Abstract
The mitotic spindle checkpoint protein Bub1 has been found to be mutated at low frequency in certain human cancers characterized by aneuploidy. Simian virus 40 large T antigen efficiently immortalizes rodent cells and occasionally transforms them to tumorigenicity. T antigen can also cause genomic instability, inducing chromosomal aberrations and aneuploidy. Here, we report an interaction between Bub1 and T antigen. T antigen coimmunoprecipitates with endogenous Bub1 and Bub3, another component of the spindle checkpoint complex. Genetic analysis demonstrates that the interaction of T antigen with Bub1 is not required for immortalization but is closely correlated with transformation. T antigen induces an override of the spindle checkpoint dependent on Bub1 binding. This interaction with proteins of the spindle checkpoint machinery suggests another role for T antigen and provides insight into its ability to cause chromosomal aberrations, aneuploidy, and transformation.
Simian virus 40 (SV40) large T antigen is a multifunctional phosphoprotein that has been extensively studied as a model system for understanding diverse and complex cellular phenomena such as nuclear transport, transcriptional regulation, eukaryotic DNA replication, and deregulation of cell growth resulting in neoplastic transformation (reviewed in refs. 1–3). T antigen's ability to deregulate cellular proliferation pathways is reflected in its very efficient immortalization of primary rodent cells (4, 5) and transformation of established rodent cell lines to tumorigenicity (6).
The ability of T antigen to deregulate cell proliferation depends on its specific interaction with a variety of host cell proteins, studies of which have led to the elucidation of many signaling pathways and identification of tumor suppressor genes (reviewed in refs. 1 and 3). The tumor suppressor protein p53, mutated or deleted in the majority of human cancers, was originally discovered as a T antigen interacting protein (7, 8). Identification of the interaction of T antigen with members of the retinoblastoma protein (pRB) family has also resulted in substantial advances in understanding their normal cellular functions as cell cycle regulatory proteins (9, 10). More recently, it was found that the extreme N terminus (amino acids 1–70) of T antigen constitutes a bona fide DnaJ domain (11) that is required for efficient viral replication and some transformation functions (1, 11–13).
Although T antigen is clearly an extremely versatile oncoprotein, it is a relatively weak transforming protein, unlike the polyomavirus middle T antigen or activated Ras proteins. Even when T antigen is delivered by a retroviral vector into almost every cell in a culture, only a few cells become transformed, suggesting that additional activities or genetic alterations may be required for full transformation (5). Perhaps germane to the process of oncogenic transformation is the fact that expression of T antigen in human cells has been shown to cause genomic instability by inducing chromosomal aberrations and aneuploidy (14–17). Interestingly, the N-terminal 147 aa of the protein, which do not contain p53 binding sequences, can induce genome destabilization as effectively as the WT protein (15). The ability of T antigen to cause such karyotypic instability in human cells has been found to correlate with its ability to deregulate normal mitotic checkpoints (17), but the exact mechanism by which T antigen causes karyotypic instability or induces endoreduplication is not known.
We have carried out a yeast two-hybrid screen using the N-terminal domain of T antigen as bait and a HeLa cDNA library as prey and identified the human Bub1 protein kinase as an interacting protein. Bub1 is involved in a checkpoint monitoring assembly of the mitotic spindle (18–20). Notably, Bub1 ensures the accurate segregation of sister chromatids at mitosis by monitoring their proper bivalent attachment to mitotic spindles. Bub1 has been found occasionally mutated in certain types of human cancer such as colorectal cancer, which is characterized by chromosomal instability and increased aneuploidy (19, 21, 22). Our genetic analysis demonstrates that the binding of T antigen to Bub1 is dispensable for immortalization but may be required for transformation. Furthermore, T antigen, but not a non-Bub1 binding mutant of T, overrides the spindle checkpoint. Our results thus suggest an important additional mechanism for the transforming activity of T antigen.
Materials and Methods
Detailed protocols for all of the methods used are in Supporting Text, which is published as supporting information on the PNAS web site.
Silencing by Small Interfering RNA (siRNA) Oligos. Oligos to target Bub1 were designed essentially as recommended by Elbashir et al. (23) and obtained from Dharmacon (Lafayette, CO). The duplex oligos were Bub1 sense (5′-GAUGCAUUUGAAGCCCAGUdTdT-3′) and Bub1 antisense (5′-ACUGGGCUUCAAAUGCAUCdTdT-3′), targeting the region nucleotides 1490–1512 relative to the human Bub1 ATG start codon.
