LIG1 | Rs3730859 | G/G | 17 | 1.00 ± 0.86 | 0.002 | 1.00 ± 0.86 | 0.05 |
| G/A | 31 | 0.70 ± 0.68 | 0.88 ± 1.06 | ||||
| A/A | 11 | 0.26 ± 0.30 | 0.43 ± 0.54 | ||||
| TP53 | Rs1641511 | A/A | 19 | 1.00 ± 0.81 | 0.01 | 1.00 ± 0.81 | 0.02 |
| A/G + G/G | 39 (6) | 0.60 ± 0.53 | 0.61 ± 0.35 | ||||
| BIK | Rs1883264 | G/G | 42 | 1.00 ± 0.87 | 0.36 | 1.00 ± 0.75 | 0.04 |
| G/C + C/C | 17 (1) | 0.86 ± 0.92 | 0.62 ± 0.31 | ||||
Logrithm transformed LIG1 expression was compared across rs3730859 genotypes using the one way ANOVA. A dominant model that combines variant homozygotes and heterozygotes was applied for rs1641511 and rs1883264 and the difference between the two genotype groups was compared using the student t test.
Number in parentheses indicates number of variant homozygotes.
PCNA was used as the endogenous control to calculate the relative quantification of gene expression. The average of gene expression in wild homozygotes was normalized to 100%.
β-Actin was used as the endogenous control to calculate the relative quantification of gene expression. The average of gene expression in wild homozygotes was normalized to 100%.
Ancestry admixture in LSC and its effect on genetic association
Hispanics in the LSC are highly admixed between Native American ancestry and European ancestry. The component of Native American ancestry in self-reported Hispanics ranges from 18.2 to 56.0% with an average of 33.7%. In contrast, the average component of Native American ancestry in self-reported Caucasians is < 1.0% with only 10 subjects having Native American ancestry >10%. The African ancestry is very low in both Hispanics (2.1%) and Caucasians (0.3%) enrolled in the LSC. In a comparison of methods for adjusting for ancestry, no substantial differences were observed between the results obtained from Poisson regression models that included self-reported ethnicity and those that included the components of the Native American ancestry and African ancestry (not shown). Furthermore, limiting the analysis to Caucasians with European ancestry > 90% identified similar SNP associations. Thus, population admixture is an unlikely explanation for the genetic association identified.
Discussion
This is the first study to comprehensively evaluate the relationship between methylation index in a large population of lung-cancer free smokers and common variation in 40 candidate genes involved in carcinogen metabolism, regulation of methylation, and DNA damage response pathways. Our findings support a role for genetic variation in LIG1, TP53, and BIK as predictors for the acquisition of gene promoter methylation in exfoliated cells in smokers’ sputum. Specifically, molecular validation through a genotype – expression correlation analysis indicates that the three most significant SNPs in LIG1, TP53, and BIK affect the expression of these three genes in cis in NHBECs. Thus, genetic variation in genes affecting DNA replication and apoptosis impacts the propensity for gene promoter methylation in the aerodigestive tract of smokers and this effect may be driven by the change in the endogenous expression of LIG1, TP53, and BIK.
