Abstract
Phosphatase and tensin homolog deleted on chromosome ten (Pten) in stromal fibroblasts suppresses epithelial mammary tumors, but the underlying molecular mechanisms remain unknown. Using proteomic and expression profiling, we show that Pten loss from mammary stromal fibroblasts activates an oncogenic secretome that orchestrates the transcriptional reprogramming of other cell types in the microenvironment. Downregulation of miR-320 and upregulation of one of its direct targets, ETS2, are critical events in Pten-deleted stromal fibroblasts responsible for inducing this oncogenic secretome, which in turn promotes tumor angiogenesis and tumor cell invasion. Expression of the Pten-miR-320-Ets2 regulated secretome distinguished human normal breast stroma from tumor stroma and robustly correlated with recurrence in breast cancer patients. This work reveals miR-320 as a critical component of the Pten tumor suppressor axis that acts in stromal fibroblasts to reprogram the tumor microenvironment and curtail tumor progression.
Stromal cells, including fibroblasts, endothelial cells and immune cells collaborate with tumor cells to promote tumor proliferation, invasion and metastasis to distant sites. Fibroblasts are particularly important because they coordinate critical interactions between the various stromal and tumor cells by modulating the composition and function of the extracellular matrix (ECM). While normal fibroblasts prevent tumor growth and invasiveness, tumor-associated fibroblasts become reprogrammed by unknown mechanisms to co-evolve with epithelial tumor cells and provide an environment conducive for tumor initiation and progression 1-5. Recently, we showed that genetic inactivation of Pten in mammary stromal fibroblasts of mice accelerates the initiation, progression and malignant transformation of mammary epithelial tumors and that this depended on the activation of the transcription factor v-ets erythroblastosis virus E26 oncogene homolog 2 (ETS2) 6. Importantly, this work demonstrated that decreased PTEN protein levels were frequently observed in the stroma of human invasive breast cancer patients and were inversely correlated with the stromal expression of activated phosphorylated AKT and ETS2. In this report we show that Pten loss reprograms mRNA and microRNA (miR) expression profiles in normal mammary stromal fibroblasts to elicit a tumor-associated fibroblast phenotype and the reprogramming of gene expression in the entire mammary gland microenvironment. While there is extensive evidence supporting a role for miRs in tumor cells 7, 8, little is known about their regulation in stromal fibroblasts and whether they participate in the communication between the different cellular compartments of the tumor microenvironment. We show that miR-320 is a critical target of Pten in stromal fibroblasts that directly controls ETS2 expression and instructs the tumor microenvironment to suppress many of the aggressive phenotypes associated with advanced stages of breast cancer, including tumor cell invasiveness and increased angiogenic networks.
Results
Pten loss in stromal fibroblasts reprograms gene expression in the mammary gland microenvironment
In order to explore the mechanism of how Pten mediated signaling in stromal fibroblasts mediates tumor suppression, we compared expression of miRs in mouse mammary fibroblasts (MMFs) containing or lacking Pten. The Fsp-cre transgene and a conditional allele of Pten previously established in our laboratory were used to carry out fibroblast-specific deletion of Pten in mammary glands of mice (Fsp-cre;PtenloxP/loxP) 9. Of 400 miRs profiled, 10 miRs conserved between mouse and human genomes were significantly downregulated at least 2-fold in Pten deleted MMFs and (Supplementary Information Table S1). Quantitative Real Time PCR (qRT-PCR) confirmed reduced expression of 9 of the 10 relevant miRs in Pten-null fibroblasts (Fig. 1a, Supplementary Information Table S1). Together, these observations indicate that Pten loss influences mRNA 6 and miR profiles in mammary stromal fibroblasts.
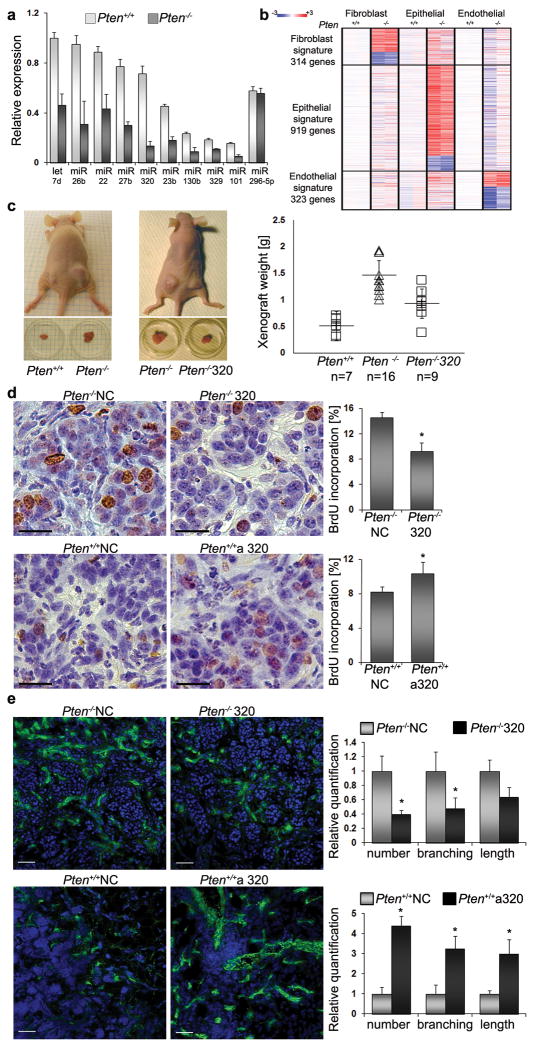
Figure 1. MiR-320 is a Pten target in fibroblasts that suppresses tumor growth.
A. Expression of selected microRNAs was validated by qRT-PCR in three sets of primary Pten+/+ or Pten-/- MMFs. Relative MiR expression level (normalized to 18S ribosomal RNA) is shown as mean ±SD. miR-296-5p is an example of a miR whose expression did not change.
B. Gene profiling was performed (n=2) on three types of cells from mammary glands with PtenLoxP/LoxP(Pten+/+) or Fsp-cre;PtenLoxP/LoxP(Pten-/-) as indicated. Heat map shows gene signature with 314 genes changed in fibroblasts, 919 genes changed in epithelial cells and 323 genes changed in endothelial cells (>2-fold and p<0.05).
C. Representative images of tumors before and after excision from mice injected with DB7 cells admixed with Pten+/+ (n=7) or Pten-/- (n=16) MMFs (left panel), and DB7 cells admixed with Pten-/- MMFs or Pten-/- cells re-expressing miR-320 (n=9) (right panel). Tumor weights were quantified after four weeks, and the data are expressed as mean ±SD. All three groups were tested by ANOVA and subsequent adjusted pairwise comparisons were performed. These comparisons were significant: Pten+/+ vs. Pten-/- MMFs; p = 5.58E -06. Pten+/+ vs. Pten-/- miR-320 MMFs; p = 0.022215. Pten-/- vs. Pten-/- miR-320 MMFs; p = 0.007976.
