Abstract
Purpose
Detection of pancreatic cancer remains high priority and effective diagnostic tools are needed for clinical applications. Many cancer cells overexpress integrin αvβ6, a cell surface receptor being evaluated as a novel clinical biomarker.
Experimental Design
To validate this molecular target, several highly stable cystine knot peptides were engineered by directed evolution to bind specifically and with high-affinity (3-6 nM) to integrin αvβ6. The binders don’t cross-react with related integrin αvβ5, integrin α5β1 or tumor-angiogenesis associated integrin, αvβ3.
Results
Positron emission tomography showed that these disulfide-stabilized peptides rapidly accumulate at tumors expressing integrin αvβ6. Clinically relevant tumor-to-muscle ratios of 7.7 ± 2.4 to 11.3 ± 3.0 were achieved within one hour after radiotracer injection. Minimization of off-target dosing was achieved by reformatting αvβ6-binding activities across various natural and pharmacokinetically-stabilized cystine knot scaffolds with different amino acid content. We demonstrate that a peptide scaffold’s primary sequence directs its pharmacokinetics. Scaffolds with high arginine or glutamic acid content suffered high renal retention of > 75 percent injected dose per gram (%ID/g). Substitution of these amino acids with renally-cleared amino acids, notably serine, led to significant decreases in renal accumulation of < 20 %ID/g 1h post injection (p < 0.05, n=3).
Conclusions
We have engineered highly stable cystine knot peptides with potent and specific integrin αvβ6 binding activities for cancer detection. Pharmacokinetic engineering of scaffold primary sequence led to significant decreases in off-target radiotracer accumulation. Optimization of binding affinity, specificity, stability and pharmacokinetics will facilitate translation of cystine knots for cancer molecular imaging.
INTRODUCTION
Integrins are a family of heterodimeric cell surface receptors that mediate cellular adhesion to extracellular matrix proteins. Integrins also serve as bi-directional signal transducers to regulate differentiation, migration, proliferation, and cell death (1, 2). Integrin αvβ3 promotes tumor neovascularization (3, 4). However in certain cancers, other integrins, such as αvβ6, become highly overexpressed on cell surfaces (2, 5, 6). Therefore, this biomarker is being validated for detection of colon, liver, ovarian, pancreatic, and squamous cell cancers (7-9). Molecular imaging of integrin αvβ6 may be used to gauge receptor expression levels to determine prognosis and guide therapy (10-12). Here, we have developed potent integrin αvβ6 binders for clinical translation as radiotracers for early cancer detection.
Previously, several integrin αvβ6 binders were identified from natural and combinatorial sources. Linear αvβ6-binding peptides derived from the coat protein of foot-and-mouth-disease viruses (FMDV) generally suffered poor in vivo stability, which raise concerns about their potential immunogenicity. 64Cu-labeled versions of FMDV peptides demonstrated extremely high renal retention, which suggests that these peptides may not be ideal translational candidates. An alternative to radio-metals is the use of radio-halogens (7, 9, 13). Phage display systems have identified several linear and disulfide-cyclized peptides that bind αvβ6 (14, 15). In one study, a radio-iodinated linear peptide HBP-1, showed rapid degradation in serum (16). One-bead-one-compound libraries have identified many binders, of which 43 18F-labeled linear peptides were tested in a high throughput live animal imaging survey (17). While some of these peptides show promising small animal data, linear peptides or even simple disulfide-bonded peptides with stability problems may discourage translation (17). Peptide fragments can be highly immunogenic thus rendering the parent peptide untranslatable (18). For these reasons, we sought to generate high-affinity binders that are very stable in physiological media, demonstrate low off target accumulation and effectively detect cancer in living subjects.
Engineered cystine knot peptides (knots) have shown promise for cancer imaging with αvβ3 as a target (19-21). The cystine knot is a rigid molecular scaffold of 3-4 kDa that owes its exceptionally-high stability to three interwoven disulfide bonds and a centrally-located beta sheet. Potent receptor-binding activities have been engineered into the scaffold’s solvent exposed loops (22). The knotted structure helps to resist degradation/denaturation in hostile biological, chemical and physical environments such as strong acids and boiling water (23). Cystine knots have shown exceptional structural stability during prolonged incubation in serum (19). Moreover, their use in humans as uterogenics has not led to reports of adverse side effects, albeit without formal published studies (24, 25). Binding potency remains high for engineered knots that we have subjected to long-term storage (> 1 year) in water at 4°C or stored in lyophilized form at room temperature. The N-terminus provides a sole primary amine for site-specific conjugation of imaging labels, bioactive cargo, or pharmacokinetic stabilizers. Collectively, these characteristics bode well for clinical translation.
In this study, we developed new cystine knots that bind integrin αvβ6 expressed on pancreatic tumors grown in mice. The binders showed single-digit nanomolar dissociation constants similar to antibodies used clinically for imaging and therapy. These new knots rapidly accumulated in αvβ6-positive tumors, which led to excellent tumor-to-normal contrast. The new knots did not accumulate in tumors that do not express integrin αvβ6. The peptides were specific for the targeted integrin αvβ6 receptors expressed on orthotopic pancreatic tumors and various xenografts used in this study. Additionally, pharmacokinetic-stabilization strategies endowed knots with rapid renal clearance, which significantly reduced off-target dosing. This study indicates a broadly-applicable way to engineer new binders and to stabilize their pharmacokinetics for enhanced clinical performance.
MATERIALS AND METHODS
The SUPPLEMENTAL INFORMATION section provides additional details.
Materials, Cell Lines, and Reagents
BxPC-3 pancreatic cancer cells were obtained from American Type Culture Collection (ATCC) and grown in RPMI 1640 media (ATCC). A431 epidermoid cancer cells, human embryonic kidney 293T cells (293), U87MG (malignant glioma) and MDA-MB-435 (breast cancer) cells were obtained from frozen lab stocks and grown in DMEM supplemented with 10% FBS and penicillin/streptomycin (Invitrogen). Recombinant human integrins αvβ6, αvβ3, αvβ5, α5β1 were purchased from R&D Systems. All other chemicals were obtained from Fisher Scientific unless otherwise specified. Yeast media, growth and induction conditions and integrin binding buffer (IBB) are previously described (22).
Library Synthesis and Screening
The open reading frames encoding cystine knot peptides were generated by overlap-extension PCR using yeast-optimized codons. Positions for randomization, denoted by “X” were constructed with NNB degenerate codons (Supplemental). PCR products were amplified using primers with overlap to the pCT yeast display plasmid upstream or downstream of the NheI and BamHI restriction sites, respectively. For each library, ~40 μg of DNA insert and 4 μg of linearized pCT vector were electroporated into the S. cerevisiae strain EBY100 by homologous recombination as previously described (26). For libraries L2 and L3, ~ 5-7×106 transformants per sub-library were combined for a total diversity of ~1×107 clones as estimated by serial dilution plating and colony counting. For libraries L2.1 and L3.1, ~2×106 transformants per sub-library were combined for a total diversity of ~4×106 clones.