Epigenetic silencing of TSGs in lung cells is an intermediate biomarker for lung cancer. Therefore, genetic variation that is strongly associated with epigenetic silencing of TSGs should also impact the risk for lung cancer. Indeed, this premise is supported by studies that systematically assessed the association between tag SNPs in LIG1 and risk for lung cancer (33–36). The criteria for selecting these studies to be summarized include large samples size (>500 pairs cases and controls), Caucasian ethnicity, pathway or genome wide association studies, and adequate SNP coverage in LIG1. These criteria were set to minimize the confounding from the publication bias and to ensure inclusion of the sufficiently powered studies. Only association with lung cancer in moderate and heavy smokers (current or former) was presented because over 90% of cohort members enrolled in the LSC are smokers with over 20 pack-year smoking history. A case-control study conducted in the Pittsburgh Lung Screening Study cohort included 722 lung cancer patients and 929 controls who were Caucasian and current or ex-cigarette smokers with > 10 pack-year smoking history and found that subjects carrying the variant homozygote for rs10500298 that is in high LD with rs3730859 (r2=0.89) in the 1k genome project CEU population have a 96% increased risk for lung cancer (P<0.0001) (33). A second case-control study conducted in the Beta-Carotene and Retinol Efficacy Trial cohort also identified rs156640 and rs156641 that is in high LD with rs3730859 (r2=0.89) in CEU as a strong risk factor for lung cancer in Caucasian smokers with >20 pack-year of smoking history (ORs=1.20, Ps<0.003, 746 cases and 1477 controls) (34). Two lung cancer GWAS studies conducted in United States populations were queried as well and subjects with homozygous rs156641 have a 22% increased risk for lung cancer in subjects with over 20 pack-year smoking history in MD Anderson GWAS study (P<0.14, 1004 cases and 926 controls) (35). However, no association was identified between LIG1 SNPs and risk for lung cancer in two US populations used in the discovery stage in the NCI GWAS study (1652 cases and 1212 controls) (36). The SNPs in TP53 and BIK associated with methylation index were also queried in the MD Anderson and NCI GWAS with no significant association to lung cancer identified (not shown). Together, these studies support a moderate association for rs3730859 in LIG1 with risk for the smoking-induced lung cancer and this effect may be mediated by its affect on the predisposition for epigenetic silencing of TSGs.
Among the three DNA ligase families (LIG1, LIG3, and LIG4) in vertebrates, LIG1 plays an essential role in the joining of Okazaki fragments during lagging strand synthesis and is also implicated in multiple DNA repair pathways, although the role of LIG1 in DNA repair can be substituted by other DNA ligases (37–40). Deficiency in LIG1 expression results in the delayed joining of Okazaki fragments, genome instability, and increased incidence of epithelial cancers and lymphoma (37–40). Expression of LIG1 in lung tumors (n=23) is reduced by 34% compared to that in paired distant normal tissues providing further support for a functional deficiency of this gene in lung carcinogenesis (P=0.028, Leng unpublished). The strong association between genetic variation in LIG1 and methylation index found in this study provides in vivo evidence that incomplete loss of LIG1 function due to SNPs could promote the epigenetic silencing of TSGs which in turn contributes to the epithelial carcinogenesis. The mechanism for the effect of LIG1 deficiency on gene methylation is unclear. A recent study found that the replication defect in 46BR.1G1 cells which are homozygote for the Arg771Trp mutation and retain only 3 to 5% of normal LIG1 activity results in the generation of endogenous single- and double-stranded DNA breaks during S phase (40). These DNA breaks behind the replication fork induce a persistent activation of the ATM/CHK2 pathway without triggering the S-phase-specific DNA damage checkpoint and lead to an altered chromatin structure needed for efficient processing of the DNA damage throughout the cell cycle. Thus, the replication timing could be disrupted in cells with LIG1 deficiency because of the inefficient joining of Okazaki fragments in newly synthesized DNA and the extensively altered chromatin structure. This may be a mechanism by which the guidance of the cytosine-DNA methytransferase to regions that are to be methylated is disrupted during DNA replication and the normally unmethylated regions of DNA become aberrantly methylated (41). Although such a scenario is clearly speculative, an in vitro malignant transformation model or in vivo tumorigenesis model using LIG1 knock out human epithelial cells or animals could be used to directly test this hypothesis.