D. Epithelial tumor cell proliferation measured by BrdU incorporation in xenografts composed of DB7 cells admixed with Pten-/- or Pten+/+ MMFs transfected with either negative control (NC), miR-320 (320, upper right panel) or anti-miR-320 (a320, lower right panel); n=4 for each group. The epithelial/fibroblast composites were harvested five days after subcutaneous growth in mice. BrdU injection was performed 3h before harvesting the tissue. Scale bars: 25μm. Data are expressed as mean ±SD *p < 0.05.
E. Angiogenesis determined using CD31 staining to detect blood vessels in nascent tumors composed of DB7 cells and the same Pten-/- or Pten+/+ MMFs transfected with negative control (NC), miR-320 (320, upper panels) or anti-miR-320 (a320, lower panels), as above. The epithelial/fibroblast mixtures were grown for five days subcutaneously in mice. Representative micrographs showing merged images are shown (CD-31 - green, DAPI - blue). Scale bars: 50μm. Data are expressed as mean ±SD *p < 0.05.
We then entertained the possibility that the specific ablation of Pten in MMFs could lead to the reprogramming of gene expression in other cell compartments of the mammary gland. Analysis of mRNA expression profiles derived from epithelial and endothelial cells purified from PtenloxP/loxP and Fsp-cre;PtenloxP/loxP mammary glands revealed distinct sets of genes that were differentially expressed dependent on the status of Pten in mammary fibroblasts (Fig. 1b; Supplementary Information Fig. S1a).
MiR320 in fibroblasts suppresses tumor cell growth in vivo
Because miRs impact wide networks of gene expression 10, we focused on miRs in mammary stromal fibroblasts that could potentially function to communicate Pten signaling to the other cells in the microenvironment. We selected miR-320 for further analysis because it was the only miR of the nine conserved murine miRs to be reported to be downregulated in human breast cancer 11-15 and because relatively little is known about its' function. In four pairs of independently isolated MMF populations, miR-320 expression was significantly reduced when Pten was ablated and was inversely correlated with expression of the critical Pten target ETS2 (Supplementary Information Fig. S1b). To test whether decreased miR-320 expression had any functional consequence on the behavior of mammary epithelial tumor cells, we ectopically expressed miR-320 in Pten-null MMFs (see Supplementary Information FigS2d) and evaluated their capacity to support epithelial tumor cell growth in xenograft assays. A mouse mammary epithelial tumor cell line expressing a variant of the polyoma virus middle T-antigen gene (DB7) 16 was tagged with dsRed and co-injected with eGFP-tagged MMFs into immunocompromised mice. When compared to the co-injection of Pten+/+ MMFs, the co-injection of Pten-/- MMFs increased tumor growth four-fold, while restoration of miR-320 expression in the Pten-/- MMFs significantly reduced tumor growth (Fig. 1c). Differences in tumor size were due almost exclusively to an increase in the epithelial tumor cell compartment, as determined by the ratio of dsRed tumor cells to eGFP MMFs visualized by confocal microscopy (Supplementary Information Fig. S2a). Importantly, tumors derived from DB7 cells co-injected with Pten-/- MMFs expressing miR-320 were less invasive and had diminished vasculature compared to tumors from DB7 cells co-injected with Pten-/- MMFs (Supplementary Information Fig. S2b, c).
In order to capture the direct effects of fibroblasts on the tumorigenicity of epithelial cells without the potential confounding effects of infiltrating host stromal cells, we transiently expressed miR-320 or anti-miR-320 in MMFs and analyzed the consequences on DB7 tumor cells at an earlier time point in tumor development. These experiments had two complementary arms. First, co-injection of DB7 cells with Pten-/- MMFs was compared to the co-injection of DB7 with Pten-/- MMFs re-expressing miR-320. In this set of experiments, tumor cell proliferation, measured by BrdU incorporation, decreased by approximately 30% with miR-320 re-expression in Pten-/- MMFs (Fig. 1d, top panels). The formation of new blood vessels in these tumors, as measured by CD31 immunostaining, was also significantly decreased by the re-expression of miR-320 in Pten-/- MMFs (Fig. 1e, top panels). In the complementary second set of experiments, DB7 cells were co-injected with Pten+/+ MMFs or with Pten+/+ MMFs knock-downed for endogenous miR-320 (via anti-miR, see Supplementary Information FigS2e). In these experiments, tumor cell proliferation increased by approximately 25% with knockdown of miR-320 in Pten+/+ MMFs (Fig. 1d, bottom panels). Similarly, the formation of new blood vessels was dramatically increased by anti-miR-320 knockdown (Fig. 1e, bottom panels). Taken together, these results uncover a cell non-autonomous tumor suppressor role for miR-320 in stromal fibroblasts.