For library screening, various concentrations of recombinant αvβ6 integrin were added to yeast suspended in IBB for various times at room temperature. Next, a 1:250 dilution of chicken anti-cMyc IgY antibody (Invitrogen) was added for 1h at 4°C. The cells were washed with ice-cold IBB and incubated with a 1:25 dilution of fluorescein-conjugated anti-αv integrin antibody (Biolegend) and a 1:100 dilution of Alexa 555-conjugated goat anti-chicken IgG secondary antibody (Invitrogen) for 0.5h at 4°C. Cells were washed in IBB and αvβ6 integrin binders were isolated using a Becton Dickinson FACS Aria III instrument. For the first round of sorting, ~2 × 107 yeast clones were screened with 100 nM αvβ6 integrin. To increase sort stringency, integrin concentrations were successively decreased to 1 nM in later sort rounds, and a diagonal sort gate was used to isolate yeast cells with enhanced integrin binding (FITC fluorescence) for a given protein expression level (Alexa 555 fluorescence). For the second diversification round, ~5 × 106 yeast clones were screened as described above with a final concentration of 300 pM αvβ6 integrin coupled with 3 days of washing in IBB at 37°C. Plasmid DNA was recovered by Zymoprep (Zymo Research), amplified in Max Efficiency DH5α E. coli cells (Invitrogen) and sequenced.
Peptide Synthesis/Biosynthesis, Folding and Radiolabeling
S02 and R01 were synthesized, folded and purified as previously described with a modification to the folding buffer, which included the addition of an equal volume of isopropanol and 800 mM guadinium hydrochloride (22). A20 was similarly prepared without folding. R02 and E02 were biosynthesized using Pichia pastoris (Supplemental). Peptides were conjugated through their N-terminus amine to DOTA-NHS and radiolabeled with 64CuCl2 as previously described (19). The radiochemical purity was determined by HPLC to be > 95%. The radiochemical yield was usually over 80%. The specific activity of the probe was ~500 Ci/mmol. Molecular masses were confirmed by MALDI-MS (ABI 5800, Supplemental Table 2).
Binding Affinity
Various concentrations of integrin αvβ6, αvβ5, αvβ3 and α5β1 were incubated with 105 yeast cells expressing R01, R02, R03, S01, S02, S03 or 2.5F in the presence of 106 un-induced yeast cells as previously described (22, 27). Prior to flow cytometry and analysis, yeast cells were processed, stained, and washed as described above in Library Synthesis and Screening.
Protein and Cell Capture Assays
Neuravidin coated wells (Pierce) were saturated with biotinylated peptides A20, R01, S02 or 2.5F. 10nM recombinant integrins or 105 cells in IBB were added to wells at room temperature for 2h. Wells were washed 3x with ice-cold IBB. Integrins were detected with mouse anti-αv or anti-α5 primary antibodies (Biolegend) and anti-mouse Alexa-488 (Cellsignal). Cells were detected with crystal violet (22). Signals were quantified with a plate reader (Tecan). For competition binding assays, integrin αvβ6 or BxPC-3 cells were preincubated with R01 or S02 for 2 hours prior to introduction into A20 coated wells.
64Cu-DOTA Peptide Stability
Aliquots of 64Cu-DOTA-peptide were incubated in an equal volume of mouse or human serum for up to 24h. Samples were acidified with TFA and centrifuged to remove precipitants. In addition, 64Cu-DOTA-peptides were extracted from full mouse bladders 1.5-2 hour after injection. Soluble fractions were filtered with a Spin-X 0.2 μm filter (Corning) when necessary. All samples were analyzed by radio-HPLC on a Dionex C4 column.
Tumor Models
Animal procedures were performed per protocol 11580 and 21637 (Stanford University Administrative Panels on Laboratory Animal Care). Female athymic nude mice, 4-6 weeks old (Charles River), were subcutaneously shoulder-injected with 107 cells suspended in 100 μL PBS. Orthotopic tumors were generated with 106 cells/20 uL Matrigel (Supplemental). Mice were used for imaging/biodistribution studies when xenografts or orthotopic tumors reached ~10 mm or ~5 mm, respectively, in diameter.
MicroPET Imaging
Tumor-bearing mice (n=3 for each probe) were injected with ~50-100 μCi (~0.15 nmol) of probe via the tail vein and imaged with a microPET R4 rodent model scanner (Siemens) using 5min static scans. Images were reconstructed by a two-dimensional ordered expectation maximum subset algorithm and calibrated as previously described (28). ROIs were drawn over the tumor on decay-corrected whole body images using ASIPro VM software (Siemens). ROIs were converted to counts/g/min, and %ID/g values were determined assuming a tissue density of 1 g/mL. No attenuation correction was performed.
Biodistribution Analysis
Anesthetized nude mice bearing xenograft/orthotopic tumors were injected with ~50-100 μCi (~0.15 nmol) of 64Cu-DOTA-peptides via tail vein, and euthanized after 1h or 24h. Tissues were removed, weighed and measured by scintillation counting (19, 29). Radiotracer uptake in tissues was reported as percent injected dose per gram (%ID/g) and represents the mean ± standard deviation of experiments performed on three mice.
RESULTS
Engineering αvβ6 Binders
Nine phage-display derived lead-motif (RTDLXXL) containing sequences were engrafted into loop-1 of an acyclized cystine knot scaffold Momordica cochinchinensis Trypsin Inhibitor-II (MCoTI-II) from squash (14, 30, 31). These chimeric-binders are referred to as the “R-knots” (R0) since MCoTI-II has high arginine content (Schematic 1). Optimal loop-1 length and motif position were determined, and enabled design of libraries X3RTDLXXLX3 and X4RTDLXXLX3 (L2 and L3 Schematic 1A, Supplemental Figure 1), which were pooled and sorted by FACS. The X3RTDLXXLX3 library dominated sort-round five (1 nM target). Frequent occurrence of arginine or lysine residues at certain loop positions suggested favorable binding interactions. Arginine was fixed at these positions in the second libraries (L2.1 and L3.1) to ensure a sole N-terminus amine for chemical labeling. The remaining loop positions were randomized (Schematic 1A). The final sort-round was conducted in 300 pM target followed by 3 days of washing at 37°C. The top three most-represented binders (1, 2 and 3) were characterized.