Antibodies. Antibodies used were: T antigen mouse monoclonals PAb416, PAb419, PAb423 and PAb100, Bub3 rabbit polyclonal (obtained from P. Sorger, Massachusetts Institute of Technology, Cambridge), and anti-phospho-histone H3 (Upstate Biotechnology, Lake Placid, NY). In coimmunoprecipitation experiments, Bub1 protein was visualized only after T antigen was immunoprecipitated with a mAb recognizing the extreme C terminus (PAb423). It was not detected when T antigen was immunoprecipitated with mAbs PAb416, PAb419, or PAb100, probably because of reduced accessibility of the binding site.
Our N-terminal and C-terminal Bub1 polyclonal antibodies were raised in rabbits by using the hBub1 amino acids 1–303 or 691–1085, respectively, fused in-frame to GST. The fusion proteins were insoluble, so inclusion bodies were purified and used for immunization by standard procedures (24). In addition, Bub1 antibodies from other sources were used to analyze the T antigen/Bub1 interaction (25, 26).
Results
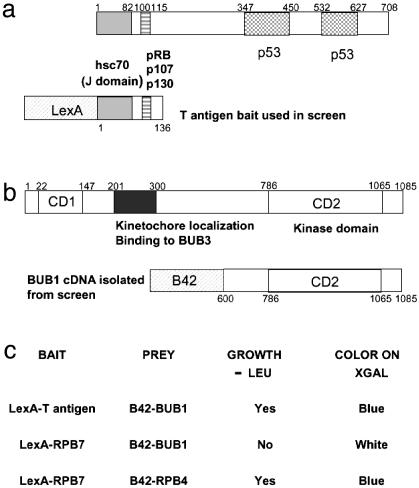
Yeast Two-Hybrid Analysis Identifies Bub1 as a T Antigen Interactor. An amino-terminal fragment of T antigen comprising nucleotides 1–408 (encoding amino acids 1–136) fused in-frame with the LexA DNA-binding domain was used as bait, as shown in Fig. 1a. The LexA-T antigen bait and a LacZ reporter plasmid were transformed into a MATα Saccharomyces cerevisiae LEU2 selection strain. The resulting strain was mated with a MATa strain, which carried a HeLa cDNA library expressed as fusion proteins with the B42 transcriptional activator, and the resulting diploids were plated on selective media lacking leucine to select for interactors. An estimated 3 × 105 diploid cells were screened and 122 LEU+ colonies were collected after incubation at 30°C for 4 days. Of these, 72 colonies showed galactose-dependent growth on media lacking leucine and galactose-dependent blue color on media containing 5-bromo-4-chloro-3-indolyl β-d-galactoside. The 42 colonies with the strongest phenotype were subjected to sequencing and blast sequence comparisons with the GenBank and European Molecular Biology Laboratory databases. This process showed that of the 42 colonies transformed and grown up 39 were β-tubulin and 2 were COPII, a vesicle coat protein. The third candidate was a single clone of the carboxyl terminus of Bub1 (encoding amino acids 600–1085), encompassing the protein's conserved kinase domain (Fig. 1b). Retransformation of clones representing each of these three interactors showed that only Bub1 was a true interactor.
Fig. 1.
The yeast two-hybrid screen. (a) Schematic representation of T antigen indicating the domains for the interaction with Hsc70, pRB, and p53. The amino-terminal 136 aa of T antigen fused to the DNA-binding domain of LexA was used as the bait. (b) Bub1 has two domains highly conserved between human, mouse, and yeast: CD1 (amino acids 22–147) and CD2 (amino acids 786–1065), which encodes the kinase domain of the protein. Also depicted is the domain required for kinetochore localization and binding to Bub3 (amino acids 201–300) (29). The cDNA isolated from the screen encodes the C-terminal 485 aa of Bub1. (c) Bub1 interacts specifically with T antigen and not with a control bait in the yeast two-hybrid system. The RPB7 and RPB4 RNA polymerase II subunits are fused to LexA and B42, respectively, and were used as positive controls for the reporter activation assays and as negative controls for the interaction with T antigen or Bub1. XGAL, 5-bromo-4-chloro-3-indolyl β-d-galactoside.