MiR-320 and PTEN expression are correlated in human breast cancer stroma but not in the tumor
We then examined stromal and epithelial expression of miR-320 in a panel of human invasive breast carcinoma and matched normal samples from 126 patients by in situ hybridization 17. The human genome contains eight annotated miR-320 genes that encode five mature miR variants; mouse has an ortholog for only one of these, miR-320a. We used the miR-320a sequence to design the locked oligonucleotide probe used for this analysis; however, all five variant miRs would likely be detected with the hybridization conditions utilized. Robust miR-320 expression was detected in the epithelial and stromal cell compartments of normal breast tissue but was consistently reduced in both cell compartments of invasive carcinoma samples (multispectral blue channel images and quantification in Fig. 2a, light microscope images in Supplementary Information Fig. S3a). Some nuclear staining for miR-320 was also evident. This nuclear staining could be explained either by probe hybridization to nuclear pre-miR forms or by nuclear re-localization of the mature miR 18-22
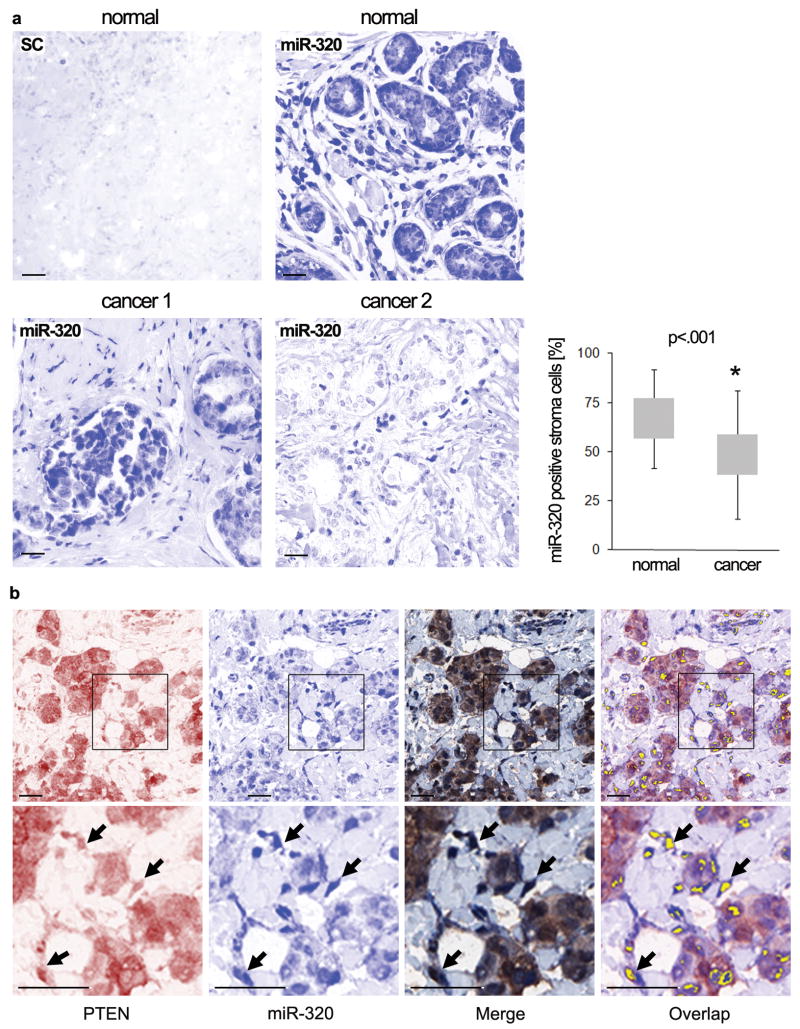
Figure 2. MiR-320 and PTEN are co-expressed in human tumor stroma.
A. Representative images of miR-320 ISH staining in paraffin imbedded, human normal (control) and breast cancer tissue; scrambled control was performed on adjacent normal breast tissue. Images were captured with the Nuance multispectral system (blue channel shown). Scale bars: 50μm. Percentage of positive cells per random field was determined in normal and carcinoma TMA sections from 126 matched patient samples (see Methods); results shown as mean ±SD on graph. P-value=1.76E-15.
B. Representative multispectral images of human breast carcinoma TMA samples stained for both miR-320 (ISH staining, blue) and PTEN (IHC, red). Upper panels -low magnification; bottom panels - high magnification. Scale bars: 50μm. Co-localization of both miR-320 and PTEN signals determined from the merged image was converted to fluorescent (yellow) signal using the Nuance multispectral system and is indicated by the arrows.
Multispectral imaging of samples stained for both PTEN and miR-320 demonstrated that their expression could be co-localized in the tumor stroma (Fig. 2b; Supplementary Information Fig. S3b). When the staining of PTEN and miR-320 were scored, a significant positive correlation between PTEN and miR-320 expression in the tumor stroma was revealed (Spearman rho=0.24, p=0.004, Supplementary Information Table S2). In contrast, there was no significant correlation between PTEN and miR-320 in the epithelial tumor cell compartment of the same TMA samples (Spearman rho=0.093, p=0.268). Interestingly, a significant inverse correlation was found between PTEN expression in the tumor and tumor stroma, while a strong positive correlation was found between miR-320 expression in the tumor and tumor stroma (Table S2), suggesting that downregulation of miR-320 in the stroma and tumor compartments may be mediated through distinct mechanisms.
Given the established relationship between PTEN function and ETS2 6, we used these tissue arrays to examine the phosphorylation status of threonine 72 in ETS2 (P-ETS2T72), which corresponds to its active state as a transcriptional activator. This analysis revealed that P-ETS2T72 levels inversely correlated with miR-320 levels in breast carcinoma samples (Spearman correlation coefficient -0.166, p=0.04, Supplementary Information Table S2). The correlative human data raised the possibility of a functional connection between PTEN and miR-320 in blocking the function of ETS2 in stromal fibroblasts in a way that might bias the mammary gland microenvironment towards tumor suppression.
MiR-320 in stromal fibroblasts suppresses tumor cell and endothelial cell proliferation and invasion in vitro
Increased proliferation and migration of epithelial cells with concurrent epithelial-mesenchymal transition (EMT) are hallmarks of malignant tumor growth 23, 24. Stromal fibroblasts are known to produce factors that signal to other cell types in the tumor microenvironment 25. To determine whether miR-320 in stromal fibroblasts influences the invasiveness of epithelial tumor cells, we developed a three-dimensional sphere invasion assay to monitor and quantify the migration of a non-invasive breast cancer tumor cell line (DB7). As monitored by microscopy, DB7 epithelial tumor cells embedded in a type I collagen matrix didn't migrate to any appreciable extent when incubated with either fresh media or conditioned media derived from wild-type MMFs (Fig. 3a, left and middle panels). In contrast, migration was dramatically increased when DB7 cells were incubated with conditioned medium from Pten-null MMFs (Fig. 3a, right panel). Interestingly, conditioned media from Pten-null MMFs elicited morphological changes in DB7 cells that resembled EMT (Fig. 3a, magnified inserts). Using this invasion assay, we could showed that re-expression of miR-320 in Pten-null MMFs attenuated the migration capacity of DB7 cells and also restored their typical cuboidal epithelial morphology (Fig. 3b). BrdU incorporation assays demonstrated that conditioned media from Pten-null MMFs overexpressing miR-320 eventually led to a modest but consistent decrease in the proliferation of DB7 cells (Fig. 3c);
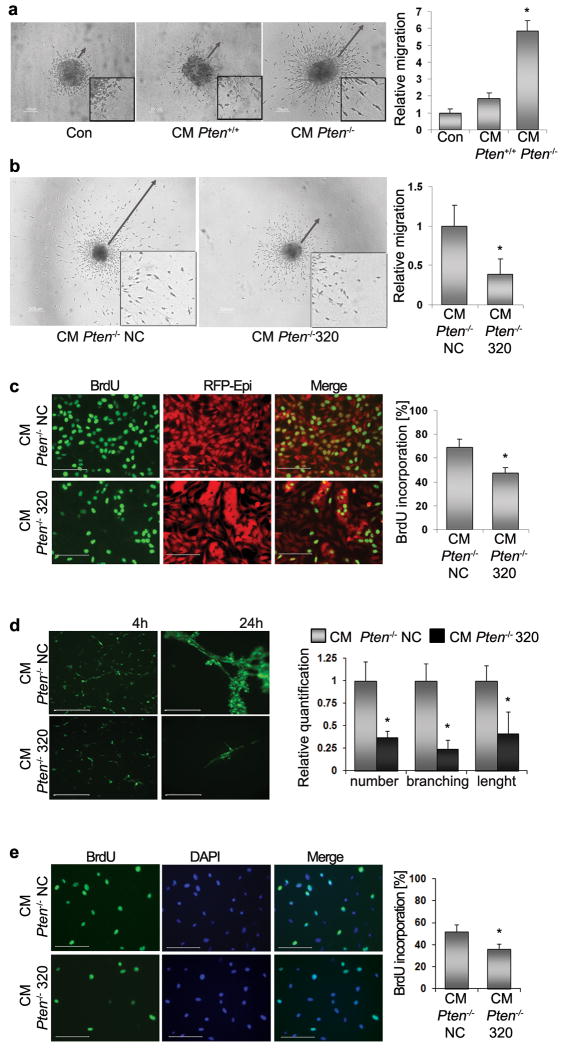
Figure 3. Pten and MiR-320 in fibroblasts control tumor cell migration and growth.
