Abstract
Although the mitochondrial genome exhibits high mutation rates, common mitochondrial DNA (mtDNA) variation has not been consistently associated with pancreatic cancer. Here, we comprehensively examined mitochondrial genomic variation by sequencing the mtDNA of participants (cases=286, controls=283) in a San Francisco Bay Area pancreatic cancer case-control study. Five common variants were associated with pancreatic cancer at nominal statistical significance (p<0.05) with the strongest finding for mt5460g in the ND2 gene (odds ratio (OR)=3.9, 95% confidence interval (CI)=1.5–10; p=0.004) which encodes an A331T substitution. Haplogroup K was nominally associated with reduced pancreatic cancer risk (OR = 0.32, CI=0.13–0.76; p=0.01) when compared with the most common haplogroup, H. A total of 19 haplogroup-specific rare variants yielded nominal statistically significant associations (p<0.05) with pancreatic cancer risk, with the majority observed in genes involved in oxidative phosphorylation. Weighted-sum statistics were used to identify an aggregate effect of variants in the 22 mitochondrial tRNAs on pancreatic cancer risk (p=0.02). While the burden of singleton variants in the HV2 and 12S RNA regions was three times higher among European haplogroup N cases than controls, the prevalence of singleton variants in ND4 and ND5 was two to three times higher among African haplogroup L cases than in controls. Together, the results of this study provide evidence that aggregated common and rare variants and the accumulation of singleton variants are important contributors to pancreatic cancer risk.
Keywords: Pancreatic cancer, mitochondrial DNA, oxidative phosphorylation, DNA sequencing, epidemiology
Introduction
Pancreatic cancer is the fourth leading cause of death from cancer among men and women in the United States and was expected to result in 43,140 new cases and 36,800 deaths in 2010 (1). Due to its aggressiveness and a lack of early detection methods, pancreatic cancer is metastatic in >50% of patients at the time of diagnosis and has a 5-year relative survival rate for all stages of <6%. The incidence of pancreatic cancer is higher in industrialized countries, and varies by age, sex, and race with >70% of pancreatic cancer cases diagnosed after age 60 (2).
Otto Warburg first hypothesized that cancer might be caused by defects in the mitochondrion based on his observation that tumors actively metabolize glucose and produce excessive lactate in the presence of oxygen (aerobic glycolysis) (3). Mitochondria produce most of the cellular energy, generate reactive oxygen species (ROS), and regulate apoptosis. The primary site of ROS and free radical production during oxidative respiration is the inner mitochondrial membrane (4–6), where the mitochondrial DNA (mtDNA) resides. If increased mitochondrial ROS production increases cancer risk, then mtDNA mutations that partially inhibit electron transport and increase ROS production might also increase cancer risk. Although a complete elucidation of the “Warburg effect” has not been achieved, several mechanisms have been proposed to explain this phenomenon (7). In turn, cancer cells have been shown to exhibit multiple alterations in mitochondrial content, structure, function, and activity (8–10).
The mtDNA is a circular double-stranded DNA molecule of 16,569 bp in humans that encodes 13 essential polypeptides of the oxidative phosphorylation (OXPHOS) system and the necessary RNA machinery for their translation within the mitochondria. MtDNA does not recombine, is maternally inherited (11), and has a unique organization in that its structural genes lack introns, intergenic spaces, and 5′ and 3′ noncoding sequences. Each human cell contains hundreds of mitochondria and thousands of copies of mtDNA with the number of copies being dependent upon the cell type. Somatic mtDNA mutations have been identified in many human tumors and are common in pancreatic cancers (12–15). Jones et al. (12) sequenced the complete mtDNA in 15 pancreatic cancer cell lines and xenografts and identified somatic mtDNA mutations and novel variants in nearly all samples. Kassauei et al. (14) sequenced the mtDNA in 15 primary pancreatic cancers and noneoplastic tissue and all pancreatic cancers demonstrated at least one somatic mutation with the number of mutations per case ranging from 1 to 14. Somatic mtDNA mutations present in primary tumors were also detected in pancreatic juice from the same patients (15).
The evolution of human mtDNA is characterized by the emergence of distinct lineages associated with the major global ethnic groups. Mitochondrial haplogroups, and in some cases specific nonsynonomous (NS) SNPs, have been correlated with cancer development (16–24) suggesting that individuals who inherit certain variants might be more prone to cancer. Results to date do not support a significant involvement of common inherited mtDNA variation as a risk factor for pancreatic cancer (25, 26). However, these previous studies did not comprehensively examine sequence level mtDNA variation including rare variants and singletons (variants unique to a single participant). Because human mtDNA has a mutation rate that is 10–20 times higher than that of nuclear DNA (27–29) and approximately one-third of sequence variants found in the general population may be functionally important(30), it is likely that most of the mtDNA variation that impacts function is rare in frequency and only detectable by direct sequencing. Population-based mtDNA resequencing (31) has identified >140 mtDNA polymorphisms with >1% allele frequency and a total of 64 tagging SNPs that efficiently capture all common variation in the coding regions exclusively. In the present study we sequenced the entire mtDNA genome (~16.5kb) using the Affymetrix Mitochondrial Resequencing Array 2.0 (MitoChip) to assess the role of common and rare mtDNA sequence variation in pancreatic cancer in nearly 600 case and control participants from a large population-based study of pancreatic cancer in the San Francisco Bay Area.
Materials and Methods
Study Participants
A population-based case-control study of 532 pancreatic cancer cases and 1,701 controls was conducted between 1994 and 2001 in the San Francisco Bay Area in California (32). Participants who were on blood thinning medications, had a bleeding disorder, had a portacath in place, or had other contraindications to blood draw were not eligible to participate in the optional laboratory portion of the study. Cases with primary adenocarcinoma of the exocrine pancreas were identified using cancer registry rapid case ascertainment. Eligible cases were newly diagnosed from 1995–1999, 21–85 years old, and a San Francisco Bay Area resident. Patient diagnoses were confirmed by participants’ physicians and by the Surveillance, Epidemiology, and End Results abstracts that included histologic confirmation of disease. Of 794 eligible cases who were alive at first contact and able to complete an interview in English, 532 (67%) participated, 19% were too ill, 9% refused, and 6% had moved or had physician-indicated contraindications to contact. Of the 532 case participants, 452 (85%) were eligible for venipuncture, 12% had moved out of the area, and 3% had a portacath or were taking blood thinning medications. Of the 452 case participants who were eligible for venipuncture, 309 (68%) provided a blood sample, 16% refused (needle phobia or physician-indicated contraindications), 7% were too ill, 3% had died, and 6% were not asked or blood draws were unsuccessful. The demographic characteristics of the 309 study cases who provided blood samples for analyses were similar to the 223 who did not (p-values >=0.30 for age at diagnosis, sex, white race and Hispanic ethnicity). Of the 309 cases who provided a blood sample, 297 had DNA available for the mtDNA analysis and 12 did not have DNA available.