Scaffold Swapping and its Effects on Binding Affinity and Specificity
A novel αvβ6-binding activity, “2”, was grafted into several arginine-rich scaffolds (R0-3), glutamic acid-rich scaffolds (E0-4) and serine-rich scaffolds (S0-3). αvβ6-binding was determined by flow cytometry (Schematic 1B, Supplemental Figure 2B and Supplemental Schematic 1) to be comparable across scaffolds; maintenance of high-affinity binding to integrin αvβ6 indicates that engineered loops and knotted scaffolds can function independently and interchangeably. Novel integrin αvβ6 binding activities called “1”, “2”, and “3”, were each tested in the context of both the R0 and S0 scaffolds for their ability to bind target. We measured the KD of R01, R02, and R03 to be 3.6 ± 0.9 nM, 3.2 ± 2.7 nM, 3.6 ± 1.6nM, respectively, by flow cytometry using soluble integrin αvβ6 (Figure 2A). Interestingly, the KD of the S01, S02 was only slightly less at 6.5 ± 2.0 nM and 6.0 ± 0.1 nM, respectively, while the KD of S03 (3.1 ± 0.5 nM) matched that of its parent (Figure 2B). These results suggest that the knotted structure is tightly maintained in engineered R0 and S0 binders, so that novel activities can be trans-grafted into other wild type or rationally-designed knots without notable loss of potency (Supplemental Figure 2B). We also tested cross-reactivity of peptides used throughout these studies to other integrins. Peptides demonstrate very low binding to integrins αvβ3, αvβ5 and α5β1 (Figure 2C). Scrambled peptides did not bind integrin αvβ6 (Supplemental Figure 2A).
Figure 2.
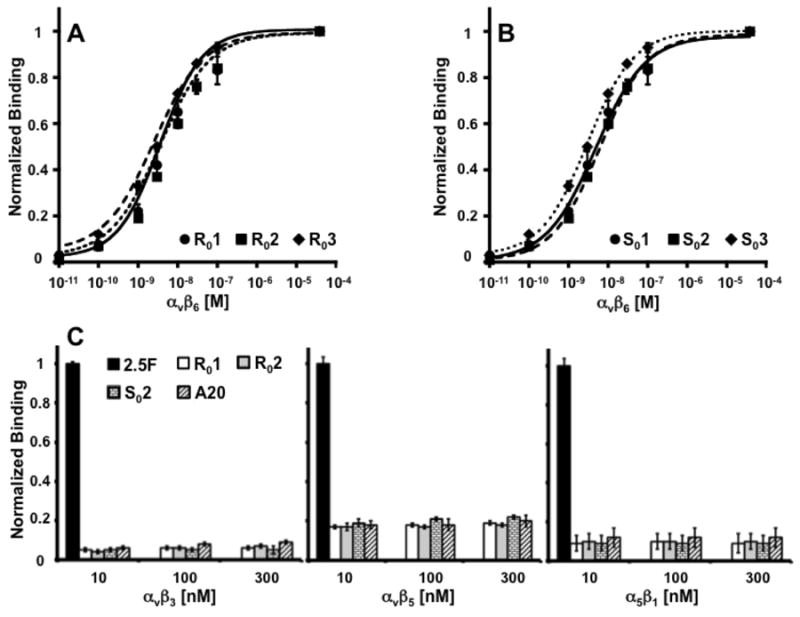
Representative binding curves of (a) R01 (circles/solid lines), R02 (squares/dashed lines) and R03 (diamonds/dotted lines). The same activities were reformatted in a serine-rich scaffold to produce (b) S01 (circles/solid lines), S02 (squares/dashed lines) and S03 (diamonds/dotted lines). By flow cytometry, all six binders demonstrated single-digit nanomolar affinities for integrin αvβ6. Data were normalized with respect to saturated fluorescence intensity (plateau) observed at the highest target concentrations. (c) Indicated peptides were also evaluated for binding to related integrins αvβ3, αvβ5 and α5β1 and normalized to binding by 2.5F (19).
Protein and Cell Capture Assays
Biotinylated peptides A20, R01, S02 and 2.5F were immobilized onto neuravidin coated plates. All 4 peptides captured recombinant integrin αvβ6, but only knottin 2.5F captured integrins αvβ3, αvβ5 and α5β1. Engineered binders show different levels of specificity for their targets (Figure 3A). Biotinylated A20, precoated onto microwell plates, engaged in competitive binding with soluble R01 or S02 for recombinant integrin αvβ6. Dose dependent inhibition indicated competition between peptides for a specific target-binding site (Figure 3B). A20, R01 and S02 also captured cells that express native integrin αvβ6 (Figure 3C). Flow cytometry showed all tested cell lines (BxPC-3, A431, U87MG, MDA-MB-435) but the 293 cells express integrin αvβ6 (Supplemental Figure 3AB). Peptides R01 and S02 blocked adhesion of BxPC-3 cells onto A20 coated wells confirming specific binding between peptides and functionally-active integrins expressed on cellular surfaces (Figure 3D). 64Cu-DOTA-labeled peptides also demonstrated binding to target expressing cells and not to 293 negative controls (Supplemental Figure 4).
Figure 3.
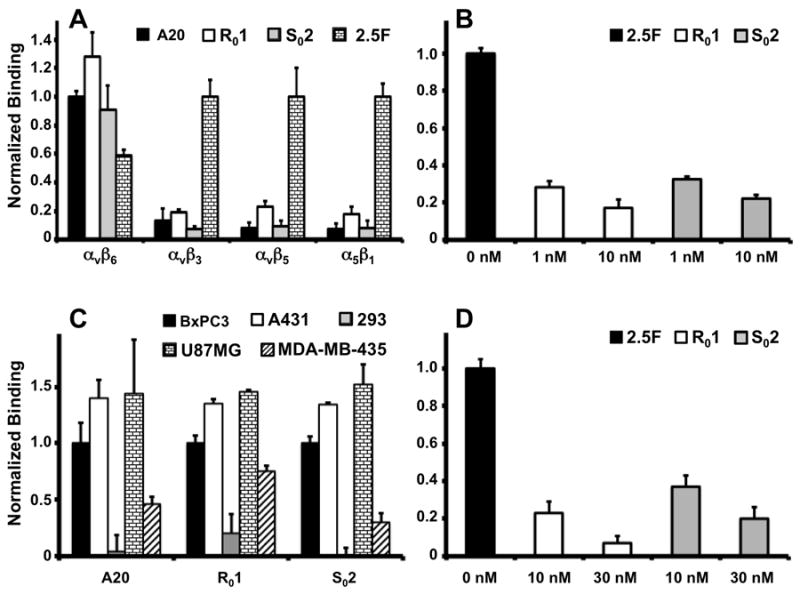
Protein and Cell Capture Assays. (a) Microwells coated with A20, R01, S02 or 2.5F are tested for binding to 10nM integrins αvβ6, αvβ3 αvβ5 or α5β1. αvβ6 binding was normalized to A20. αvβ3 αvβ5 and α5β1 binding were normalized to 2.5F. (b) R01 and S02 inhibit binding of 10nM αvβ6 to A20 coated wells. Data are normalized to non-competitive binding. (c) Various cell lines are tested for binding to A20, R01 or S02 coated wells. Data are normalized to BxPC-3 cell-binding. (d) R01 and S02 inhibit capture of 105 BxPC-3 cells. Data are normalized to non-competitive binding.