A. Representative images of DB7 mammospheres in the presence of DMEM/FBS medium (Con) (n=6) or conditioned medium (CM) produced by either Pten+/+ (n=4) or Pten-/- (n=6) MMFs (left). Scale bars: 100μm. Quantification of migratory zones indicated by arrows (right) expressed as mean area of migration ±SD. *p < 0.01. The inserts in all panels are magnified ×2.5.
B. Representative images of DB7 mammospheres in the presence of conditioned medium from Pten-/- MMFs expressing miR negative control (Pten-/- NC) (n=12) or miR-320 precursor (Pten-/-320) (n=12). Scale bars: 200μm. Quantification of migratory zones indicated by arrows (right) expressed as mean area of migration ±SD. *p < 0.01.
C. Representative images of BrdU incorporation in DB7 cells in the presence of conditioned medium from MMFs expressing miR negative control (Pten-/- NC) (n=4) or with miR-320 precursors (Pten-/-320) (n=4) (left). Scale bars: 100μm. Data expressed as mean ±SD *p < 0.01.
D. Representative images of tube-like formation of endothelial cells grown on Matrigel in the presence of conditioned medium from MMFs expressing miR negative control (Pten-/- NC) (n=3) or with miR-320 precursor (Pten-/-320) (n=3). Quantification of tube formation (right) expressed as mean ±SD. *p < 0.01. Scale bars: 50μm.
E. Representative images of BrdU incorporation by endothelial cells in the presence of conditioned medium from MMFs expressing miR negative control (Pten-/-NC) (n=3) or with miR-320 precursors (Pten-/-320) (n=3) (left). Data expressed as mean ±SD *p < 0.01. Scale bars: 100μm.
Because co-injection of DB7 cells with MMFs lacking Pten or miR-320 resulted in highly vascular tumors, we examined whether miR-320 in MMFs might directly influence the behavior of endothelial cells using 3-dimensional tube formation assays. Conditioned media from Pten-null MMFs stimulated the invasion of endothelial cells into matrigel and enhanced their arrangement into branched, tube-like structures, while conditioned media from miR-320-expressing Pten-null MMFs mitigated these effects (Fig. 3d). Moreover, conditioned media from Pten-null MMFs overexpressing miR-320 led to a decrease in the proliferation of endothelial cells compared to control (Fig. 3e). Thus, we conclude that miR-320 in stromal fibroblasts profoundly influences the behavior of other cell types in the tumor microenvironment through the action of secreted factors.
Stromal miR-320 regulates a tumor-specific secretome
We used a proteomic approach to identify the miR-320-regulated factors secreted by fibroblasts that modulate tumor and endothelial cell function. Mass spectrometry analysis of unfractionated, conditioned media from Pten-null MMFs, expressing either miR-320 or a negative control miR, identified 51 secreted proteins that were differentially represented, with 32 unique proteins present in the conditioned media from control Pten-null MMFs and 19 proteins unique to the conditioned media from miR-320 expressing Pten-null MMFs (Supplementary Information Fig. S4a-b and Table S3).
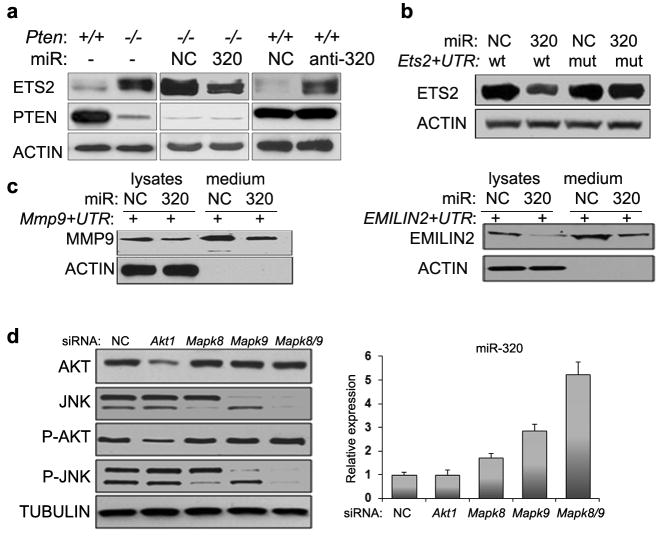
Western blot analysis on a subset of these proteins confirmed their differential secretion and also identified quantitative changes in the level of three additional secreted proteins that were not reproducibly detected by mass spectrometry, giving rise to a miR-320 responsive secretome profile of 54 factors (Fig. 4a and Supplementary Information Fig. 7a, Table S3). Three main groups of secreted factors were identified in the 54-factor secretome. In the first group, secreted factors such as matrix metalloproteinase-9, matrix metalloproteinase-2, bone morphogenetic protein-1, lysyl oxidase homolog-2 (MMP9, MMP2, BMP1, LOXL2 respectively) and EMILIN2, were increased in Pten-/- MMFs. Re-introduction of miR320 into the cells resulted in decreased expression of these secreted factors back to near-normal levels (Fig. 4a, top panel), suggesting they may represent direct targets of miR-320. In the second group, secreted factors such as Thrombospondin-1(THBS1) and Secreted frizzled-related protein-1(SFRP1), were downregulated in Pten-/- MMFs and re-introduction of miR320 restored their expression (Fig. 4a, middle panel), and thus likely represent indirect targets of miR-320. Importantly, overexpression of anti-miR320 (see Supplementary Information Fig. S2e) led to an increase in the secretion of MMP9, MMP2, LOXL2 and EMILIN2 and a decrease in the secretion of THBS1 and SFRP1 (Fig. 4a, top and middle panels). The third group of factors, represented by cathepsin B (CTSB), was dramatically increased in Pten-/- MMFs but remained unaffected by miR-320 re-expression (Fig. 4a, bottom panels). Conditioned media had no effect on the expression of the miR-320 targets in DB7 cells (Supplementary Information Fig. S4c).