Evading apoptosis is a hallmark of cancer and occurs during the progression from premalignant lesions to carcinoma in the lung (42). Apoptosis occurs through two mechanisms: the death receptor FAS/FAS ligand (also called extrinsic) pathway and the mitochondrial (DNA damage–induced and p53-mediated, also called intrinsic) pathway (43). The intrinsic apoptosis pathway plays a key role in eliminating lung epithelial cells with extensive DNA damage upon carcinogen exposure (43). Thus, the association between functional SNPs in TP53 and BIK and methylation index support the concept that the TP53 mediated apoptosis pathway could prevent the acquisition of heritable genetic and epigenetic changes through eliminating lung cells with extensive DNA damage. The loss of TP53 function by gene mutation is a common genetic alteration found in human cancers and over 50% of lung cancer cases carry somatic mutation of TP53 (44). TP53 mutation or reduced level of wild-type TP53 protein was associated with apoptosis suppression during progression from premalignant lesions to carcinoma in the lung (42). Thus, rs1641511 that was associated with a 40% reduction of TP53 transcription could render lung cells more resistant to apoptosis upon extensive DNA damage in smokers, which in turn could result in an increased methylation index. Although rs1641511 is geographically closer to ATP1B2, the gene downstream of TP53, LD analysis using the Phase 2 HapMap CEU data and the 1000 genome project identify one SNP (rs72829457) around 3kb downstream of TP53 in moderate LD with rs1641511 (D'=1, r2=0.5). Moreover, the deletion/insertion polymorphisms that usually account for 15–20% of the entire genetic variation, are not available from the above two databases. Thus, it is possible that the missing deletion/insertion polymorphisms in high LD with rs1641511 are causal for the reduced expression of TP53.
BIK (Bcl2-interacting killer) is the founding member of the pro-apoptotic BH3-only proteins (45). BIK functions as a death sensor to mediate activation of the mitochondrial apoptosis pathway as shown in response to oncogenic stress signals or DNA damage in epithelial cancer cell lines (45). BIK’s role in cancer development may vary by tumor type. Inactivation of BIK in some primary tissues occurs frequently by chromosomal deletion in renal cell carcinomas (40%), colorectal cancers (22%), and gliomas (42%), but only occasionally in head and neck cancers (5%; 45). Somatic mutations that lead to amino acid changes were also detected in 8% of patients with B cell lymphomas (45). Epigenetic silencing was also proposed as a potential mechanism to silence BIK expression in specific tumor types, although this mechanism was only indirectly tested based on BIK re-expression in tumor cell lines treated with DNA demethylating agent and/or histone deacetylase inhibitor (45). Rs1883264 associated with reduced BIK expression in NHBECs is actually protective for gene methylation in smokers. This seems to be in conflict with the pro-apoptotic role of BIK identified in epithelial cancer cell lines. However, BIK is over expressed in lung tumors (n=23) versus paired distant normal tissues (P=0.015, Leng unpublished). Furthermore, CpG sites in the BIK promoter and exon1 are not methylated in lung adenocarcinomas interrogated on the Illumina HumanMethylation 27K BeadChip (Leng unpublished). Consistent with these findings in lung cancer, BIK over expression was also reported in breast tumors (46). In addition, poor prognosis of non-small cell lung cancers correlated with high expression of BIK (47). These results support different mechanisms for BIK regulation in cancer biology by tumor site and carcinogen exposure pattern. Thus, the protective effect of rs1883264 on gene methylation in smokers identified in our studies is consistent with the increased expression of BIK in lung tumors. Additional mechanistic studies are required to understand the regulation of BIK biology in cigarette smoke-induced lung cancer.
These studies identified the DNA replication and apoptosis pathways as new determinants for acquiring gene methylation in lung cells from smokers. Lung cancer GWAS studies successfully identified several loci in the human genome that are associated with risk for lung cancer at 10−8 significance level (35,36). However, these validated genetic variants only explain a small fraction of lung cancer heritability. Some of the missing heritability likely exists in the genetic variants with less significant P values. The detection of promoter methylation of TSGs in exfoliated cells in sputum as an assessment of the extent of field cancerization in the lung can be used as a functional readout to help identify the true association from the candidate SNPs with GWAS P values > 10−8. Thus, the genetic variants in LIG1 provide a proof-of-concept that SNPs strongly associated with risk for gene methylation in lung cells from smokers can be moderately associated with risk for lung cancer and should be incorporated into risk assessment models for lung cancer.