Among 1588 eligible controls identified by random digit dial, 1066 (67%) participated in the study, 29% refused and 4% were too ill. Among 1211 eligible Medicare controls identified from random sampling of the Health Care Financing Administration lists and recruited by mail, 635 (52%) participated in the study, 31% refused, 12% could not be located/had moved and 5% were too ill. Of the 1701 control participants, 1634 (96%) were eligible for venipuncture (lived in the area). Of the 1634 controls eligible for venipuncture, 964 gave blood (59%), 10% were not asked for a blood sample as the laboratory portion of the study had closed to recruitment, 11% refused for no specific reason, 15% refused for reasons of needle phobia, confidentiality, bad veins or other reasons, 4% were too ill or blood draws were unsuccessful and 1% were lost to follow-up. Control participants who provided a blood sample were similar by age and Hispanic ethnicity (all p-values >=0.81) but were more likely to be white (p=0.002) and male (p=0.002) when compared to those who did not provide blood. We had previously selected 301 random controls with sufficient DNA for inclusion in a genome wide association study and in order to create a comprehensive dataset we selected these same 301 controls for mtDNA analysis. A total of 286 pancreatic cancer cases and 283 controls yielded sequence data of sufficient quality for analysis (>95% success rate).
Interviews
Detailed in-person interviews were conducted in the homes of the participants or at a location of their choice. The University of California San Francisco Committee on Human Research approved the study protocols and procedures. Written informed consent was obtained from each study participant before interview. Race/ethnicity was based on self-report and was broadly defined as Caucasian, black/African-American, Asian, Hispanic (black or white), or “other race/ethnicity”.
Mitochondrial DNA sequencing
The entire mitochondrial genome was first amplified in two long-range PCR reactions. Mitochondrial fragments were amplified and prepared for array hybridization according to the Affymetrix protocol for GeneChip CustomSeq Resequencing Array. Briefly, long-ranged PCR reactions were performed using LA PCR Kit (Takara Bio U.S.A., Madison, WI) for each sample using two sets of overlapping primers. 5 μL of 5ng/μL template genomic DNA were mixed with 0.1μL HS LA Taq polymerase (5U/μL), 2.5 μL 10X LA Taq buffer, 4μL 2.5 mM dNTPs, 5μL of 3 μM primer pair, and 8.4μL dH20 for a total reaction volume of 25 μL. The PCR program was as follows: (1) 1 cycle, 94°C for 2 minutes; (2) 30 cycles, 94°C for 15 seconds and 68°C for 16 minutes; (3) 1 cycle, 94°C for 21 minutes; (4) hold, 4°C. The resulting PCR products were assessed qualitatively by 1% agarose gel electrophoresis. The PCR product was purified using a Clonetech Clean-Up plate. The purified DNA was quantified by PicoGreen and for selected samples, confirmed by NanoDrop measurements. The amplicons were pooled at equi-molar concentrations. Chemical fragmentation was performed and products confirmed to be in the size range of 20 – 200 bp by 20% polyacrylamide gel electrophoresis with SYBR Gold staining. The IQ-EX control template, a 7.5 kb plasmid DNA, was used as positive control. The samples were labeled with TdT and hybridized to the array in a 49°C rotating hybridization oven at 16 hours. Finally, streptavidin phycoerythrin (SAPE), and then antibody staining was performed. The microarrays were processed in the GeneChip Fluidic Station and the GeneChip Scanner. Signal intensity data was output for all four alleles (“a”, “c”, “g”, and “t”), permitting quantitative estimates of allelic contribution. The allelic contribution was assessed using the raw data from the individual signal intensities by deriving the ratio of expected allele (REA), which is the log ratio of the raw signal intensity of the expected allele at any site (as defined by the mtDNA reference sequence) to the average raw signal intensity of the other three alleles, at each site for every individual.
The MitoChip uses standard Affymetrix GeneChip Sequence Analysis Software (GSEQ). The output of the GeneChip DNA Analysis (GDAS) included a report of the individual and total number of SNPs as well as a case-by-case list of genotype variations as determined by comparison to the mitochondrial DNA reference sequence. Most of the 15,452 mtDNA loci tiled on the array were duplicated via independent probe sets allowing a test of within-chip reproducibility. Two previous studies (15, 33) using the MitoChip reported within chip error rates of 0.0025% and 0.00021%. DAT files with raw pixel data were generated and used as input for grid alignment. CEL files generated were analyzed in batches using GeneChip Sequencing Analysis Software (GSEQ) 4.1 (Affymetrix, Santa Clara, CA). All CEL files from each plate were analyzed as a batch. Base calls were extracted and used for downstream analyses. Samples with calls rates of less than 95% were discarded. For samples passing initial filtering, ResqMi 1.2(34) was used for re-analysis of bases originally called as “N” by GSEQ. Analysis was performed by custom Perl scripts. Data were extracted from genic regions as defined by NCBI annotations for the revised Cambridge Reference Sequence (rCRS; NC_012920.1).
Quality Control
Twenty samples were repeated for concordance testing. Laboratory personnel were blinded to QC and case–control status and all 20 QC samples had >98% sequence concordance (the majority of discordant calls resulted from positions successfully called in one but called as “N” in another).
Statistical analysis
To examine mtDNA sequence variation for associations with pancreatic cancer we analyzed variants from the following categories: Common haplogroups and individual variants (minor allele frequency [MAF] ≥5%); rare variants (MAF <5%); and singletons (occurring in a single participant– in the case of haplogroup-specific analyses, singletons are variants occurring in a single participant within the haplogroup). Unconditional logistic regression was used to obtain odds ratios (ORs) as estimates of relative risks (hereafter called risk) and 95% confidence intervals (CIs) for analyses involving haplogroups and common variants. Allele frequencies and rare variants (excluding singletons) were compared between cases and controls using χ2 tests. The major European mitochondrial haplogroups were defined using variants identified from PhyloTree (35) and included subgroups H, V, J, T, U, K, (B, F), and (A, I, W, X, Y). To account for confounding by ancestry we derived eigenvectors using principal components analysis (PCA) using the complete mtDNA genotype data (36). This method has been demonstrated to outperform haplogroup-stratified or adjusted association analyses with no loss in power for detection of true associations (36). All models were adjusted for age in 5-year groups, sex and the first 6 eigenvectors of mitochondrial genetic ancestry derived from principal component analysis. Models examining individual common variants were restricted to haplogroup N since the number of cases and controls are equivalent. We did not examine associations for haplogroups L and M since the sample sizes are small and the numbers of cases and controls reflect the proportions of African- and Asian-American participants in the study.