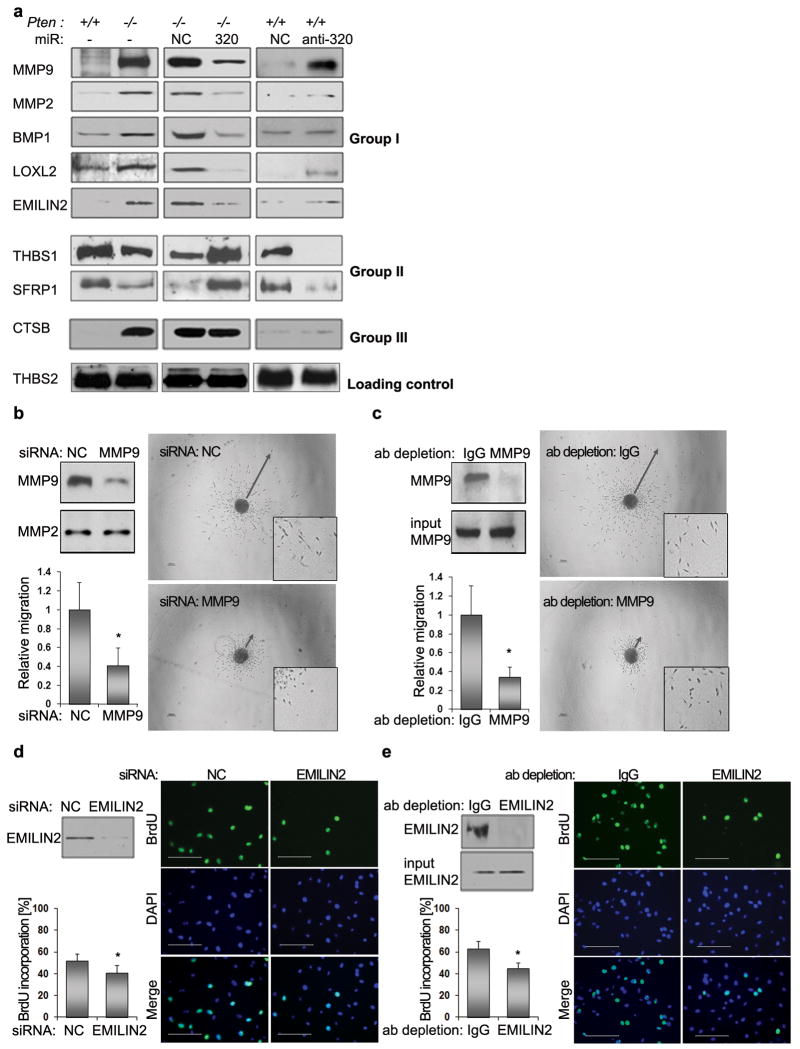
Figure 4. MiR-320 regulates the secretome of MMFs.
A. Pten+/+ and Pten-/- MMFs were left untreated (lane 1 and 2) or were transiently transfected with miR negative control (NC), miR-320 precursors (320) or anti-miR-320 precursor (anti-320), respectively. Conditioned media was examined by Western blotting with antibodies against the indicated protein; THBS2 serves as internal control. Full length blots are presented in Supplementary Figure 7a.
B. Representative images of DB7 mammospheres in the presence of conditioned medium from Pten-/- MMFs expressing siRNA negative control (NC) (n=4) or siRNA against Mmp9 (n=4). MMP9 down-regulation was verified by Western blot using MMP9 antibody. MMP2 was used as a loading control. Scale bars are 200μm. The inserts in all panels are magnified ×2.5. Quantification of migratory zones (indicated by arrows) expressed as mean area of migration ±SD. *p < 0.01 (bar graphs).
C. Representative images of DB7 mammospheres in the presence of conditioned medium from Pten-/- MMFs pre-cleared with IgG (n=4) or IgG pre-coupled with α-MMP9 (n=4). Western blot using MMP9 antibody in pre-cleared and input control samples were performed as a control. Scale bars are 200μm. The inserts in all panels are magnified ×2.5. Quantification of migratory zones (indicated by arrows) expressed as mean area of migration ±SD. *p < 0.01 (bar graphs).
D. Representative images of BrdU incorporation in endothelial cells in the presence of conditioned medium from Pten-/- MMFs expressing siRNA negative control (NC) (n=3) or siRNA against Emilin2 (n=3). Emilin2 down-regulation was verified by Western blot using EMILIN2 antibody. Scale bars are 100μm. Data expressed as mean ±SD *p < 0.01.
E. Representative images of BrdU incorporation in endothelial cells in the presence of conditioned medium from Pten-/- MMFs pre-cleared with IgG (n=3) or IgG pre-coupled with α-EMILIN2 (n=4). Western blot using EMILIN2 antibody in pre-cleared and input control samples were performed as a control. Scale bars are 100μm. Data expressed as mean ±SD *p < 0.01.
Two members of the Pten-regulated secretome were selected for further analysis, MMP9 and EMILIN2, which are thought to be involved in the control of epithelial cell migration26 and angiogenesis 27. Blocking MMP9 expression from stromal fibroblasts by either Mmp9-specific siRNA, or by treating conditioned media with an MMP9-specific antibody, blunted the ability of conditioned media derived from Pten-null MMFs to promote the migration of DB7 cells (Fig. 4b, c). Similarly, blocking EMILIN2 expression by either Emilin2-specific siRNA or antibody reduced the ability of Pten-null MMF conditioned media to stimulate endothelial cell proliferation (Fig. 4d, e). Thus, these results indicate that miR-320 regulates a fibroblast secretome required for remodeling of the ECM and vascular network.
Ets2 is a direct target of miR-320
Of the stromal-secreted proteins identified, only seven were predicted by miR/target recognition software to represent direct targets of miR-320 (Supplementary Information Table S3), indicating that regulation of this secretome may predominantly be an indirect consequence of miR-320 downregulation. Interestingly, Ets2 was among the genes predicted to be directly regulated by miR-320 (Supplementary Information Fig. S5a). Moreover, promoter/enhancer sequences in 20 genes of the 54-factor secretome (37%) contain ETS2-binding elements that are conserved across species 28-33 (Supplementary Information Table S4), including Mmp9, which is a known bona fide transcriptional target of ETS2 6. Based on the observed correlative relationship between PTEN, ETS2 and miR-320 expression in both MMFs and breast cancer samples, and the above in silico predictions, we entertained the possibility that ETS2 may be acting downstream of miR-320 to regulate the expression of a large fraction of the 54-factor secretome.