Scaffold Reformatting: Serum and Metabolic Stability of Peptides
Most wild-type and engineered knots resist degradation/denaturation in physiological media such as serum and urine (19, 23, 29). However, loop engineering can compromise stability. For example, R02 was ~80% degraded during 24h serum incubation (Figure 4A). Comparatively, 64Cu-DOTA-labeled R01 demonstrated much greater stability; approximately 20% degradation occurred during 24h serum incubation. These two peptides share identical primary sequences in loops 2 through 5 of their structural frames (Schematic 1). In contrast, 64Cu-DOTA-S02 and -E02 demonstrated exceptionally high (>95%) stability during 24h serum incubation (Figure 4BC). For completeness, the linear control 64Cu-DOTA-A20 was >90% degraded after 24h incubation in serum (Figure 4D). Urine samples drawn from mice 1.5-2h after injection showed ~20% degradation of 64Cu-DOTA-R01 and <5% degradation of 64Cu-DOTA S02, our current lead translational candidates (Supplemental Figure 4AB). These results demonstrate that non-binding portions of peptide scaffolds may be used to increase in vivo stability
Figure 4.
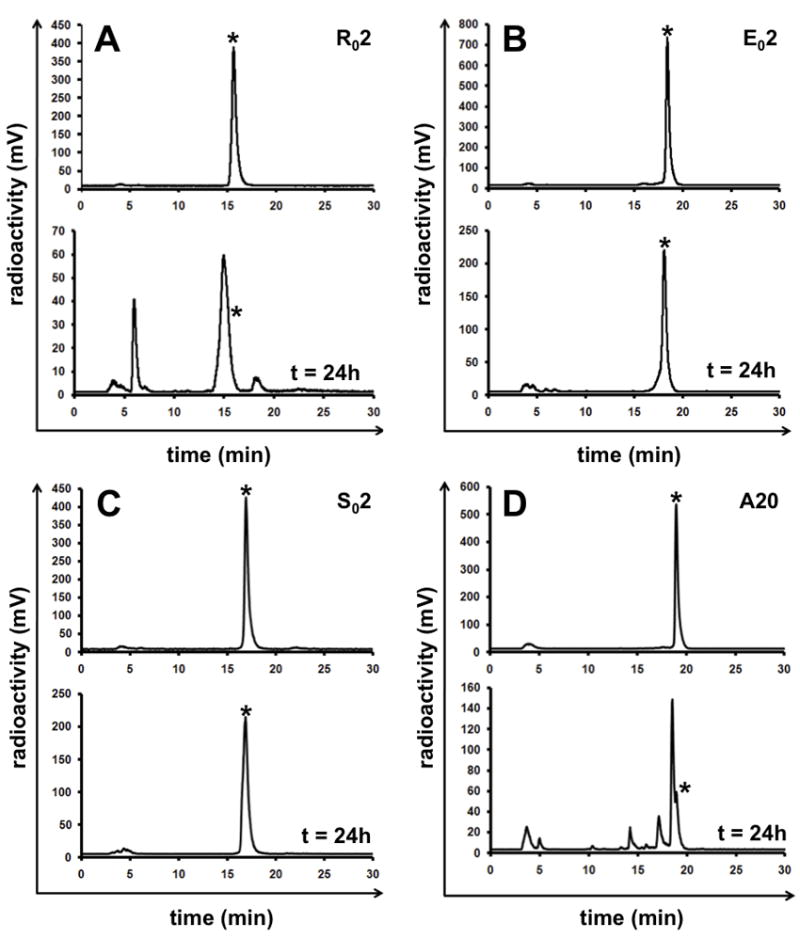
Radio-HPLC analysis of 64Cu-DOTA labeled (a) R02, (b) E02, (c) S02 and (d) A20 prior to (upper panel) and after 24h incubation (lower panel) in mouse serum at 37°C. Asterisk indicates HPLC retention time of intact radiotracer.
MicroPET Imaging and Biodistribution
Radio-labeled versions of knots were evaluated by PET in mice bearing integrin αvβ6-expressing BxPC-3 (pancreatic cancer) or A431 (epidermoid cancer) xenografts, and αvβ6-negative 293 tumors. Mice were injected with ~75 uCi of 64Cu-DOTA labeled peptides. R02, E02, S02, R01 and the positive control A20, rapidly accumulated in αvβ6-positive A431 tumors and generated excellent tumor-to-muscle contrast ratios of 6-11 at 1h post injection (p.i., Figure 5AB). Absolute uptake in A431 tumors was highest for the two R0-based binders R01 (~5 %ID/g) and R02 (~4 %ID/g) compared to E02 (~1.5 %ID/g), S02 (~2 %ID/g) and A20 (~2 %ID/g) at 1h p.i. (Figures 5AB).
Figure 5.
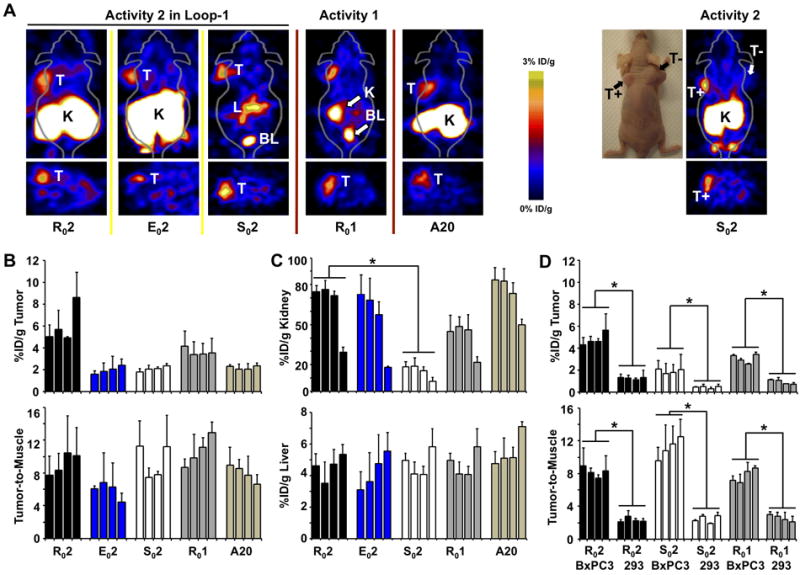
MicroPET images of (a) A431 xenografts are shown at 1h post injection of 64Cu-DOTA labeled R02, E02 and S02 and Ro1. The positive control is indicated by A20. On the far right, a BxPC-3 αvβ6-positive (T+) and αvβ6-negative 293 (T-) double xenograft injected with So2 is shown both as a photograph and microPET image of the same mouse. (b,c) Quantification of microPET images shown in panel a. (d) Side-by-side comparison of R02, S02 and R01 in BxPC-3 xenografts and 293 xenografts (* P < 0.05). (b-d) Four bars indicate 1,2,4 and 24h post injection.
PET studies of BxPC-3 pancreatic xenografts confirmed these results. Importantly, significantly less radiotracer accumulated in αvβ6-negative 293 xenografts (p <0.05, n = 3 mice, Figure 5D). Comparatively, tumor uptake of R02, measured 4.3 ± 0.7 %ID/g in BxPC-3 xenografts compared to 1.3 ± 0.1%ID/g in 293 xenografts. S02 and R01 corroborated these results (Figure 5D). Collectively, these results show that the αvβ6-specific probes selectively bind αvβ6-positive (T+) tumors (A431 and BxPC-3) and do not accumulate in the αvβ6-negative (T-) 293 tumors (Figure 5AD). Unfortunately, conclusive identification of orthotopic BxPC-3 tumors by PET remained elusive.