Variants also were grouped and tested jointly to assess the contribution of multiple variants to the pancreatic cancer risk using weighted-sum statistics computed as described in Madsen and Browning (37). The weighted-sum method is designed for resequencing data where rare mutations are observed directly and generally has higher power than alternative methods for these types of analyses (37). The weighted-sum method is based on the common disease-rare variant hypothesis wherein variants with lower frequencies in the unaffected individuals were weighted more heavily; approximately 70% of all rare missense mutations are reported to be deleterious (38). 1000 permutations of case-control status were performed to obtain one-sided p-values, testing the hypothesis that most rare mutations are deleterious and associated with disease status. Variants from genes encoding the four mtDNA-encoded OXPHOS complexes, rRNA, tRNA and hypervariable regions were assessed using the weighted sum method (37) among haplogroup N participants only. All weighted-sums were computed using custom Perl scripts. For singletons, the total number of gene or region specific variants was compared between pancreatic cancer cases and controls using Fisher’s exact test as was the number of individuals harboring singleton variants unique to cases or controls. Due to the potential for confounding by mtDNA ancestry, all singleton analyses were performed for each major haplogroup L, M, and N. Bonferroni correction for multiple testing took into account the number of haplogroups (n=8) and genes/regions examined for singleton analysis (n=17), and the number of common and rare variants discovered and analyzed (detailed in Results).
In-silico prediction methods were employed to examine mtDNA nucleotide conservation and the impact of NS coding substitutions on amino acid protein sequence. PhastCons (39) is a hidden Markov model-based method that estimates the probability that each nucleotide belongs to an evolutionary conserved element. Based on a multi-species sequence alignment the method considers the conservation of sites flanking the base of interest when producing base-by-base conservation scores. The phastCons scores range from 0 to 1 and represent probabilities of negative selection. PhyloP (40) separately measures conservation at individual nucleotides, ignoring the effects of their neighbors. Also based on a multi-species sequence alignment, this method is more appropriate for evaluating signatures of selection at particular nucleotides. PhyloP scores represent -log p-values under a null hypothesis of neutral evolution. Sites predicted to be conserved are assigned positive scores and sites predicted to be fast-evolving are assigned negative scores. For phastCons and phyloP, a higher value indicates a more conserved position. The effects of NS coding substitutions (amino acid changes) on protein function was assessed using PolyPhen2 (41). PolyPhen2 is a tool that predicts the possible impact of an amino acid substitution on the structure and function of a human protein. Predictions are based on information that includes eight sequence-based and three structure-based features characterizing the substitution. As a final output, PolyPhen2 calculates a posterior probability that a mutation is damaging and qualitatively reports it as benign, possibly damaging, or probably damaging.
Results
Sequencing of 16,543 mtDNA bases (positions 12–16,555) from 286 pancreatic cancer cases and 283 controls participants yielded a cumulative total of 2,169 variants including: 66 common variants (MAF ≥5%); 251 low frequency variants (MAF 1–5%); and 1,859 rare variants (MAF <1%) including 1,393 haplogroup-specific (L, M or N) singletons unique to either cases or controls. Distributions of age, education level, race, ethnicity, sex, smoking status, and major haplogroups for cases and controls are presented in Table 1. The case and control participants were similar by white/non-white race (chi-square, p=0.74) and Hispanic ethnicity (chi-square, p=0.58). In general, cases were somewhat less educated, were more likely to be current smokers, and a greater proportion were African-American. Haplogroup distributions largely overlapped with self-identified race for African-Americans (97%) and European-Americans (96%). Self-identified Asian-Americans were distributed between haplogroups M (55%) and N (43%). While major haplogroup M is largely unique to Asia, the minor Asian haplogroups are descended from both major haplogroups M (e.g. haplogroups C, D, and G) and N (haplogroups A, B, and F). Participants that self-identified as Hispanic were distributed between haplogroups N (75%) and M (19%). The results for the Asian and Hispanic participants are not unexpected since the mtDNA traces the maternal lineage exclusively and may not reflect an admixed genetic or self-identified ancestry.
Table 1.
Characteristics of pancreatic cancer cases and controls in a population based study in the San Francisco Bay Area, California (1995–1999)
| Cases N = 286 |
Controls N = 283 |
|
|---|---|---|
| Age (years) | 65 (11) | 64 (12) |
| Mean (s.d.) | n (%) | n (%) |
| Education (years) | ||
| 1–12 | 120 (42) | 91 (32) |
| > 12–16 | 111 (39) | 120 (42) |
| > 16 | 55 (19) | 72 (25) |
| Self-reported race/ethnicity | ||
| White, non-hispanic | 226 (79) | 223 (79) |
| White, hispanic | 16 (6) | 20 (7) |
| Black | 22 (8) | 11 (4) |
| Asian | 17 (6) | 22 (8) |
| other | 5 (2) | 7 (2) |
| Sex | ||
| Men | 153 (54) | 152 (54) |
| Women | 133 (46) | 131 (46) |
| Cigarette Smoking | ||
| Never smoked | 82 (29) | 106 (39) |
| Former smoker | 131 (48) | 129 (48) |
| Current smoker | 64 (23) | 35 (13) |
| Halogroup N | 246 (87) | 248 (88) |
| Haplogroup L | 27 (10) | 14 (5) |
| Haplogroup M | 11 (4) | 21 (7) |
Common haplogroups and individual variants
Risk of pancreatic cancer among 8 European sub-haplogroups is reported in Table 2. No haplogroup met statistical significance after adjustment for multiple comparisons (8 haplogroups, critical α=0.006). Carriers of haplogroup K had a nominally reduced pancreatic cancer risk compared with the most common European haplogroup H (odds ratio (OR)=0.32, 95% confidence interval (CI)=0.13–0.76, p=0.01). There also were no individual common variants that met statistical significance after multiple comparisons adjustment (66 common variants detected by sequencing [MAF ≥5%], critical α=0.0008). Of the 66 common variants, Five reached nominal statistical significance (P<0.05) and two yielded a strong (statistically non-significant) association with pancreatic cancer risk: mt5460g (p=0.004) and mt1811g (p=0.008). The mt5460g>a variant associated with an increased risk of pancreatic cancer (OR=3.9, 95% (CI)=1.5–10; p=0.004) encodes an A331T substitution in the ND2 gene. All analyses were adjusted for age, sex and 6 eigenvectors of mitochondrial genetic ancestry derived from principal component analysis. Restricting the haplogroup and common variant analyses to self-identified white non-Hispanic did not alter the results (data not shown).