Consistent with this hypothesis, reintroduction of miR-320 in Pten-null MMFs led to a decrease in ETS2 protein levels and conversely, overexpression of the anti-miR-320 in wild-type fibroblasts increased ETS2 protein to levels found in Pten-/- MMFs (Fig. 5a, Supplementary Information Fig. S7b); no significant effect on Akt, Jnk and Erk signaling pathways, known to be upstream activators of ETS26, were observed by the manipulations of miR-320 levels (Supplementary Information Fig. S5b). To test if miR-320 directly regulates ETS2, we examined the ectopic expression of an Ets2 full-length cDNA with an authentic 3′ UTR containing the predicted miR-320 target sequences. Co-transfection of miR-320 along with this full length Ets2 cDNA vector resulted in a marked decrease in ectopic ETS2 protein levels. Mutation of the predicted miR-320 target sequences in the Ets2 3′ UTR abolished this effect (Fig. 5b, Supplementary Information Fig. S5c, S7b). Moreover, introduction of the Ets2 3′ UTR into a luciferase reporter gene was sufficient to modulate reporter expression in response to miR-320 (Supplementary Information Fig. S5d).
Figure 5. Identification and characterization of Ets2, Mmp9 and Emiln2 as bona fide targets of miR-320.
A. Pten+/+ and Pten-/- MMFs were left untreated (lane 1 and 2) or were transiently transfected with miR negative control (NC), miR-320 precursors (320) or anti-miR-320 (anti-320), respectively. Nuclear extracts were blotted with anti-ETS2 antibody or anti-PTEN antibody; anti-β-actin antibody was used as a loading control.
B. COS7 cells were transfected with full length cDNA (5′-UTR/ORF/3′-UTR) of: Ets2 (Ets2+UTR) wild type (wt) or mutated in miR-320 seeding region (mut) expression vector and co-transfected with either miR negative control (NC) or miR-320 precursors (320). Cell lysates or conditioned medium were blotted with anti-ETS2, antibody; anti-β-actin antibody was used as a loading control.
C. COS7 cells were transfected with full length cDNA (5′-UTR/ORF/3′-UTR) of: Mmp9 (Mmp9+UTR) (left panel) or Emilin2 (Emilin2+UTR) (right panel) and co-transfected with either miR negative control (NC) or miR-320 precursors (320). Cell lysates or conditioned medium were blotted with anti-MMP9 or anti-EMILIN2 antibody; anti-β-actin antibody was used as a loading control.
D. Pten-/- MMFs were transiently transfected either with siRNA negative control (NC) or siRNA against Akt1, Mapk8/Jnk1, Mapk9/Jnk2 or the combination of Mapk8/Jnk1 and Mapk9/Jnk2. Akt and Jnk phosphorylation status was verified by Western blot using anti-phospho specific antibodies, and compared to total kinase levels using non-discriminating antibodies. Tubulin was used as a loading control. Expression of miR-320 was validated by qRT-PCR (n=3). Mean relative miR expression level ±SD is shown. For A, B, C, D full length blots are presented in Supplementary Figure 7b-d.
In silico analysis also predicted Mmp9 and Emilin2 to be direct targets of miR-320. Employing a similar experimental strategy as described above, we could show that Mmp9 and Emilin2 are directly regulated by miR-320 via their 3′ UTR target sequences (Fig. 5c, Supplementary Information Fig. S5c, S7c; luciferase reporter gene assays in Supplementary Information Fig. S5d). Exogenous introduction of miR-320 into Pten-null MMFs also led to decreased Ets2, Mmp9, and Emilin2 mRNA levels (Supplementary Information Fig. S5e). These results are consistent with a dual mechanism for the regulation of Mmp9 expression that involves transcriptional control by ETS2 6 and posttranscriptional control by miR-320.
Loss of Pten leads to activation of the PI3-Kinase/AKT pathway, which is believed to be the pathway responsible for the majority of phenotypes associated with loss of Pten in human tumors 34. However, Pten loss has also been shown to activate additional signaling pathways, including Akt-independent activation of the JNK pathway 35, 36. Based on preliminary results using small molecule inhibitors for Pten regulated pathways (data not shown), we focused on Akt1, Jnk1/Mapk8 and Jnk2/Mapk9 using the more specific approach of siRNA mediated knockdown (Fig. 5d, Supplementary Information Fig. S7d). These experiments demonstrated that knockdown of either Jnk1/Mapk8 or Jnk2/Mapk9 in Pten-null fibroblasts led to a 2-3 fold increase in miR-320 expression, while knockdown of both led to a 5-fold increase in miR-320 expression. In contrast, knockdown of Akt1 had no significant effect on miR-320 expression.
A mouse stromal miR-320 profile predicts human breast cancer patient outcome
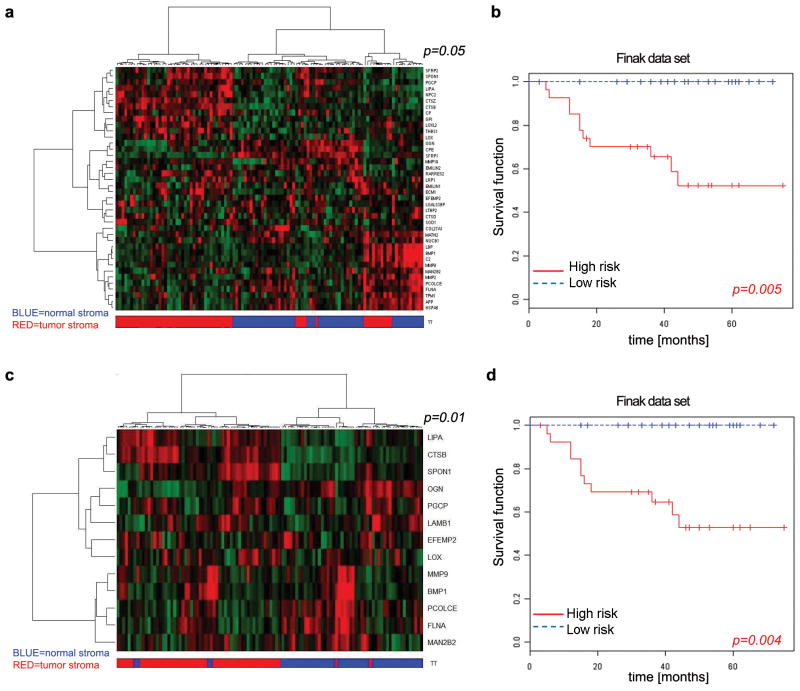
To explore the clinical relevance of our findings, we used the mouse derived 54-secretome profile identified above to query stroma-specific expression profiles of breast cancer patients 37. Human homologues of the mouse 54-factor secretome profile were differentially expressed in tumor versus adjacent normal stroma 38, leading to a partial segregation of tumor stroma and normal stroma that attained statistical significance (Fig. 6a and Supplementary Information Table S5). Importantly, the 54-factor secretome profile correlated with clinical outcome based on stromal gene expression in breast cancer patients 37 (Fig. 6b). Given the role of Ets2 in regulating the expression of a subset of these 54 genes (20/54), we also used this miR-320/Ets2 regulated 20-gene subset to query the same stroma-specific expression profiles derived from breast cancer patients. As shown in Figure 6c, the 20 gene Ets2 subset differentiated tumor stroma from adjacent normal stroma even more effectively than the complete 54-gene list (Fig. 6c and Supplementary Information Table S5) and robustly correlated with patient outcome (Fig. 6d). The 54- or the 20-gene secretome profiles, when used to query additional expression profiles derived from whole tumors and linked to breast cancer patient outcome data 39, also correlated with patient outcomes (Supplementary Information Fig. S6a, b).