Biodistribution analysis of R01 and S02 in BxPC-3 xenograft tumors showed 4.13±1.01 and 1.80± 0.50 %ID/g 1h p.i.. These data closely matched corresponding PET data (Table 1A and Figure 5D). Importantly, S02 effectively accumulated in BxPC-3 orthotopic tumors (1.85±0.11 %ID/g, 1h p.i.) compared to normal pancreas (~0.2-0.3 %ID/g, 1h p.i., Table 1). However, substantial amounts of S02 accumulated in liver, stomach, intestines and kidneys as measured by biodistribution analysis and seen by PET imaging (Table 1, Figure 5).
Table 1.
Biodistribution of 64Cu-DOTA-R01, and -S02, in pancreatic cancer models, measured by tissue resection and scintillation counting are reported as (a) mean %ID/g ± SD (n=3) in various tissues, and (b) their associated tumor-to-normal tissue ratios at 1 hour and 24 hours post injection are reported as mean ± SD (n=3). Orthotopic tumors are represented by “o*”
| A. Biodistribution of 64Cu-DOTA-labeled peptides in αvβ6-positive BxPC3 pancreatic tumor models.
| ||||||
|---|---|---|---|---|---|---|
| Tissue | Data are presented as mean %ID/g ± SD | |||||
| 64Cu-DOTA-R01 | 64Cu-DOTA-S02 | 64Cu-DOTA-S02 (*) | ||||
| 1h | 24h | 1h | 24h | 1h | 24h | |
| Tumor | 4.13±1.01 | 3.31±0.20 | 1.80 ±0.50 | 1.46±0.43 | 1.85±0.11 (*) | n.d. |
| Muscle | 0.49±0.21 | 0.33±0.14 | 0.24±0.14 | 0.14±0.02 | 0.21±0.02 | n.d. |
| Blood | 0.46±0.18 | 0.52±0.09 | 0.29±0.07 | 0.23±0.03 | 0.22±0.03 | n.d. |
| Heart | 0.39±0.08 | 0.67±0.12 | 0.22±0.04 | 0.47±0.03 | 0.32±0.01 | n.d. |
| Kidney | 68.12±9.69 | 33.75±1.77 | 26.53±12.26 | 10.69±1.63 | 31.49±2.47 | n.d. |
| Liver | 2.28±0.40 | 4.69±0.75 | 3.03±1.10 | 2.79±0.65 | 2.70±0.53 | n.d. |
| Lung | 2.78±0.59 | 2.34±080 | 1.03±0.29 | 1.22±0.50 | 0.95±0.09 | n.d. |
| Spleen | 0.52±0.18 | 0.67±0.11 | 0.27±0.12 | 0.36±0.13 | 0.39±0.09 | n.d. |
| Pancreas | 0.57±0.10 | 0.73±0.28 | 0.17±0.02 | 0.25±0.02 | orthotopic (*) | n.d. |
| Stomach | 2.31±0.12 | 2.71±0.42 | 0.79±0.19 | 0.98±0.27 | 1.51±0.11 | n.d. |
| Intestine | 1.49±0.2 | 2.05±0.38 | 1.280.26 | 0.92±0.27 | 1.05±0.11 | n.d. |
| Brain | 0.07±0.01 | 0.15±0.03 | 0.04±0.01 | 0.07±0.01 | 0.06±0.01 | n.d. |
| Bone | 0.26±0.08 | 0.510.24 | 0.09±0.01 | 0.19±0.06 | 0.23±0.01 | n.d. |
| Skin | 0.99±0.14 | 1.04±0.72 | 0.34±0.09 | 0.66±0.15 | 0.46±0.32 | n.d. |
| B. Tumor-to-Normal Tissue Ratios | ||||||
|---|---|---|---|---|---|---|
| Ratio | 64Cu-DOTA-R01 | 64Cu-DOTA-S02 | 64Cu-DOTA-S02 (*) | |||
| 1h | 24h | 1h | 24h | 1h | 24h | |
| T/Muscle | 8.89±2.00 | 6.52±0.83 | 6.22±0.93 | 6.36±1.20 | 8.54±1.23 | n.d. |
| T/Blood | 9.72±3.63 | 10.81±3.31 | 9.98±5.95 | 10.68±3.44 | 8.86±1.38 | n.d. |
| T/Liver | 1.91±0.85 | 0.72±0.11 | 0.67±0.32 | 0.53±0.19 | 0.70±0.09 | n.d. |
| T/Lung | 1.48±0.12 | 1.54±0.53 | 1.89±0.81 | 1.26±0.28 | 1.95±0.28 | n.d. |
| T/Spleen | 8.34±2.14 | 5.05±0.73 | 7.22±2.26 | 4.23±1.62 | 6.81±2.54 | n.d. |
| T/Pancreas | 7.18±0.55 | 4.96±1.75 | 10.89±3.95 | 5.96±2.20 | orthotopic(*) | n.d. |
| T/Kidneys | 0.06±0.02 | 0.10±0.01 | 0.07±0.03 | 0.14±0.06 | 0.06±0.01 | n.d. |
R02 and R01 accumulated to greater levels in A431 tumors compared to E02 and S02. However, these arginine-rich binders cleared slowest from surrounding muscle tissue. Biodistribution analysis indicated ~0.6 %ID/g muscle at 1h for the R02, while approximately half (~0.3 %ID/g) was observed for E02 and S02 (Supplemental Table 1). Therefore, tumor-to-muscle contrast was comparable for each of the engineered knots.
Renal retention varied by binding activity and scaffold (Figure 5C). R01 ranged from ~45 %ID/g (1h) to ~20 %ID/g (24h), whereas R02 ranged from ~75 %ID/g (1h) to ~28 %ID/g (24h). This difference is attributed to the higher arginine content of activity “2”. However, transfer of activity “2” to the carboxyl-rich scaffold (E02) did not decrease kidney signal (~ 72 %ID/g (1h) and ~16 %ID/g (24h)). These results suggest that in addition to arginine residues, the kidneys also actively retain carboxyl-containing residues. Importantly, when the “2” activity was tested in the S0 scaffold (S02), significantly lower renal retention was measured (~18 %ID/g (1h) and ~7 %ID/g (24h)). The large differences in renal uptake suggest that scaffold pharmacokinetics may be optimized to evade renal reuptake and allow a binder to pass freely into the bladder. Arginine is one of the most versatile amino acids in animal cells, serving as a precursor for the synthesis of proteins, nitric oxide, urea, polyamines, proline, glutamate, creatine and agmatine (32). Therefore, arginine recycling by the kidneys is beneficial to the organism. By substituting pharmacokinetically favorable amino acids in the diversity-tolerant cystine knot framework, non-specific uptake by off-target tissues may be reduced.