Table 2.
Odds Ratios (OR) and 95% confidence intervals (CI) for pancreatic cancer associated with haplogroup N subgroups and common mtDNA variants among haplogroup N participants, San Francisco Bay Area, California (1995–1999)
| Haplogroup N subgroups | Cases N = 246 n (%) |
Controls N = 248 n (%) |
OR (95 % CI)a | P-valuec | |
|---|---|---|---|---|---|
| H | 107 (44) | 101 (40) | 1.0 (Ref.) | ||
| V | 8 (4) | 8 (3) | 0.92 (0.3 – 2.8) | 0.88 | |
| J | 27 (11) | 24 (9) | 1.03 (0.52 – 2.0) | 0.94 | |
| T | 24 (10) | 27 (11) | 0.6 (0.31 – 1.2) | 0.13 | |
| U | 31 (13) | 35 (14) | 0.67 (0.37 – 1.2) | 0.17 | |
| K | 10 (4) | 24 (9) | 0.32 (0.13 – 0.76) | 0.01 | |
| B, F | 8 (3) | 9 (3) | 0.86 (0.3 – 2.42) | 0.77 | |
| A, I, W, X, Y | 22 (8) | 27 (9) | 0.89 (0.45 – 1.8) | 0.73 | |
| Region/geneb | Site, allele | ||||
| Complex I | |||||
| ND2 mt5460g | 20 (8) | 6 (2) | 3.9 (1.5 – 10) | 0.004 | |
| Complex IV | |||||
| COIII mt9698c | 10 (4) | 24 (9) | 0.44 (0.20 – 0.97) | 0.02 | |
| 16S | mt1811g | 20 (7) | 40 (14) | 0.51 (0.29 – 0.91) | 0.008 |
| tRNA | mt12307g | 16 (7) | 34 (14) | 0.45 (0.24 – 0.85) | 0.02 |
| HV2 | mt150t | 13 (5) | 26 (11) | 0.70 (0.49 – 0.99) | 0.03 |
All analyses were adjusted for age, sex and 6 eigenvectors of mitochondrial genetic ancestry derived from principal component analysis.
Nominally significant results for 5 common variants out of 66 total common variants detected.
Multiple comparisons adjusted p-value (α=0.05) for 8 haplogroups (α=0.006) and 66 common variants (α=0.0008).
Rare variants
Of 710 low frequency (MAF 1–5%) and rare variants (MAF <1%) detected by sequencing (excluding singletons), none met statistical significance after adjustment for multiple comparisons (critical α=0.0001). A total of 19 haplogroup-specific variants yielded nominal statistically significant associations (α<0.05) with pancreatic cancer risk (Table 3). All were from either haplogroup N or L. Of these, 13 were detected from complex I, III, IV and V coding regions, 2 from the 12S RNA and 4 from the hypervariable (HV) or non-coding regions. Two of the coding region variants resulted in NS substitutions: L555Q (mt14000) in ND5 and K6N (mt14763) in CytB. The L555Q substitution in ND5 was predicted to be probably damaging when assessed using PolyPhen2. The mt14763 variant underlying the K6N substitution shows strong evidence of belonging to a conserved sequence element (PhastCons = 1.00, PhyloP=4.47).
Table 3.
Rare, haplogroup specific mtDNA variants associated with pancreatic cancer, San Francisco Bay Area, California (1995–1999)
| Region | Gene | Site | Cases | Controls | Haplogroup | Common allele | Rare allele | Amino Acid position | PhastCons | PhyloP | p-value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Complex I | ND4L | 10586 | 7 | 1 | L | tcg | tca | S39S | 0.00 | −7.02 | 0.046 |
| ND5 | 12810 | 6 | 0 | L | tga | tgg | W158W | 0.00 | −0.58 | 0.046 | |
| ND5 | 12954 | 4 | 0 | N | gct | gcc | A206A | 0.00 | −2.44 | 0.040 | |
| ND5 | 13485 | 6 | 0 | L | ata | atg | M383M | 0.00 | −4.18 | 0.046 | |
| ND5 | 14000 | 6 | 0 | L | cta | caa | L555Qa | 0.00 | −0.50 | 0.046 | |
| Complex III | CytB | 14763 | 0 | 3 | L | aaa | aca | K6Na | 1.00 | 4.47 | 0.003 |
| CytB | 14869 | 0 | 3 | L | ctg | cta | L41L | 0.00 | −0.59 | 0.01 | |
| CytB | 15784 | 7 | 1 | N | cct | ccc | P346P | 0.00 | −3.64 | 0.040 | |
| Complex IV | COI | 5951 | 6 | 0 | L | gga | ggg | G16G | 0.32 | −1.60 | 0.046 |
| COI | 6071 | 6 | 0 | L | gtt | gtc | V56V | 0.00 | −8.27 | 0.046 | |
| COI | 6260 | 1 | 6 | N | gag | gaa | E119E | 0.98 | −0.11 | 0.050 | |
| COIII | 9548 | 8 | 2 | N | ggg | gga | G114G | 0.00 | −6.52 | 0.050 | |
| Complex V | ATP6 | 9072 | 6 | 0 | L | tca | tcg | S182S | 0.00 | −1.59 | 0.046 |
| rRNA | 12S | 951 | 1 | 6 | N | g | a | 0.00 | −2.22 | 0.050 | |
| 12S | 961 | 3 | 0 | N | t | g | 0.00 | −4.81 | 0.050 | ||
| HV2 | 193 | 6 | 0 | N | c | t | 0.73 | 0.18 | 0.040 | ||
| 296 | 8 | 0 | L | a | g | 0.01 | −0.12 | 0.046 | |||
| 316 | 6 | 0 | L | g | a | 0.03 | −1.32 | 0.046 | |||
| Noncoding | 16527 | 10 | 1 | N | c | t | 0.00 | −1.40 | 0.020 |
Nonsynonomous variant predicted to have a damaging effect on the resulting protein using PolyPhen2 (41).
Weighted-Sum Statistics
Multiple variants across the combined 22 mtDNA tRNA regions were statistically significantly associated with pancreatic cancer as determined by permutation of case-control status (one-sided p=0.02). Our results also suggest that there was an excess of variants in Complex III (CytB) among pancreatic cancer cases although the effect was not statistically significant (one-sided p=0.06).