Supplementary Material
Acknowledgments
We thank Kieu C. Do, Amanda M. Bernauer, and Cynthia L. Thomas at Lovelace Respiratory Research Institute and Autumn Gaitherdavis at University of Pittsburgh School of Medicine for their technical assistance in conducting methylation-specific PCR assay, gene expression assay, and culture of human bronchial epithelial cells.
Grant support
This work was primarily supported by National Cancer Institute (NCI) R01 CA097356 and the State of New Mexico as a direct appropriation from the Tobacco Settlement Fund to S.A.B. Support for previous lung cancer case control studies was through NCI U19CA148127 and R01CA121197 to C.I.A., NCI P50-CA090440 to J.M.S., individual contracts from the NCI to the University of Colorado Denver (NO1-CN-25514), Georgetown University (NO1-CN-25522), the Pacific Health Research Institute (NO1-CN-25515), the Henry Ford Health System (NO1-CN-25512), the University of Minnesota, (NO1-CN-25513), Washington University (NO1-CN-25516), the University of Pittsburgh (NO1-CN-25511), the University of Utah (NO1-CN-25524), the Marshfield Clinic Research Foundation (NO1-CN-25518), the University of Alabama at Birmingham (NO1-CN-75022), Westat, Inc. (NO1-CN-25476), and the University of California, Los Angeles (NO1-CN-25404), American Cancer Society, and the Intramural Research Program of the National Institutes of Health, NCI, Division of Cancer Epidemiology and Genetics.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org)
References
- 1.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: The impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953;6:963–968. doi: 10.1002/1097-0142(195309)6:5<963::aid-cncr2820060515>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 3.Strong MS, Incze J, Vaughan CW. Field cancerization in the aerodigestive tract--its etiology, manifestation, and significance. J Otolaryngol. 1984;13:1–6. [PubMed] [Google Scholar]
- 4.Belinsky SA. Gene-promoter hypermethylation as a biomarker in lung cancer. Nat Rev Cancer. 2004;4:707–717. doi: 10.1038/nrc1432. [DOI] [PubMed] [Google Scholar]
- 5.Palmisano WA, Divine KK, Saccomanno G, Gilliland FD, Baylin SB, Herman JG, et al. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res. 2000;60:5954–5958. [PubMed] [Google Scholar]
- 6.Belinsky SA, Klinge DM, Dekker JD, Smith MW, Bocklage TJ, Gilliland FD, et al. Gene promoter methylation in plasma and sputum increases with lung cancer risk. Clin Cancer Res. 2005;11:6505–6511. doi: 10.1158/1078-0432.CCR-05-0625. [DOI] [PubMed] [Google Scholar]
- 7.Belinsky SA, Liechty KC, Gentry FD, Wolf HJ, Rogers J, Vu K, et al. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. Cancer Res. 2006;66:3338–3344. doi: 10.1158/0008-5472.CAN-05-3408. [DOI] [PubMed] [Google Scholar]
- 8.Hang B. Formation and repair of tobacco carcinogen-derived bulky DNA adducts. J Nucleic Acids. 2010 doi: 10.4061/2010/709521. doi:10.4061/2010/709521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leng S, Stidley CA, Willink R, Bernauer A, Do K, Picchi MA, et al. Double-strand break damage and associated DNA repair genes predispose smokers to gene methylation. Cancer Res. 2008;68:3049–3056. doi: 10.1158/0008-5472.CAN-07-6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stidley CA, Picchi MA, Leng S, Willink R, Crowell RE, Flores KG, et al. Multivitamins, folate, and green vegetables protect against gene promoter methylation in the aerodigestive tract of smokers. Cancer Res. 2010;70:568–574. doi: 10.1158/0008-5472.CAN-09-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tellez CS, Juri DE, Do K, Bernauer AM, Thomas CL, Damiani LA, et al. EMT and stem cell-like properties associated with miR-205 and miR-200 epigenetic silencing are early manifestations during carcinogen-induced transformation of human lung epithelial cells. Cancer Res. 2011;71:3087–3097. doi: 10.1158/0008-5472.CAN-10-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuozzo C, Porcellini A, Angrisano T, Morano A, Lee B, Di Pardo A, et al. DNA damage, homology-directed repair, and DNA methylation. PLoS Genet. 