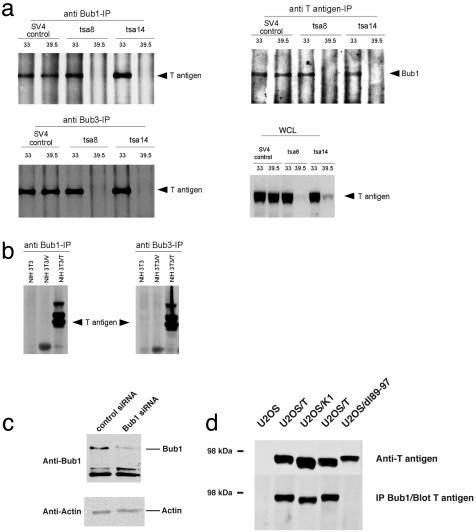
T Antigen and Bub1 Interact as Evidenced by Coimmunoprecipitation. To confirm our yeast two-hybrid result, we demonstrated the interaction of T antigen with Bub1 by using full-length endogenous proteins in mammalian cell lysates prepared from a wide variety of cell lines such as rat tsa, mouse NIH 3T3, and human U2OS cells. tsa8 and tsa14 cells are conditionally immortal cell lines derived by immortalizing rat embryo fibroblasts (REFs) with the thermolabile tsA58 T antigen (27). These cell lines grow continuously at the permissive temperature (33°C) but undergo irreversible growth arrest in the G1 and G2 phases of the cell cycle upon shift to the nonpermissive temperature (39.5°C), where the tsA58 T antigen is rapidly inactivated (27) (Fig. 2a). T antigen and Bub1 could be reciprocally coimmunoprecipitated only at the permissive temperature from tsa cells (Fig. 2a). At the nonpermissive temperature, tsA58 T antigen was absent, thus confirming antibody specificity. As a control for antibody affinity and stability of the protein–protein interaction at 33°C and 39.5°C, SV4 cells were used. These were derived by immortalization of REFs with WT T antigen and can proliferate continuously at both temperatures. T antigen and Bub1 interacted at both 33°C and 39.5°C in SV4 cells. Control immunoprecipitations with an irrelevant antibody demonstrated the coimmunoprecipitation was specific (data not shown).
Fig. 2.
T antigen coimmunoprecipitates (IP) with Bub1 and Bub3 in various cell lines. (a) T antigen/Bub1 and T antigen/Bub3 coimmunoprecipitations in the tsa cell lines. tsa8 and tsa14 are two cell lines derived by using the thermolabile tsA58 T antigen, and SV4 is a control cell line derived by using WT T antigen. WCL denotes whole-cell lysates from tsa8, tsa14, or SV4 cells. (b) T antigen/Bub1 and T antigen/Bub3 coimmunoprecipitations in NIH 3T3 cells. NIH 3T3/V is a pooled culture of clones transfected with pBluescript plasmid, and NIH 3T3/T is a pooled culture of clones transfected with SV40 early region DNA. In the NIH 3T3/T cells, T antigen has two additional higher molecular mass forms probably corresponding to different modification states of the protein. (c) An siRNA specifically targeting Bub1 confirms the specificity of the Bub1 antibody in Western blotting. A control siRNA or a specific siRNA oligo duplex targeting Bub1 was transfected into U2OS cells, and the cell lysates were analyzed by immunoblotting with Bub1 antibody. Equal amounts of total protein from the lysate were loaded in each lane as demonstrated by the actin immunoblot. (d) T antigen/Bub1 coimmunoprecipitation in U2OS cells. Stable U2OS lines expressing an empty vector, T antigen (two independent cell clones), K1, or dl89–97 mutant T antigens were used for a coimmunoprecipitation assay. The cell lines were also assessed for T antigen expression levels by immunoblotting.
T antigen also coimmunoprecipitated with Bub1 from NIH 3T3 cells that ectopically express T antigen (NIH 3T3/T) (Fig. 2b). As controls, parental NIH 3T3 and NIH 3T3 expressing an empty vector (NIH 3T3/V) were used. We also examined the interaction in human U2OS cells stably expressing T antigen (U2OS/T). In these cells we first verified the specificity of the Bub1 antibody by using siRNA technology. The level of endogenous Bub1 as judged by immunoblotting was significantly decreased upon transfection with a specific siRNA duplex targeting Bub1 (Fig. 2c). Subsequently, we showed that T antigen coimmunoprecipitates with Bub1 in U2OS/T cells (Fig. 2d). Taken together, these data show that Bub1 and T interact as assayed by coimmunoprecipitation using either Bub1 or T antigen antibody, and that the interaction can be detected in a wide variety of cell lines.