Figure 6. The miR-320 secretome profile separates human breast normal and cancer stroma and predicts patient outcome.
A. Heat map displaying the differential expression in human tumor versus normal stroma of the 40 human orthologs from the 54-factor mouse secretome that were retrieved from the McGill stromal microarray (GSE4823). The p-value indicates the ability of the 40-gene signature to partition normal and tumor stroma compared to 10,000 random permutations (see Methods).
B. Expression of the 40 secretome gene signature present in GSE9014 data set correlates with poor patient outcomes. Kaplan Meir curves of high and low-risk groups based on expression of the 40 gene secretome signature. The permutation p-value of the log-rank test statistic between risk groups is based on 1000 permutations.
C. Heat map displaying the differential expression in tumor versus normal stroma of 13 human orthologs from the 20 ETS2-target genes that were retrieved from the McGill breast cancer stroma microarray, (GSE4823). The p-value indicates the ability of the mouse 17-gene signature to partition normal and tumor stroma as above (see Methods).
D. Expression of the subset of secretome genes directly regulated by ETS2 (13/20 genes in GSE4823) correlates with poor patient outcomea. Kaplan Meir survival curves of high and low-risk groups based on expression of the 13 gene ETS2-related secretome signature. The permutation p-value of the log-rank test statistic between risk groups is based on 1000 permutations.
Discussion
In the current study, we identified a Pten-miR-320-Ets2 tumor suppressor axis in stromal fibroblasts that modulates the inter-cellular communication within the tumor microenvironment and is responsible for pathological and molecular events observed in malignant human breast cancer. Loss of Pten in stromal fibroblasts results in down-regulation of miR-320 and the reprogramming of mRNA expression profiles in neighboring endothelial and epithelial cells of the mammary gland. We show that by influencing the behavior of multiple cell types, miR-320 in stromal fibroblasts is a critical Pten-regulated determinant for the suppression of epithelial tumors. While identifying a critical function for the miR-320-Ets2 pathway in the stroma, our results don't show that the miR-320-Ets2 pathway is strictly stromal specific. The pathway could also be active in tumor epithelial tumor cells that have low PTEN expression, although the PTEN IHC results suggest that in the majority of human breast cancer patients the pathway is unlikely to be active in both cell compartments.
Proteomic analysis of fibroblast conditioned medium identified a miR-320-regulated tumor-promoting secretome that when activated by loss of Pten incites profound changes in endothelial and epithelial cell phenotypes typical of malignant tumors. While some of the mRNAs encoding these secreted factors are targeted by miR-320 directly, for example EMILIN2, most are regulated indirectly through transcriptional control by ETS2, which is itself an essential direct target of miR-320. Together, these findings expose a miR-320 regulatory switch in normal fibroblasts that operates in a cell autonomous fashion to inhibit the expression of a tumor-promoting secretome and in a cell non-autonomous fashion to block expression programs in other cell types in the microenvironment that together suppress tumor cell growth and invasiveness. Remarkably, bioinformatic analyses demonstrated that a miR-320 secretome signature could distinguish normal from tumor stroma and could be utilized to robustly predict outcome in breast cancer patients, underlining the potential clinical impact of the stromal Pten-miR-320 regulatory axis on human breast cancer.
In conclusion, our results extend the concept of miR function beyond the tumor cell boundary by defining a complex network of communication within the tumor microenvironment that is necessary for tumor growth and spread. In spite of the overwhelming complexity, modulation of a few key regulatory nodes such as miR-320 may be sufficient to impede the most malignant properties of tumor cells, suggesting novel strategies for developing agents that disrupt select tumor microenvironment networks.
Supplementary Material
Acknowledgments
We thank Kara Batte for technical assistance with microRNAs platform, Rumeysa Biyik for bioinformatics assistance, Abdel-Rasoul Mahmoud for statistical assistance, the Ohio State University Human Tissue Resource Network and the Ohio State University Comprehensive Cancer Center Microarray, Nucleic Acids, Proteomic Shared Facilities for technical assistance. This work was funded by the National Institutes of Health to M.C.O. (R01CA053271, P01CA097189) and to G.L. (R01CA85619, R01HD47470, P01CA097189) by the Komen Breast Cancer Foundation and Evelyn Simmers Charitable Trust to M.C.O.
Footnotes
Author Contributions: M.C.O. and G.L. designed and supervised this study, analyzed data, and helped write and edit the manuscript.
A.B. J.G. designed and performed experiments, collected and analyzed data, and co-wrote the paper.
J.A.W. H.M. R.S. and F.L. technically assisted with experiments, and collected and analyzed data.
M.O.N. assisted with fluorescent and confocal microscopy and immunohistochemistry
L.Y. and S.A.F. contributed to the statistical analyses of data and writing the manuscript.
A.M. S.C. M.H. M.P. T.P. contributed to the analysis and comparison of mouse and human profile data
M.G.P. and C.B.M. contributed to the analysis microRNA data and writing the manuscript.
C.K.M. L.Y. G.N. T.R. contributed to the histopathological analysis of human samples and writing the manuscript.
S.E.L. and E.A.C. contributed to the data analysis and writing the manuscript.