Singletons
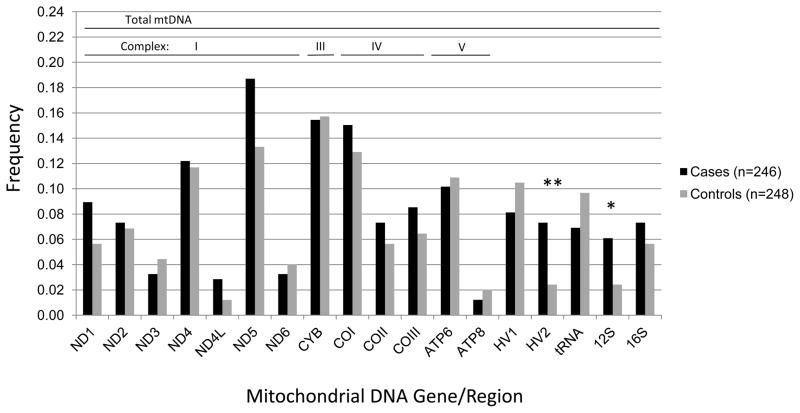
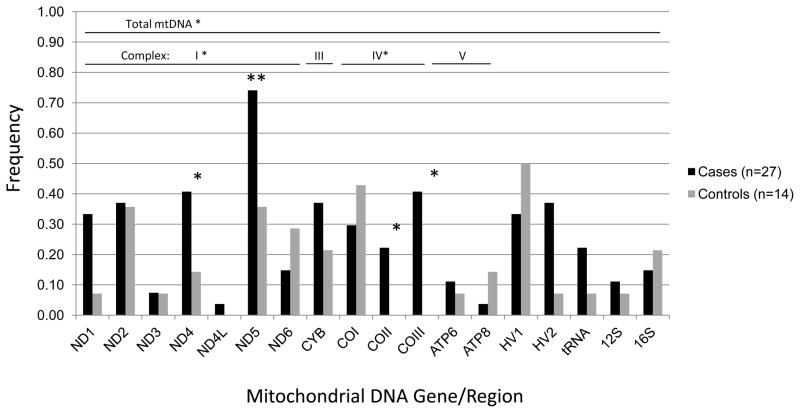
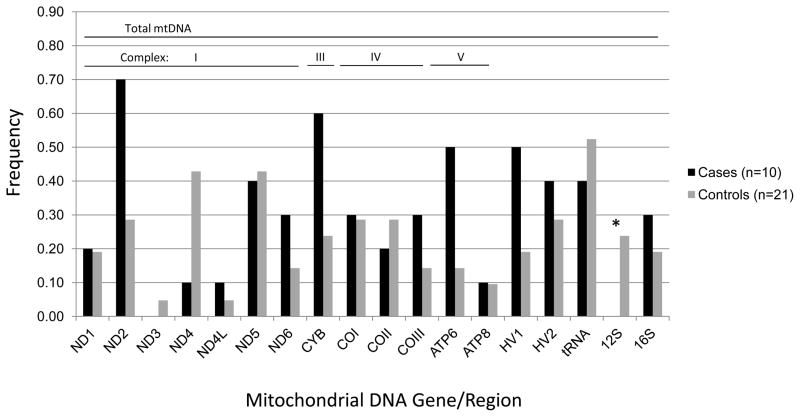
A total of 1,393 singleton mtDNA variants unique to cases or controls were identified across the coding, tRNA, rRNA and HV regions for the three major haplogroups L, M, and N. Two of these genes/regions harbored a significantly higher burden of singleton variants in cases compared with controls after adjustment for multiple comparisons (18 mtDNA genes/regions, critical α=0.003). Specifically, the number of singleton variants among haplogroup N cases vs. controls was statistically significantly higher in the HV 2 region (p=0.006) and nominally higher in the 12S RNA (p=0.03) region. The frequency of HV 2 and 12S RNA singletons was three times higher in cases than controls: HV 2 region, 9% cases and 3% controls; 12S RNA 7% cases and 2% controls (Figure 1). In haplogroup L, the number of singletons was nominally higher among cases for the entire mtDNA (p=0.004), and for complexes I (p=0.03) and IV (p=0.05). Haplogroup L cases had a statistically significant greater number of singleton variants in the ND5 gene (p<0.001) and nominally greater numbers in the ND4 (p=0.02), COII (p=0.04), and COIII (p=0.007) genes compared with controls. The frequency of ND4 and ND5 singletons was two to three times higher in cases than controls: ND4, 41% cases versus 14% controls; ND5, 74% cases versus 36% controls (Figure 2). In addition, singleton variants were observed among haplogroup L cases only for the COII and COIII genes. Among haplogroup M participants, cases had statistically significant fewer singleton variants in the 12S RNA (p=0.03) region compared with controls (Figure 3).
Figure 1.
Frequency of mtDNA singleton variants among haplogroup N pancreatic cancer cases and controls, San Francisco Bay Area, California (1995–1999).
Figure 2.
Frequency of mtDNA singleton variants among haplogroup L pancreatic cancer cases and controls, San Francisco Bay Area, California (1995–1999).
Figure 3.
Frequency of mtDNA singleton variants among haplogroup M pancreatic cancer cases and controls, San Francisco Bay Area, California (1995–1999).
Of the 1,393 singleton variants identified in the three major haplogroups, 625 were located within the four mtDNA-encoded OXPHOS complexes with 221 resulting in NS coding substitutions (Supplementary Tables 1–5). An excess of ND2 NS substitutions was observed among cases compared with controls (p=0.02). Among the complex V genes, ATP6 and ATP8, approximately 25% of NS substitutions in controls were predicted to be non-conserved whereas all NS substitutions in cases were conserved (p=0.02).
Discussion
In this study, we sequenced the entire mtDNA in a large population-based case-control study of pancreatic cancer to examine the role of haplogroups and common genetic variants, rare sequence variants, and singletons. We observed inverse associations with pancreatic cancer risk for participants from European haplogroup K when compared with the most common European haplogroup H. The haplogroup K association was not seen in a previous study of pancreatic cancer that included replication (26). The low frequency of haplogroup K participants in this study and lack of consistency with the earlier study likely mean that our finding is a false positive result. Common haplogroup N variants (MAF ≥5%) in Complex I (ND2), Complex IV (COIII), 16S, tRNA, and HV2 genes/regions also yielded nominally significant associations with pancreatic cancer risk. This includes an A331T substitution in the ND2 gene that was present in 20 cases (8%) and 6 controls (2%).