If T antigen and Bub1 are biological partners, T antigen might coprecipitate with other proteins known to complex with Bub1. Therefore, the interaction of T antigen with Bub3, another component of the spindle assembly checkpoint, was also tested. Bub3 is found in a complex with Bub1 in yeast, mouse, and human cells (18, 26, 28, 29). The binding of Bub3 to Bub1 is believed to be critical for the localization of Bub1 to the kinetochore (29). We found that T antigen coimmunoprecipitated with Bub3, both in tsa (Fig. 2a) and NIH 3T3/T cells (Fig. 2b), suggesting that T antigen and Bub1/Bub3 are components of the same protein complex.
Genetic Analysis of the T Antigen/Bub1 Interaction. To identify the Bub1 binding site on T antigen, we used mutants that have previously been isolated within the amino-terminal 136 aa. Initial experiments were conducted with U2OS lines stably expressing T antigen or various mutants. As shown in Fig. 2d, a deletion mutant of T antigen, dl89–97 (13), failed to bind Bub1 in a coimmunoprecipitation assay, whereas WT T antigen showed significant binding. K1, a mutant (30) which fails to bind pRB family members, retained binding to Bub1 (Fig. 2d).
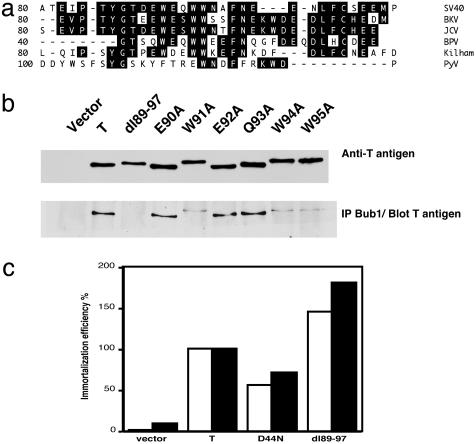
These analyses demonstrated that the binding between Bub1 and T antigen required one or more amino acids within residues 89–97 of T antigen. To more precisely map the interaction site, point mutants were generated. Selection of the residues for mutational analysis was guided by their conservation between different polyomavirus family members. As shown in Fig. 3a, a WEXWW motif found within residues 89–97 was conserved between all large T antigens with the exception of mouse polyomavirus T antigen. We concentrated our efforts on mutating every residue of the conserved motif, as well as neighboring amino acids, to alanine by site-directed mutagenesis (alanine scanning mutagenesis). Mutants E90A, W91A, E92A, Q93A, W94A, and W95A were generated for testing in binding assays. pBabe-puro vectors for expression of the mutants were prepared, packaged into retroviruses, and used to infect Rat-1 cells for production of pools of stable cell lines after puromycin selection. Each stable cell line was lysed and immunoprecipitated with Bub1 antibody followed by immunoblotting with T antigen antibody. Whole-cell lysates were analyzed in parallel to assess the amount of T antigen used for the immunoprecipitation. As shown in Fig. 3b, the coimmunoprecipitation analysis revealed that in Rat-1 cells T antigen also associated with Bub1. The deletion mutant dl89–97 was defective for this interaction, consistent with our previous data in U2OS cells (Fig. 2d). Interestingly, each of the tryptophan substitution mutants (W91A, W94A, and W95A) was largely defective for binding Bub1, whereas mutants E90A, E92A, and Q93A were able to bind Bub1 at levels approaching WT. This result emphasizes the key role played by the conserved tryptophan residues within the WEXWW motif for binding Bub1. Perhaps not surprisingly, the single tryptophan substitution mutants retained some binding, whereas the much broader deletion mutant dl89–97 was almost totally defective. Interestingly, the T antigens encoded by the tryptophan substitution mutants, like the dl89–97 mutant, displayed an aberrant, more retarded mobility on SDS/PAGE that could be caused by changes in modification.
Fig. 3.
Genetic analysis of the T antigen/Bub1 interaction. (a) Sequence alignment of the polyomavirus large T antigens reveals a conserved sequence motif WEXWW. The region between T antigen amino acids 80–110 of SV40, human BK polyomavirus (BK), human JC polyomavirus (JC), Kilham strain of polyomavirus (Kilham), amino acids 40–65 of bovine polyomavirus (BPV), and amino acids 100–125 of murine polyomavirus (PyV) was aligned by using the dnastar program. The sequence motif W(D/E)XWW (amino acids 91–95) is conserved between SV40, BKV, JCV, BPV, and Kilham T antigens, but no obvious homologous sequence is found in the same region of PyV. (b) T antigen mutants W91A, W94A, and W95A are reduced in Bub1 binding. Rat-1 stable lines expressing T antigen point mutants between amino acids 90 and 95 were generated by retroviral infection, and Bub1 binding was assessed in a coimmunoprecipitation assay. As controls, cell lines expressing an empty vector, WT T antigen, or dl89–97 T antigen were included. (Upper) T antigen expression levels. (c) All T antigen mutants analyzed are proficient in immortalization of REFs. An immortalization assay in REFs was conducted by using retroviruses that transduced empty vector, WT T antigen, or each of the mutant T antigens D44N or dl89–97. The immortalization efficiency was calculated relative to that of WT T antigen. Filled and open bars represent duplicate experiments.