DISCUSSION
Integrin αvβ6 is being explored as a clinical target due to its elevated expression in several cancers (2). In the current study, yeast surface display and high throughput screening by FACS produced many high-affinity binders that selectively accumulate in αvβ6-positive pancreatic and epidermoid cancer models, but not in αvβ6-negative tumors actively growing in mice. By grafting the loop known as activity “2” (RSLARTDLDHLRGR) across different cystine knot scaffolds with biased amino acid content (R0, E0 and S0), we achieved significantly lower renal signal without compromising αvβ6 binding affinity or specificity. This study suggests that primary sequence composition drives pharmacokinetics, so that it may be possible to optimize tumor uptake, tumor-to-muscle contrast and off-target accumulation in tissues by fine-tuning the non-target-binding portions of the scaffold in addition to engineering target-binding loops.
Absolute 64Cu-DOTA signal intensity at the tumor was greatest for the two R0 binders, R01 and R02 (~ 4.5 %ID/g at 1h p.i.). 64Cu-DOTA-labeled E02, So2 and A20 generated approximately half the tumor signal intensity compared to the R-rich scaffolds. Given the similar affinities of the engineered knots (Figure 2AB), we rationalize that R0s’ higher tumor signal benefitted from the inherent “stickiness” of arginine. However, independent of a scaffold’s compositional differences, each of the activity “2” knots (R02, E02, and S02) demonstrated similarly useful tumor-to-normal contrast enabling precise delineation of integrin αvβ6-positive tumors (T+) from αvβ6-negative tumors (T-) shown in Figure 5.
For clinical imaging of pancreatic cancer, S02 has emerged as our lead candidate. 64Cu-DOTA- S02 demonstrated an excellent tumor-to-background ratio (~10 at 1 h), rapid renal clearance (~25 %ID/g 1h p.i. to < 10 %ID/g after 24h), and high serum stability (>95% after 24h). In contrast, 64Cu-DOTA-R02 rapidly degrades in serum (< 20% intact 24h) and accumulates to a much higher degree in the kidneys (~75 %ID/g 1h). 64Cu-DOTA-E02 also suffers from prolonged and high renal retention (~75 %ID/g 1h). Therefore, to take advantage of S0s’ lower renal background, we are performing affinity maturation to achieve higher tumor uptake comparable to R0 binders. We are also exploring 18F labeling strategies since the pharmacokinetics of these binders support a shorter lived isotope for clinical translation.
Biodistribution studies indicated that 64Cu-DOTA-S02 also effectively accumulated in orthotopic pancreatic tumors (~1.8 %ID/g 1h p.i.) compared to normal pancreas (~0.2-0.3 %ID/g). However, tumor-to-liver/kidney/gut ratios were suboptimal (Table 1AB). The proximity of these PET-avid tissues is ~1-3mm from the pancreas and is beyond the spatial resolution of the PET scanner. Therefore, definitive identification of orthotopic tumors by PET was limited by closely-packed mouse organs. However, human scanning should not suffer from these limitations.
The engineered knots exhibit specific binding to integrin αvβ6. They do not bind related integrins αvβ3, αvβ5 or α5β1, and the scrambled (RDTXXL) versions do not bind αvβ6 even at high concentrations (Figures 2C and 3A, Supplemental Figure 2A). Low uptake of αvβ6-specific radiotracers by 293 xenografts correlated to their limited expression of both the αv and β6 integrin subunits (Supplemental Figure 3A, Figure 5). In a very clear example, 64Cu-DOTA-S02 specificity was demonstrated in a mouse bearing both BxPC-3 and 293 xenografts (far right panel Figure 5A). Here, signal intensity for the αvβ6-positive tumor (T+, left shoulder) was ~2 %ID/g, vs. ~0.5 %ID/g for the αvβ6-negative tumor (T-, right shoulder) at 1h p.i.. The “stickier” R0 binders also demonstrated selectivity for integrin αvβ6 expressing cells/tumors and recombinant protein (Figures 2, 3, 5 and Supplemental Figure 3). However, muscle wash-out was slower for the R0s, so that contrast was comparable across all binders tested (Figure 5BD).
Renal clearance can potentially be controlled and optimized. Our current study provides insight about the significant differences in renal uptake reported for peptides previously engineered to bind integrin αvβ3, a vascular target (19). Knottins 2.5D and 2.5F, based on the cystine knot Ecballium elaterium Trypsin Inhibitor (EETI-II), contain a minimal number of arginines. The renal signals of 64Cu-DOTA-2.5D and -2.5F measured < 10 %ID/g from 1-24h. In contrast, 64Cu-DOTA-AgRP-7C, derived from the agouti related peptide, which contains many complex nitrogen- and oxygen-containing residues, demonstrated significantly higher renal retention of > 65 %ID/g at the same time points (29). Renal uptake of radio-metal labeled affibodies has also been reported to be very high (> 100 %ID/g) at multiple time points for binders 002, 1907 and 2377 (33-35). Affibody sequence inspection reveals the presence of many K, D/E and Q/N residues, which can contribute to high renal retention (36-38). The kidneys appear to actively survey for the more-complex, charge-rich amino acids (D/E/K/R) while allowing rapid clearance of the simpler non-charged amino acids (e.g., A,G,S). The development of stable, pharmacokinetically-optimized scaffolds is highly desirable. Our results suggest one way to gain precise control of binder pharmacokinetics.
In summary, cystine knots are inherently stable scaffolds that are potentially well-suited for clinical molecular imaging. Loop-grafting and directed evolution experiments generated single-digit nanomolar binders. PET imaging studies produced clinically useful tumor-to-normal contrast within 1h. We demonstrated that simple scaffold swapping could enhance knot stability and stabilize pharmacokinetics to evade renal surveillance and approach renal-stealth. Importantly, most knots are very well-behaved during long-term storage. Collectively, these data suggest that cystine knots warrant further exploration for clinical translation.
Supplementary Material
Figure 1.
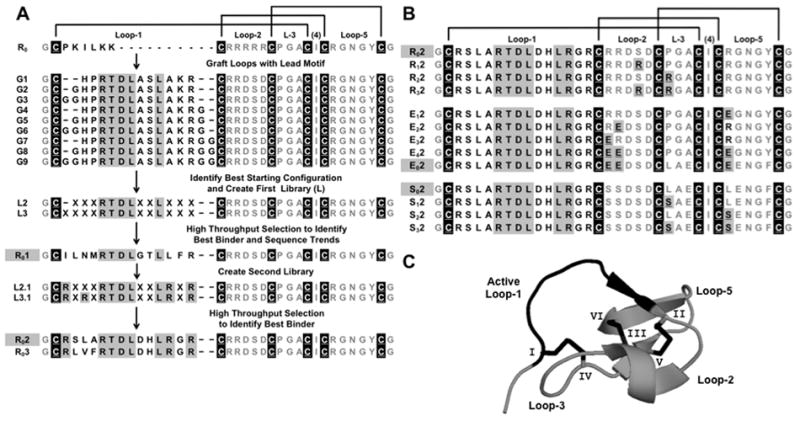
Schematic Outline of Engineering Process. (a) The primary sequence of the wild type trypsin inhibitor cystine knot framework (R0) includes six cystine residues (white underlined black-back letters, and disulfide connectivities as black lines) and five loops. Grafts one to nine (G1–G9) each contain the RTDLXXL motif (gray-back letters). Libraries L2 and L3 were derived from highest affinity grafts, G2 and G3. High-affinity binder R01 emerged after 5 sort rounds. Libraries, L2.1 and L3.1, fixed arginine residues to prevent the occurrence of lysine at those positions. Binders R02 and R03 emerged from sorting. (b) The arginine-rich (R0), glutamic acid-rich (E0) and serine-rich (S0) frameworks show the number and positions of amino acids (underlined gray-back letters) that were substituted. The names of peptides that were fully characterized in vitro and validated in vivo are shown in gray-back letters. (c) The structure of McoTI-II (R0, PDB 2IT8) and its active loop (black) are stabilized by three knotted disulfide bonds (black) formed by six cystine residues (I-VI).