Research to identify genetic factors that contribute to complex phenotypes such as the development of cancer must be sensitive to the ways that genes and genetic perturbations operate. For example, it is now widely recognized that common genetic variants play a much smaller role in mediating phenotypic expression and disease risk than initially thought (42–45) and that identification of causative variants requires comprehensive resequencing of genomic loci in multiple subjects (46). In this study, we identified numerous low frequency (MAF 1–5%) and rare variants (MAF <1%) from haplogroups N and L that were associated with pancreatic cancer risk. Nearly seventy percent of these variants were observed in the OXPHOS complexes including two NS variants from the ND5 and CytB genes. The ND5 substitution was predicted to have a damaging effect on the resulting protein whereas the CytB substitution showed evidence of belonging to a conserved sequence element. However, focusing on NS variants may not be overly informative as NS SNPs and synonymous SNPs (S SNPs) share similar likelihood and effect size for disease association and S SNPs are just as likely to be involved in disease mechanisms (47). The remaining variants occurred in the 12S RNA, HV and non-coding regions. Several of the coding and non-coding variants were predicted to belong to evolutionary conserved regions and may play important roles in mtDNA copy number (48) and genome transcription (49, 50) possibly causing severe alterations in mitochondrial function (17, 50–53). In the present study, mt296 was observed in 38% of haplogroup L cases and no controls, possibly reflecting a risk factor related to mtDNA copy number. Previous studies have described a variant located at mt295 that defines Caucasian haplogroup J and that has been found to change mitochondrial copy number (48), which may partially account for observations that haplogroup J is over-represented in long-lived people and centenarians from several populations (54–56).
Because collections of rare variants within genes or genomic regions are likely to influence phenotypic expression in important ways (45), examining the collective frequency of rare or singleton variants may reveal the role of specific genes in disease etiology. Our results provide evidence for a significant aggregate effect of sequence variants in the 22 mitochondrial tRNAs for pancreatic cancer risk and suggest that Complex III (CytB) gene variants may also play a role. The burden of singleton variants among European haplogoup N cases was three times higher than in controls for the HV2 and 12S RNA regions, suggesting that these mtDNA regions may contribute to risk among persons of European descent. Further, the burden of singleton variants among the African haplogroup L cases was higher than in controls for the entire mtDNA, in particular for OXPHOS complexes I and IV. More specifically, among the complex I genes ND4 and ND5, rare variants were 2–3 times more frequent in cases than in controls whereas in the complex IV COIII gene singleton variants occurred in 41% of cases and in no controls. Interestingly, these results are consistent with a study of prostate cancer where germline mtDNA COI (complex IV) missense mutations were reported in 11% of prostate cancer cases compared with 2% of the no-cancer controls (17). This may be of particular importance as incidence and mortality of pancreatic cancer are 48% and 37% higher, respectively, in African-Americans relative to European-Americans (57), and cannot be attributed to racial differences in currently known risk factors (58).
The results of this study suggest that aggregated common and rare variants and the accumulation of singleton variants are important contributors to pancreatic cancer risk. This study had a number of strengths, including: complete mtDNA sequencing allowing for an unbiased assessment of mitochondrial genomic variation; a well-characterized case-control study of pancreatic cancer; and an analytic approach that includes both aggregated and accumulated sequence variants. A few weaknesses are also acknowledged, including: small sample sizes for the African and Asian ancestry samples; low power to detect affect of individual variants; possible survival and selection bias, and no validation study. Demographic characteristics of the study cases who provided blood samples for analyses were similar to those who did not whereas control participants who provided a blood sample were more likely to be white men. While the 13 mtDNA-encoded OXPHOS genes are essential to mitochondrial energy production and are considered the most functionally important (59), hundreds of nuclear DNA-encoded and dozens of mtDNA-encoded bioenergetics genes are distributed throughout both genomes (30, 60). Future studies of mitochondrial genetic variation will therefore need to account for a complex set of interactions involving the nuclear and mitochondrial genomes (61).
Supplementary Material
Acknowledgments
Grant Support
Support provided in part by grants CA133080 (G.J. Tranah, PI), CA59706, CA108370, CA109767, CA89726 (E.A. Holly, PI) from the National Cancer Institute, National Institutes of Health, by the Rombauer Pancreatic Cancer Research Fund and by the Hasbun Fund. The collection of cancer incidence data for the UCSF study was supported by the California Department of Public Health as part of the statewide cancer reporting program; the National Cancer Institute’s Surveillance, Epidemiology and End Results Program under contract N01-PC-35136 to the Northern California Cancer Center; and the Centers for Disease Control and Prevention’s National Program of Cancer Registries.
Footnotes
The author’s declare no conflicts of interest relevant to this manuscript’s subject.