2007;3:e110. doi: 10.1371/journal.pgen.0030110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCabe MT, Lee EK, Vertino PM. A multifactorial signature of DNA sequence and polycomb binding predicts aberrant CpG island methylation. Cancer Res. 2009;69:282–291. doi: 10.1158/0008-5472.CAN-08-3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feltus FA, Lee EK, Costello JF, Plass C, Vertino PM. Predicting aberrant CpG island methylation. Proc Natl Acad Sci U S A. 2003;100:12253–12258. doi: 10.1073/pnas.2037852100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Estécio MR, Gallegos J, Vallot C, Castoro RJ, Chung W, Maegawa S, et al. Genome architecture marked by retrotransposons modulates predisposition to DNA methylation in cancer. Genome Res. 2010;20:1369–1382. doi: 10.1101/gr.107318.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 18.Leng S, Bernauer AM, Hong C, Do KC, Yingling CM, Flores KG, et al. The A/G allele of rs16906252 predicts for MGMT methylation and is selectively silenced in premalignant lesions from smokers and in lung adenocarcinomas. Clin Cancer Res. 2011;17:2014–2023. doi: 10.1158/1078-0432.CCR-10-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rønneberg JA, Tost J, Solvang HK, Alnaes GI, Johansen FE, Brendeford EM, et al. GSTP1 promoter haplotypes affect DNA methylation levels and promoter activity in breast carcinomas. Cancer Res. 2008;68:5562–5571. doi: 10.1158/0008-5472.CAN-07-5828. [DOI] [PubMed] [Google Scholar]
- 20.Boumber YA, Kondo Y, Chen X, Shen L, Guo Y, Tellez C, et al. An Sp1/Sp3 binding polymorphism confers methylation protection. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000162. e1000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerkel K, Spadola A, Yuan E, Kosek J, Jiang L, Hod E, et al. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet. 2008;40:904–908. doi: 10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Rohde C, Reinhardt R, Voelcker-Rehage C, Jeltsch A. Non-imprinted allele-specific DNA methylation on human autosomes. Genome Biol. 2009;10:R138. doi: 10.1186/gb-2009-10-12-r138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sigurdson AJ, Jones IM, Wei Q, Wu X, Spitz MR, Stram DA, et al. Prospective analysis of DNA damage and repair markers of lung cancer risk from the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Carcinogenesis. 2011;32:69–73. doi: 10.1093/carcin/bgq204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haiman CA, Hsu C, de Bakker PI, Frasco M, Sheng X, Van Den Berg D, et al. Comprehensive association testing of common genetic variation in DNA repair pathway genes in relationship with breast cancer risk in multiple populations. Hum Mol Genet. 2008;17:825–834. doi: 10.1093/hmg/ddm354. [DOI] [PubMed] [Google Scholar]
- 25.Conti DV, Lee W, Li D, Liu J, Van Den Berg D, Thomas PD, et al. Pharmacogenetics of Nicotine Addiction and Treatment Consortium. Nicotinic acetylcholine receptor beta2 subunit gene implicated in a systems-based candidate gene study of smoking cessation. Hum Mol Genet. 2008;17:2834–2848. doi: 10.1093/hmg/ddn181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bruse S, Sood A, Petersen H, Liu Y, Leng S, Celedón JC, et al. Hispanic smokers have lower odds of COPD and less decline in lung function than non-Hispanic whites. American Journal of Respiratory and Critical Care Medicine. doi: 10.1164/rccm.201103-0568OC. Article in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Z, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009;37(Web Server issue):W600–W605. doi: 10.1093/nar/gkp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montecucco A, Biamonti G, Savini E, Focher F, Spadari S, Ciarrocchi G. DNA ligase I gene expression during differentiation and cell proliferation. Nucleic Acids Res. 1992;20:6209–6214. doi: 10.1093/nar/20.23.