References
- 1.Mueller MM, Fusenig NE. Friends or foes - bipolar effects of the tumour stroma in cancer. Nat Rev Cancer. 2004;4:839–849. doi: 10.1038/nrc1477. [DOI] [PubMed] [Google Scholar]
- 2.Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature. 2004;432:332–337. doi: 10.1038/nature03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 4.Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Sotgia F, et al. Caveolin-1-/- null mammary stromal fibroblasts share characteristics with human breast cancer-associated fibroblasts. Am J Pathol. 2009;174:746–761. doi: 10.2353/ajpath.2009.080658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trimboli AJ, et al. Pten in stromal fibroblasts suppresses mammary epithelial tumours. Nature. 2009;461:1084–1091. doi: 10.1038/nature08486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Leva G, Croce CM. Roles of small RNAs in tumor formation. Trends Mol Med. 16:257–267. doi: 10.1016/j.molmed.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trimboli AJ, et al. Direct evidence for epithelial-mesenchymal transitions in breast cancer. Cancer Res. 2008;68:937–945. doi: 10.1158/0008-5472.CAN-07-2148. [DOI] [PubMed] [Google Scholar]
- 10.Bader AG, Brown D, Winkler M. The promise of microRNA replacement therapy. Cancer Res. 70:7027–7030. doi: 10.1158/0008-5472.CAN-10-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci U S A. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ichimi T, et al. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer. 2009;125:345–352. doi: 10.1002/ijc.24390. [DOI] [PubMed] [Google Scholar]
- 13.Schepeler T, et al. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008;68:6416–6424. doi: 10.1158/0008-5472.CAN-07-6110. [DOI] [PubMed] [Google Scholar]
- 14.Mattie MD, et al. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan LX, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–2360. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borowsky AD, et al. Syngeneic mouse mammary carcinoma cell lines: two closely related cell lines with divergent metastatic behavior. Clin Exp Metastasis. 2005;22:47–59. doi: 10.1007/s10585-005-2908-5. [DOI] [PubMed] [Google Scholar]
- 17.Nuovo GJ. In situ detection of precursor and mature microRNAs in paraffin embedded, formalin fixed tissues and cell preparations. Methods. 2008;44:39–46. doi: 10.1016/j.ymeth.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 18.Jorgensen S, Baker A, Moller S, Nielsen BS. Robust one-day in situ hybridization protocol for detection of microRNAs in paraffin samples using LNA probes. Methods. 52:375–381. doi: 10.1016/j.ymeth.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Nuovo G, Lee EJ, Lawler S, Godlewski J, Schmittgen T. In situ detection of mature microRNAs by labeled extension on ultramer templates. Biotechniques. 2009;46:115–126. doi: 10.2144/000113068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. RNA. 2006;12:1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Politz JC, Hogan EM, Pederson T. MicroRNAs with a nucleolar location. RNA. 2009;15:1705–1715. doi: 10.1261/rna.1470409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim DH, Saetrom P, Snove O, Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci U S A. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 24.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 25.Hughes CC. Endothelial-stromal interactions in angiogenesis. Curr Opin Hematol. 2008;15:204–209. doi: 10.1097/MOH.0b013e3282f97dbc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orlichenko LS, Radisky DC. Matrix metalloproteinases stimulate epithelial-mesenchymal transition during tumor development. Clin Exp Metastasis. 2008;25:593–600. doi: 10.1007/s10585-008-9143-9. [DOI] [PubMed] [Google Scholar]
- 27.Mongiat M, et al. The extracellular matrix glycoprotein elastin microfibril interface located protein 2: a dual role in the tumor microenvironment. Neoplasia. 12:294–304. doi: 10.1593/neo.91930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aoyama T, et al. Structure and chromosomal assignment of the human lectin-like oxidized low-density-lipoprotein receptor-1 (LOX-1) gene. Biochem J. 1999;339(Pt 1):177–184. [PMC free article] [PubMed] [Google Scholar]
- 29.Yan S, Berquin IM, Troen BR, Sloane BF. Transcription of human cathepsin B is mediated by Sp1 and Ets family factors in glioma. DNA Cell Biol. 2000;19:79–91. doi: 10.1089/104454900314591. [DOI] [PubMed] [Google Scholar]
- 30.Cironi L, et al. IGF1 is a common target gene of Ewing's sarcoma fusion proteins in mesenchymal progenitor cells. PLoS One. 2008;3:e2634. doi: 10.1371/journal.pone.0002634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He HJ, Kole S, Kwon YK, Crow MT, Bernier M. Interaction of filamin A with the insulin receptor alters insulin-dependent activation of the mitogen-activated protein kinase pathway. J Biol Chem. 2003;278:27096–27104. doi: 10.1074/jbc.M301003200. [DOI] [PubMed] [Google Scholar]
- 32.de Kerchove D'Exaerde A, et al. Expression of mutant Ets protein at the neuromuscular synapse causes alterations in morphology and gene expression. EMBO Rep. 2002;3:1075–1081. doi: 10.1093/embo-reports/kvf220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomarev SI, Nakaya N. Olfactomedin domain-containing proteins: possible mechanisms of action and functions in normal development and pathology. Mol Neurobiol. 2009;40:122–138. doi: 10.1007/s12035-009-8076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hollander MC, Blumenthal GM, Dennis PA. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat Rev Cancer. 11:289–301. doi: 10.1038/nrc3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rong Y, et al. Epidermal growth factor receptor and PTEN modulate tissue factor expression in glioblastoma through JunD/activator protein-1 transcriptional activity. Cancer Res. 2009;69:2540–2549. doi: 10.1158/0008-5472.CAN-08-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vivanco I, et al. Identification of the JNK signaling pathway as a functional target of the tumor suppressor PTEN. Cancer Cell. 2007;11:555–569. doi: 10.1016/j.ccr.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 37.Finak G, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 38.Finak G, et al. Gene expression signatures of morphologically normal breast tissue identify basal-like tumors. Breast Cancer Res. 2006;8:R58. doi: 10.1186/bcr1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pawitan Y, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7:R953–964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hunter MP, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Godlewski J, et al. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125–9130. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- 43.Godlewski J, et al. MicroRNA-451 regulates LKB1/AMPK signaling and allows adaptation to metabolic stress in glioma cells. Mol Cell. 37:620–632. doi: 10.1016/j.molcel.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nowicki MO, et al. Lithium inhibits invasion of glioma cells; possible involvement of glycogen synthase kinase-3. Neuro Oncol. 2008;10:690–699. doi: 10.1215/15228517-2008-041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rothhammer T, Bataille F, Spruss T, Eissner G, Bosserhoff AK. Functional implication of BMP4 expression on angiogenesis in malignant melanoma. Oncogene. 2007;26:4158–4170. doi: 10.1038/sj.onc.1210182. [DOI] [PubMed] [Google Scholar]
- 46.Srinivasan R, et al. Erk1 and Erk2 regulate endothelial cell proliferation and migration during mouse embryonic angiogenesis. PLoS One. 2009;4:e8283. doi: 10.1371/journal.pone.0008283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chong JL, et al. E2f1-3 switch from activators in progenitor cells to repressors in differentiating cells. Nature. 2009;462:930–934. doi: 10.1038/nature08677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bronisz A, et al. Microphthalmia-associated transcription factor interactions with 14-3-3 modulate differentiation of committed myeloid precursors. Mol Biol Cell. 2006;17:3897–3906. doi: 10.1091/mbc.E06-05-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holm S. A simple sequentially rejective multiple test procedure. Scandinavian Journal of Statistics. 1979;6:65–70. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.