Literature Cited
- 1.American Cancer Society. Facts and Figures 2010. Atlanta, GA: American Cancer Society; 2010. [Google Scholar]
- 2.Lowenfels AB, Maisonneuve P. Epidemiology and prevention of pancreatic cancer. Jpn J Clin Oncol. 2004;34:238–244. doi: 10.1093/jjco/hyh045. [DOI] [PubMed] [Google Scholar]
- 3.Warburg OH. The metabolism of tumours. R.R. Smith; New York, New York: 1931. [Google Scholar]
- 4.Hruszkewycz AM, Bergtold DS. Oxygen radicals, lipid peroxidation and DNA damage in mitochondria. Basic Life Sci. 1988;49:449–456. doi: 10.1007/978-1-4684-5568-7_69. [DOI] [PubMed] [Google Scholar]
- 5.Hruszkewycz AM. Lipid peroxidation and mtDNA degeneration. A hypothesis. Mutat Res. 1992;275:243–248. doi: 10.1016/0921-8734(92)90028-n. [DOI] [PubMed] [Google Scholar]
- 6.Richter C, Park JW, Ames BN. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc Natl Acad Sci U S A. 1988;85:6465–6467. doi: 10.1073/pnas.85.17.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kroemer G. Mitochondria in cancer. Oncogene. 2006;25:4630–4632. doi: 10.1038/sj.onc.1209589. [DOI] [PubMed] [Google Scholar]
- 8.Cuezva JM, Krajewska M, de Heredia ML, Krajewski S, Santamaria G, Kim H, Zapata JM, Marusawa H, Chamorro M, Reed JC. The bioenergetic signature of cancer: a marker of tumor progression. Cancer Res. 2002;62:6674–6681. [PubMed] [Google Scholar]
- 9.Carew JS, Huang P. Mitochondrial defects in cancer. Mol Cancer. 2002;1:9. doi: 10.1186/1476-4598-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rossignol R, Gilkerson R, Aggeler R, Yamagata K, Remington SJ, Capaldi RA. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 2004;64:985–993. doi: 10.1158/0008-5472.can-03-1101. [DOI] [PubMed] [Google Scholar]
- 11.Giles RE, Blanc H, Cann HM, Wallace DC. Maternal inheritance of human mitochondrial DNA. Proc Natl Acad Sci U S A. 1980;77:6715–6719. doi: 10.1073/pnas.77.11.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones JB, Song JJ, Hempen PM, Parmigiani G, Hruban RH, Kern SE. Detection of mitochondrial DNA mutations in pancreatic cancer offers a “mass”-ive advantage over detection of nuclear DNA mutations. Cancer Res. 2001;61:1299–1304. [PubMed] [Google Scholar]
- 13.Navaglia F, Basso D, Fogar P, Sperti C, Greco E, Zambon CF, Stranges A, Falda A, Pizzi S, Parenti A, Pedrazzoli S, Plebani M. Mitochondrial DNA D-loop in pancreatic cancer: somatic mutations are epiphenomena while the germline 16519 T variant worsens metabolism and outcome. Am J Clin Pathol. 2006;126:593–601. doi: 10.1309/GQFCCJMH5KHNVX73. [DOI] [PubMed] [Google Scholar]
- 14.Kassauei K, Habbe N, Mullendore ME, Karikari CA, Maitra A, Feldmann G. Mitochondrial DNA mutations in pancreatic cancer. Int J Gastrointest Cancer. 2006;37:57–64. doi: 10.1007/s12029-007-0008-2. [DOI] [PubMed] [Google Scholar]
- 15.Maitra A, Cohen Y, Gillespie SE, Mambo E, Fukushima N, Hoque MO, Shah N, Goggins M, Califano J, Sidransky D, Chakravarti A. The Human MitoChip: a high-throughput sequencing microarray for mitochondrial mutation detection. Genome Res. 2004;14:812–819. doi: 10.1101/gr.2228504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Booker LM, Habermacher GM, Jessie BC, Sun QC, Baumann AK, Amin M, Lim SD, Fernandez-Golarz C, Lyles RH, Brown MD, Marshall FF, Petros JA. North American white mitochondrial haplogroups in prostate and renal cancer. J Urol. 2006;175:468–472. doi: 10.1016/S0022-5347(05)00163-1. discussion 472–463. [DOI] [PubMed] [Google Scholar]
- 17.Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, Lim S, Issa MM, Flanders WD, Hosseini SH, Marshall FF, Wallace DC. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci U S A. 2005;102:719–724. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bai RK, Leal SM, Covarrubias D, Liu A, Wong LJ. Mitochondrial genetic background modifies breast cancer risk. Cancer Res. 2007;67:4687–4694. doi: 10.1158/0008-5472.CAN-06-3554. [DOI] [PubMed] [Google Scholar]
- 19.Mosquera-Miguel A, Alvarez-Iglesias V, Carracedo A, Salas A, Vega A, Carracedo A, Milne R, de Leon AC, Benitez J, Carracedo A, Salas A. Is mitochondrial DNA variation associated with sporadic breast cancer risk? Cancer Res. 2008;68:623–625. doi: 10.1158/0008-5472.CAN-07-2385. author reply 624. [DOI] [PubMed] [Google Scholar]
- 20.Fang H, Shen L, Chen T, He J, Ding Z, Wei J, Qu J, Chen G, Lu J, Bai Y. Cancer type-specific modulation of mitochondrial haplogroups in breast, colorectal and thyroid cancer. BMC Cancer. 2010;10:421. doi: 10.1186/1471-2407-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu VW, Wang Y, Yang HJ, Tsang PC, Ng TY, Wong LC, Nagley P, Ngan HY. Mitochondrial DNA variant 16189T>C is associated with susceptibility to endometrial cancer. Hum Mutat. 2003;22:173–174. doi: 10.1002/humu.10244. [DOI] [PubMed] [Google Scholar]
- 22.Darvishi K, Sharma S, Bhat AK, Rai E, Bamezai RN. Mitochondrial DNA G10398A polymorphism imparts maternal Haplogroup N a risk for breast and esophageal cancer. Cancer Lett. 2007;249:249–255. doi: 10.1016/j.canlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 23.Canter JA, Kallianpur AR, Parl FF, Millikan RC. Mitochondrial DNA G10398A polymorphism and invasive breast cancer in African-American women. Cancer Res. 2005;65:8028–8033. doi: 10.1158/0008-5472.CAN-05-1428. [DOI] [PubMed] [Google Scholar]
- 24.Setiawan VW, Chu LH, John EM, Ding YC, Ingles SA, Bernstein L, Press MF, Ursin G, Haiman CA, Neuhausen SL. Mitochondrial DNA G10398A variant is not associated with breast cancer in African-American women. Cancer Genet Cytogenet. 2008;181:16–19. doi: 10.1016/j.cancergencyto.2007.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halfdanarson TR, Wang L, Bamlet WR, de Andrade M, McWilliams RR, Cunningham JM, Petersen GM. Mitochondrial genetic polymorphisms do not predict survival in patients with pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:2512–2513. doi: 10.1158/1055-9965.EPI-08-0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Bamlet WR, de Andrade M, Boardman LA, Cunningham JM, Thibodeau SN, Petersen GM. Mitochondrial genetic polymorphisms and pancreatic cancer risk. Cancer Epidemiol Biomarkers Prev. 2007;16:1455–1459. doi: 10.1158/1055-9965.EPI-07-0119. [DOI] [PubMed] [Google Scholar]