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mebratu YA, Schwalm K, Smith KR, Schuyler M, Tesfaigzi Y. Cigarette smoke suppresses Bik to cause epithelial cell hyperplasia and mucous cell metaplasia. Am J Respir Crit Care Med. 2011;183:1531–1538. doi: 10.1164/rccm.201011-1930OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buch SC, Diergaarde B, Nukui T, Day RS, Siegfried JM, Romkes M, et al. Comprehensive survey of genetic variability in nucleotide and base excision repair pathway and cell cycle control genes in relation to lung cancer susceptibility. Molecular Carcinogenesis. doi: 10.1002/mc.20858. Article in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakoda Lori C., Loomis Melissa M., Doherty Jennifer A., Barnett Matt J., Julianto Liberto, Neuhouser Marian L., Thornquist Mark D., Weiss Noel S., Goodman Gary E., Chen Chu. Germline variation in nucleotide excision repair genes and lung cancer risk in smokers. Paper presented at 102nd annual meeting of the American Association for Cancer Research; April 2011; Orlando, FL. [Google Scholar]
- 35.Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1 . Nat Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Landi MT, Chatterjee N, Yu K, Goldin LR, Goldstein AM, Rotunno M, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harrison C, Ketchen AM, Redhead NJ, O'Sullivan MJ, Melton DW. Replication failure, genome instability, and increased cancer susceptibility in mice with a point mutation in the DNA ligase I gene. Cancer Res. 2002;62:4065–4074. [PubMed] [Google Scholar]
- 38.Bentley DJ, Harrison C, Ketchen AM, Redhead NJ, Samuel K, Waterfall M, et al. DNA ligase I null mouse cells show normal DNA repair activity but altered DNA replication and reduced genome stability. J Cell Sci. 2002;115:1551–1561. doi: 10.1242/jcs.115.7.1551. [DOI] [PubMed] [Google Scholar]
- 39.Prigent C, Satoh MS, Daly G, Barnes DE, Lindahl T. Aberrant DNA repair and DNA replication due to an inherited enzymatic defect in human DNA ligase I. Mol Cell Biol. 1994;14:310–317. doi: 10.1128/mcb.14.1.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soza S, Leva V, Vago R, Ferrari G, Mazzini G, Biamonti G, et al. DNA ligase I deficiency leads to replication-dependent DNA damage and impacts cell morphology without blocking cell cycle progression. Mol Cell Biol. 2009;29:2032–2041. doi: 10.1128/MCB.01730-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rountree MR, Bachman KE, Herman JG, Baylin SB. DNA methylation, chromatin inheritance, and cancer. Oncogene. 2001;20:3156–3165. doi: 10.1038/sj.onc.1204339. [DOI] [PubMed] [Google Scholar]
- 42.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 43.Tesfaigzi Y. Roles of apoptosis in airway epithelia. Am J Respir Cell Mol Biol. 2006;34:537–547. doi: 10.1165/rcmb.2006-0014OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pfeifer GP, Besaratinia A. Mutational spectra of human cancer. Hum Genet. 2009;125:493–506. doi: 10.1007/s00439-009-0657-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chinnadurai G, Vijayalingam S, Rashmi R. BIK, the founding member of the BH3-only family proteins: mechanisms of cell death and role in cancer and pathogenic processes. Oncogene. 2008;27 Suppl 1:S20–S29. doi: 10.1038/onc.2009.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.García N, Salamanca F, Astudillo-de la Vega H, Curiel-Quesada E, Alvarado I, Peñaloza R, et al. A molecular analysis by gene expression profiling reveals Bik/NBK overexpression in sporadic breast tumor samples of Mexican females. BMC Cancer. 2005;5:93. doi: 10.1186/1471-2407-5-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu Y, Lemon W, Liu PY, Yi Y, Morrison C, Yang P, et al. A gene expression signature predicts survival of patients with stage I non-small cell lung cancer. PLoS Med. 2006;3:e467. doi: 10.1371/journal.pmed.0030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.