STATEMENT OF TRANSLATIONAL RELEVANCE.
We describe the engineering and validation of new peptide-radiotracers for integrin αvβ6, a cell surface biomarker overexpressed in many cancers. Currently, radiotracers that specifically detect pancreatic cancers are clinically unavailable. We validated targeting of integrin αvβ6 using pancreatic and epidermoid cancer xenografts or orthotopic models. The new peptides show high-affinity and specificity for integrin αvβ6, with no cross reactivity to related integrins αvβ3, αvβ5 and α5β1. Uptake of the radiotracers by integrin αvβ6-expressing tumors was rapid and high (~2-5 percent injected dose per gram, 1 hour post injection). Peptide pharmacokinetics has been optimized to minimize off-target background. In contrast to linear peptides, these disulfide-stabilized radiotracers are much more stable in serum, blood and urine and may therefore be less immunogenic. The cystine knot class of peptides has an extremely long shelf life in dry or solvated forms. The sole N-terminus amine enables site-specific coupling of radiometals or radiohalogens. Together, these characteristics indicate potential for clinical translation.
References
- 1.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–87. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 2.Bandyopadhyay A, Raghavan S. Defining the role of integrin alphavbeta6 in cancer. Curr Drug Targets. 2009;10:645–52. doi: 10.2174/138945009788680374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drake CJ, Cheresh DA, Little CD. An antagonist of integrin alpha v beta 3 prevents maturation of blood vessels during embryonic neovascularization. J Cell Sci. 1995;108(Pt 7):2655–61. doi: 10.1242/jcs.108.7.2655. [DOI] [PubMed] [Google Scholar]
- 4.Varner JA, Brooks PC, Cheresh DA. REVIEW: the integrin alpha V beta 3: angiogenesis and apoptosis. Cell Adhes Commun. 1995;3:367–74. doi: 10.3109/15419069509081020. [DOI] [PubMed] [Google Scholar]
- 5.Ahmed N, Pansino F, Clyde R, Murthi P, Quinn MA, Rice GE, et al. Overexpression of alpha(v)beta6 integrin in serous epithelial ovarian cancer regulates extracellular matrix degradation via the plasminogen activation cascade. Carcinogenesis. 2002;23:237–44. doi: 10.1093/carcin/23.2.237. [DOI] [PubMed] [Google Scholar]
- 6.Sipos B, Hahn D, Carceller A, Piulats J, Hedderich J, Kalthoff H, et al. Immunohistochemical screening for beta6-integrin subunit expression in adenocarcinomas using a novel monoclonal antibody reveals strong up-regulation in pancreatic ductal adenocarcinomas in vivo and in vitro. Histopathology. 2004;45:226–36. doi: 10.1111/j.1365-2559.2004.01919.x. [DOI] [PubMed] [Google Scholar]
- 7.Hausner SH, DiCara D, Marik J, Marshall JF, Sutcliffe JL. Use of a peptide derived from foot-and-mouth disease virus for the noninvasive imaging of human cancer: generation and evaluation of 4-[18F]fluorobenzoyl A20FMDV2 for in vivo imaging of integrin alphavbeta6 expression with positron emission tomography. Cancer Res. 2007;67:7833–40. doi: 10.1158/0008-5472.CAN-07-1026. [DOI] [PubMed] [Google Scholar]
- 8.Hausner SH, Abbey CK, Bold RJ, Gagnon MK, Marik J, Marshall JF, et al. Targeted in vivo imaging of integrin alphavbeta6 with an improved radiotracer and its relevance in a pancreatic tumor model. Cancer Res. 2009;69:5843–50. doi: 10.1158/0008-5472.CAN-08-4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hausner SH, Kukis DL, Gagnon MK, Stanecki CE, Ferdani R, Marshall JF, et al. Evaluation of [64Cu]Cu-DOTA and [64Cu]Cu-CB-TE2A chelates for targeted positron emission tomography with an alphavbeta6-specific peptide. Mol Imaging. 2009;8:111–21. [PMC free article] [PubMed] [Google Scholar]
- 10.Hazelbag S, Kenter GG, Gorter A, Dreef EJ, Koopman LA, Violette SM, et al. Overexpression of the alpha v beta 6 integrin in cervical squamous cell carcinoma is a prognostic factor for decreased survival. J Pathol. 2007;212:316–24. doi: 10.1002/path.2168. [DOI] [PubMed] [Google Scholar]
- 11.Bates RC, Bellovin DI, Brown C, Maynard E, Wu B, Kawakatsu H, et al. Transcriptional activation of integrin beta6 during the epithelial-mesenchymal transition defines a novel prognostic indicator of aggressive colon carcinoma. J Clin Invest. 2005;115:339–47. doi: 10.1172/JCI23183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elayadi AN, Samli KN, Prudkin L, Liu YH, Bian A, Xie XJ, et al. A peptide selected by biopanning identifies the integrin alphavbeta6 as a prognostic biomarker for nonsmall cell lung cancer. Cancer Res. 2007;67:5889–95. doi: 10.1158/0008-5472.CAN-07-0245. [DOI] [PubMed] [Google Scholar]
- 13.Bittle JL, Houghten RA, Alexander H, Shinnick TM, Sutcliffe JG, Lerner RA, et al. Protection against foot-and-mouth disease by immunization with a chemically synthesized peptide predicted from the viral nucleotide sequence. Nature. 1982;298:30–3. doi: 10.1038/298030a0. [DOI] [PubMed] [Google Scholar]
- 14.Kraft S, Diefenbach B, Mehta R, Jonczyk A, Luckenbach GA, Goodman SL. Definition of an unexpected ligand recognition motif for alphav beta6 integrin. J Biol Chem. 1999;274:1979–85. doi: 10.1074/jbc.274.4.1979. [DOI] [PubMed] [Google Scholar]
- 15.Hsiao JR, Chang Y, Chen YL, Hsieh SH, Hsu KF, Wand CF, et al. Cyclic alphavbeta6-targeting peptide selected from biopanning with clinical potential for head and neck squamous cell carcinoma. Head Neck. 2010;32:160–72. doi: 10.1002/hed.21166. [DOI] [PubMed] [Google Scholar]
- 16.Nothelfer EM, Zitzmann-Kolbe S, Garcia-Boy R, Kramer S, Herold-Mende C, Altmann A, et al. Identification and characterization of a peptide with affinity to head and neck cancer. J Nucl Med. 2009;50:426–34. doi: 10.2967/jnumed.108.058123. [DOI] [PubMed] [Google Scholar]