- 27.Wallace DC, Stugard C, Murdock D, Schurr T, Brown MD. Ancient mtDNA sequences in the human nuclear genome: a potential source of errors in identifying pathogenic mutations. Proc Natl Acad Sci U S A. 1997;94:14900–14905. doi: 10.1073/pnas.94.26.14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neckelmann N, Li K, Wade RP, Shuster R, Wallace DC. cDNA sequence of a human skeletal muscle ADP/ATP translocator: lack of a leader peptide, divergence from a fibroblast translocator cDNA, and coevolution with mitochondrial DNA genes. Proc Natl Acad Sci U S A. 1987;84:7580–7584. doi: 10.1073/pnas.84.21.7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merriwether DA, Clark AG, Ballinger SW, Schurr TG, Soodyall H, Jenkins T, Sherry ST, Wallace DC. The structure of human mitochondrial DNA variation. J Mol Evol. 1991;33:543–555. doi: 10.1007/BF02102807. [DOI] [PubMed] [Google Scholar]
- 30.Wallace DC, Fan W, Procaccio V. Mitochondrial energetics and therapeutics. Annu Rev Pathol. 2010;5:297–348. doi: 10.1146/annurev.pathol.4.110807.092314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saxena R, de Bakker PI, Singer K, Mootha V, Burtt N, Hirschhorn JN, Gaudet D, Isomaa B, Daly MJ, Groop L, Ardlie KG, Altshuler D. Comprehensive association testing of common mitochondrial DNA variation in metabolic disease. Am J Hum Genet. 2006;79:54–61. doi: 10.1086/504926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holly EA, Eberle CA, Bracci PM. Prior history of allergies and pancreatic cancer in the San Francisco Bay area. Am J Epidemiol. 2003;158:432–441. doi: 10.1093/aje/kwg174. [DOI] [PubMed] [Google Scholar]
- 33.Coon KD, Valla J, Szelinger S, Schneider LE, Niedzielko TL, Brown KM, Pearson JV, Halperin R, Dunckley T, Papassotiropoulos A, Caselli RJ, Reiman EM, Stephan DA. Quantitation of heteroplasmy of mtDNA sequence variants identified in a population of AD patients and controls by array-based resequencing. Mitochondrion. 2006;6:194–210. doi: 10.1016/j.mito.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 34.Symons S, Weber K, Bonin M, Nieselt K. ResqMi - a Versatile Algorithm and Software for Resequencing Microarrays. Proceedings of the German Conference on Bioinformatics; Dresden, Germany. 2008. pp. 10–20. [Google Scholar]
- 35.van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30:E386–394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 36.Biffi A, Anderson CD, Nalls MA, Rahman R, Sonni A, Cortellini L, Rost NS, Matarin M, Hernandez DG, Plourde A, de Bakker PI, Ross OA, Greenberg SM, Furie KL, Meschia JF, Singleton AB, Saxena R, Rosand J. Principal-component analysis for assessment of population stratification in mitochondrial medical genetics. Am J Hum Genet. 2010;86:904–917. doi: 10.1016/j.ajhg.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madsen BE, Browning SR. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009;5:e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am J Hum Genet. 2007;80:727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, Weinstock GM, Wilson RK, Gibbs RA, Kent WJ, Miller W, Haussler D. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10:241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 44.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schork NJ, Murray SS, Frazer KA, Topol EJ. Common vs. rare allele hypotheses for complex diseases. Curr Opin Genet Dev. 2009;19:212–219. doi: 10.1016/j.gde.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen R, Davydov EV, Sirota M, Butte AJ. Non-synonymous and synonymous coding SNPs show similar likelihood and effect size of human disease association. PLoS ONE. 5:e13574. doi: 10.1371/journal.pone.0013574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suissa S, Wang Z, Poole J, Wittkopp S, Feder J, Shutt TE, Wallace DC, Shadel GS, Mishmar D. Ancient mtDNA genetic variants modulate mtDNA transcription and replication. PLoS Genet. 2009;5:e1000474. doi: 10.1371/journal.pgen.1000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fernandez-Silva P, Enriquez JA, Montoya J. Replication and transcription of mammalian mitochondrial DNA. Exp Physiol. 2003;88:41–56. doi: 10.1113/eph8802514. [DOI] [PubMed] [Google Scholar]
- 50.Penta JS, Johnson FM, Wachsman JT, Copeland WC. Mitochondrial DNA in human malignancy. Mutat Res. 2001;488:119–133. doi: 10.1016/s1383-5742(01)00053-9. [DOI] [PubMed] [Google Scholar]
- 51.Hochhauser D. Relevance of mitochondrial DNA in cancer. Lancet. 2000;356:181–182. doi: 10.1016/S0140-6736(00)02475-2. [DOI] [PubMed] [Google Scholar]
- 52.Kang D, Hamasaki N. Alterations of mitochondrial DNA in common diseases and disease states: aging, neurodegeneration, heart failure, diabetes, and cancer. Curr Med Chem. 2005;12:429–441. doi: 10.2174/0929867053363081. [DOI] [PubMed] [Google Scholar]
- 53.Yoneyama H, Hara T, Kato Y, Yamori T, Matsuura ET, Koike K. Nucleotide sequence variation is frequent in the mitochondrial DNA displacement loop region of individual human tumor cells. Mol Cancer Res. 2005;3:14–20. [PubMed] [Google Scholar]
- 54.Niemi AK, Hervonen A, Hurme M, Karhunen PJ, Jylha M, Majamaa K. Mitochondrial DNA polymorphisms associated with longevity in a Finnish population. Hum Genet. 2003;112:29–33. doi: 10.1007/s00439-002-0843-y. [DOI] [PubMed] [Google Scholar]
- 55.Ross OA, McCormack R, Curran MD, Duguid RA, Barnett YA, Rea IM, Middleton D. Mitochondrial DNA polymorphism: its role in longevity of the Irish population. Exp Gerontol. 2001;36:1161–1178. doi: 10.1016/s0531-5565(01)00094-8. [DOI] [PubMed] [Google Scholar]
- 56.De Benedictis G, Rose G, Carrieri G, De Luca M, Falcone E, Passarino G, Bonafe M, Monti D, Baggio G, Bertolini S, Mari D, Mattace R, Franceschi C. Mitochondrial DNA inherited variants are associated with successful aging and longevity in humans. Faseb J. 1999;13:1532–1536. doi: 10.1096/fasebj.13.12.1532. [DOI] [PubMed] [Google Scholar]
- 57.http://seer.cancer.gov/csr/1975_2005.
- 58.Arnold LD, Patel AV, Yan Y, Jacobs EJ, Thun MJ, Calle EE, Colditz GA. Are racial disparities in pancreatic cancer explained by smoking and overweight/obesity? Cancer Epidemiol Biomarkers Prev. 2009;18:2397–2405. doi: 10.1158/1055-9965.EPI-09-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wallace DC. Colloquium paper: bioenergetics, the origins of complexity, and the ascent of man. Proc Natl Acad Sci U S A. 2010;107 (Suppl 2):8947–8953. doi: 10.1073/pnas.0914635107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wallace DC. Why do we still have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu Rev Biochem. 2007;76:781–821. doi: 10.1146/annurev.biochem.76.081205.150955. [DOI] [PubMed] [Google Scholar]
- 61.Tranah GJ. Mitochondrial-nuclear epistasis: Implications for human aging and longevity. Ageing Res Rev. 2011;10:238–252. doi: 10.1016/j.arr.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.