Bub1 Binding Is Dispensable for T Antigen-Mediated Immortalization. The immortalization potential of T antigen mutants was determined by retroviral infection of secondary REFs, followed by puromycin selection. After 14 days of puromycin selection, representative dishes were stained and counted to determine the immortalization efficiency of each mutant. At least six colonies were isolated for each mutant T antigen and expanded to determine whether they would establish cell lines. The results for colony formation efficiency from two independent experiments, depicted in Fig. 3c, demonstrate that both the D44N and the dl89–97 mutants readily formed colonies. In addition, colonies isolated for each of these mutants readily established cell lines that could be serially subcultured, demonstrating that they are both able to immortalize REFs. The dl89–97 mutant was slightly more efficient than WT T antigen in colony formation in repeated experiments (Fig. 3c). Furthermore, all of the point mutants were as efficient or better than WT T antigen when tested for REF immortalization (data not shown). Taken together, these findings demonstrate that the dl89–97 mutant and the point mutants are not universally defective, as would be expected if their structure was globally disrupted.
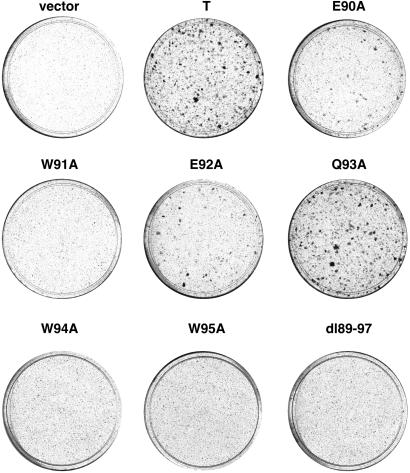
Interaction of T Antigen with Bub1 Is Closely Correlated with Transformation. Because Bub1 mutations had previously been identified in certain human cancers, we wanted to test whether interaction of Bub1 with T antigen might contribute to its transforming activity. One assay that measures T antigen transforming activity is based on its ability to overcome contact inhibition and form dense foci in Rat-1 cells (30). Hence, a cDNA expression vector encoding WT T antigen, deletion mutant dl89–97, or each of the point mutants was transfected into Rat-1 cells. Three weeks later dense foci were visualized by staining with crystal violet. A representative experiment is shown in Fig. 4. Strikingly, the mutant dl89–97 and each of the tryptophan substitution mutants, W91A, W94A, and W95A, were severely defective for focus formation, whereas mutants E90A and E92A showed a partial defect, and mutant Q93A was unaffected relative to WT T antigen. These results correlate with the Bub1 binding data, where dl89–97, W91A, W94A, and W95A have the most severe binding defect and Q93A the least, if any at all. This finding suggests that interaction with Bub1 may be required for efficient T antigen-induced focus formation in Rat-1 cells.
Fig. 4.
T antigen mutants defective in Bub1 binding fail to transform. cDNA expression vectors encoding either WT or mutant T antigen were transfected into low-passage Rat-1 cells. An empty expression vector was included as a negative control. Three weeks later, dense foci were visualized by crystal violet staining. Similar results were obtained in at least three experiments.