- 17.Gagnon MK, Hausner SH, Marik J, Abbey CK, Marshall JF, Sutcliffe JL. High-throughput in vivo screening of targeted molecular imaging agents. Proc Natl Acad Sci U S A. 2009;106:17904–9. doi: 10.1073/pnas.0906925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tangri S, LiCalsi C, Sidney J, Sette A. Rationally engineered proteins or antibodies with absent or reduced immunogenicity. Curr Med Chem. 2002;9:2191–9. doi: 10.2174/0929867023368647. [DOI] [PubMed] [Google Scholar]
- 19.Kimura RH, Cheng Z, Gambhir SS, Cochran JR. Engineered knottin peptides: a new class of agents for imaging integrin expression in living subjects. Cancer Res. 2009;69:2435–42. doi: 10.1158/0008-5472.CAN-08-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miao Z, Ren G, Liu H, Kimura RH, Jiang L, Cochran JR, et al. An engineered knottin peptide labeled with 18F for PET imaging of integrin expression. Bioconjug Chem. 2009;20:2342–7. doi: 10.1021/bc900361g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willmann JK, Kimura RH, Deshpande N, Lutz AM, Cochran JR, Gambhir SS. Targeted contrast-enhanced ultrasound imaging of tumor angiogenesis with contrast microbubbles conjugated to integrin-binding knottin peptides. J Nucl Med. 51:433–40. doi: 10.2967/jnumed.109.068007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimura RH, Levin AM, Cochran FV, Cochran JR. Engineered cystine knot peptides that bind alphavbeta3, alphavbeta5, and alpha5beta1 integrins with low-nanomolar affinity. Proteins. 2009;77:359–69. doi: 10.1002/prot.22441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colgrave ML, Craik DJ. Thermal, chemical, and enzymatic stability of the cyclotide kalata B1: the importance of the cyclic cystine knot. Biochemistry. 2004;43:5965–75. doi: 10.1021/bi049711q. [DOI] [PubMed] [Google Scholar]
- 24.Gran L. Oxytocic principles of Oldenlandia affinis. Lloydia. 1973;36:174–8. [PubMed] [Google Scholar]
- 25.Gran L, Sletten K, Skjeldal L. Cyclic peptides from Oldenlandia affinis DC. Molecular and biological properties. Chem Biodivers. 2008;5:2014–22. doi: 10.1002/cbdv.200890184. [DOI] [PubMed] [Google Scholar]
- 26.Swers JS, Kellogg BA, Wittrup KD. Shuffled antibody libraries created by in vivo homologous recombination and yeast surface display. Nucleic Acids Res. 2004;32:e36. doi: 10.1093/nar/gnh030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lipovsek D, Lippow SM, Hackel BJ, Gregson MW, Cheng P, Kapila A, et al. Evolution of an interloop disulfide bond in high-affinity antibody mimics based on fibronectin type III domain and selected by yeast surface display: molecular convergence with single-domain camelid and shark antibodies. J Mol Biol. 2007;368:1024–41. doi: 10.1016/j.jmb.2007.02.029. [DOI] [PubMed] [Google Scholar]
- 28.Wu Y, Zhang X, Xiong Z, Cheng Z, Fisher DR, Liu S, et al. microPET imaging of glioma integrin {alpha}v{beta}3 expression using (64)Cu-labeled tetrameric RGD peptide. J Nucl Med. 2005;46:1707–18. [PubMed] [Google Scholar]
- 29.Jiang L, Kimura RH, Miao Z, Silverman AP, Ren G, Liu H, et al. Evaluation of a (64)Cu-labeled cystine-knot peptide based on agouti-related protein for PET of tumors expressing alphavbeta3 integrin. J Nucl Med. 51:251–8. doi: 10.2967/jnumed.109.069831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heitz A, Hernandez JF, Gagnon J, Hong TT, Pham TT, Nguyen TM, et al. Solution structure of the squash trypsin inhibitor MCoTI-II. A new family for cyclic knottins. Biochemistry. 2001;40:7973–83. doi: 10.1021/bi0106639. [DOI] [PubMed] [Google Scholar]
- 31.Cemazar M, Joshi A, Daly NL, Mark AE, Craik DJ. The structure of a two-disulfide intermediate assists in elucidating the oxidative folding pathway of a cyclic cystine knot protein. Structure. 2008;16:842–51. doi: 10.1016/j.str.2008.02.023. [DOI] [PubMed] [Google Scholar]
- 32.Wu G, Morris SM., Jr Arginine metabolism: nitric oxide and beyond. Biochem J. 1998;336(Pt 1):1–17. doi: 10.1042/bj3360001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tolmachev V, Velikyan I, Sandstrom M, Orlova A. A HER2-binding Affibody molecule labelled with 68Ga for PET imaging: direct in vivo comparison with the 111In-labelled analogue. Eur J Nucl Med Mol Imaging. 37:1356–67. doi: 10.1007/s00259-009-1367-7. [DOI] [PubMed] [Google Scholar]
- 34.Miao Z, Ren G, Liu H, Jiang L, Cheng Z. Small-animal PET imaging of human epidermal growth factor receptor positive tumor with a 64Cu labeled affibody protein. Bioconjug Chem. 21:947–54. doi: 10.1021/bc900515p. [DOI] [PubMed] [Google Scholar]
- 35.Tolmachev V, Rosik D, Wallberg H, Sjoberg A, Sandstrom M, Hansson M, et al. Imaging of EGFR expression in murine xenografts using site-specifically labelled anti-EGFR 111In-DOTA-Z EGFR:2377 Affibody molecule: aspect of the injected tracer amount. Eur J Nucl Med Mol Imaging. 37:613–22. doi: 10.1007/s00259-009-1283-x. [DOI] [PubMed] [Google Scholar]
- 36.Hansson M, Ringdahl J, Robert A, Power U, Goetsch L, Nguyen TN, et al. An in vitro selected binding protein (affibody) shows conformation-dependent recognition of the respiratory syncytial virus (RSV) G protein. Immunotechnology. 1999;4:237–52. doi: 10.1016/s1380-2933(98)00026-8. [DOI] [PubMed] [Google Scholar]
- 37.Friedman M, Nordberg E, Hoiden-Guthenberg I, Brismar H, Adams GP, Milsson FY, et al. Phage display selection of Affibody molecules with specific binding to the extracellular domain of the epidermal growth factor receptor. Protein Eng Des Sel. 2007;20:189–99. doi: 10.1093/protein/gzm011. [DOI] [PubMed] [Google Scholar]
- 38.Hogbom M, Eklund M, Nygren PA, Nordlund P. Structural basis for recognition by an in vitro evolved affibody. Proc Natl Acad Sci U S A. 2003;100:3191–6. doi: 10.1073/pnas.0436100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