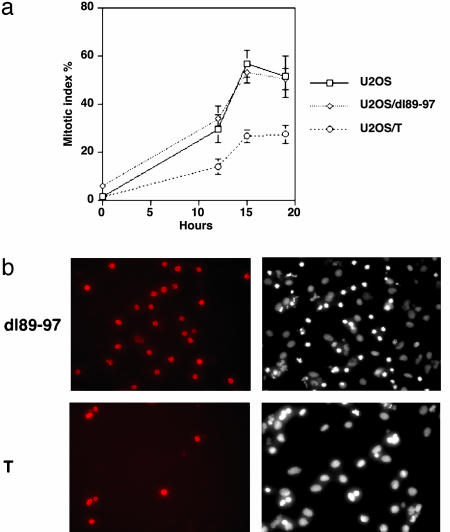
T Antigen Compromises the Spindle Checkpoint. Progression through the spindle assembly checkpoint depends on the presence of a functional mitotic spindle. Disruption of the spindle by microtubule-depolymerizing drugs such as nocodazole causes the cells to arrest in mitosis (20, 31). If the interaction between T antigen, Bub1, and Bub3 is functional, T antigen expression might alter the normal response of the cells to nocodazole. In fact, a hallmark of mitotic spindle checkpoint-defective cells is a reduced ability to arrest in response to microtubule-depolymerizing drugs (20, 31). To test this idea directly, we examined the mitotic index of U2OS, U2OS expressing T, or the dl89–97 mutant after treatment with 50 ng/ml nocodazole for 12, 15, or 19 h. The mitotic index was calculated by counting the proportion of cells with condensed chromatin after visualizing the DNA by using Hoechst 33342 staining. As shown in Fig. 5a, the mitotic index in T antigen-expressing cells was at all time points significantly lower than that of U2OS or U2OS expressing the dl89–97 mutant. To confirm these results, we also determined the mitotic index by staining the cells with antibody to phospho-histone H3, a marker of mitotic cells. After counting nonmitotic cells by visualizing their DNA with 4′,6-diamidino-2-phenylindole staining, the proportion of phopho-histone H3-positive cells was then calculated. Based on this approach, 39.9% of the U2OS cells were mitotic versus 40.8% of the U2OS/dl89–97 and only 14.6% of the U2OS/T when the cells were challenged with nocodazole for 15 h. A representative field of phospho-histone H3-positive cells is shown in Fig. 5b for either U2OS/T or U2OS/dl89–97. Both assays for measuring the mitotic index, that is, staining with Hoechst or for phosphohistone H3, yielded very similar results and indicated that WT T antigen, but not the Bub1 binding mutant dl89–97, substantially decreased the mitotic index by overriding the checkpoint. Bypass of the spindle checkpoint should also be accompanied by increased endoreduplication in the presence of nocodazole (20). As shown in Table 1, we observed that 45.7% of the U2OS/T cells underwent endoreduplication (>4 N DNA content), whereas in U2OS and U2OS/dl89–97 cells only 24.8% and 20.6% underwent endoreduplication after nocodazole treatment. The decreased mitotic index and increased propensity for endoreduplication are both consistent with a T antigen-induced override of the spindle checkpoint dependent on Bub1 binding.
Fig. 5.
T antigen compromises the spindle checkpoint. (a) The mitotic index of U2OS, U2OS/dl89–97, or U2OS/T cells was assessed after 12, 15, or 19 h of nocodazole treatment. A minimum of 300 cells was scored for each time point in each of three independent experiments. Error bars indicate the SD. (b) T antigen decreases the frequency of phospho-histone H3-positive cells, but the dl89–97 mutants do not. (Left) The mitotic cells as visualized by phosphohistone H3 staining. (Right) Nuclear DNA visualized by Hoechst 33342 staining.
Table 1. T antigen enhances endoreduplication under spindle damaging conditions.
| Cell line | Nocodazole, ng/ml | % >4N cells |
|---|---|---|
| U2OS | 0 | 14.1 |
| U2OS | 100 | 24.8 |
| U2OS/dl89-97 | 100 | 20.6 |
| U2OS/T | 100 | 45.7 |
Parental U2OS or U2OS lines stably expressing T antigen or the dl89-97 mutant were treated with nocodazole for 48 h, and the extent of endoreduplication was determined by fluorescence-activated cell sorting analysis. As a control, untreated U2OS cells were analyzed.
Discussion
We have exploited a yeast two-hybrid screen to search for cellular proteins that interact with the amino terminus of SV40 T antigen. Here, we show that T antigen interacts specifically with the mitotic spindle checkpoint protein Bub1. However, our data do not exclude the possibility that the interaction between T antigen and Bub1 could be indirect. Other tumor viral proteins, such as the human papilloma virus E6 and E7, have also been found to disrupt the mitotic spindle checkpoint and promote endoreduplication when cells are challenged with nocodazole, but the mechanistic basis remains to be determined (32, 33). There is only one previously reported example of a viral protein that interacts with a spindle checkpoint protein. The human T cell leukemia virus type I Tax gene product was found to target the Mad1 checkpoint protein for degradation, thus eliciting endoreduplication and a multinucleated cell phenotype (34). We have confirmed the T antigen/Bub1 interaction by reciprocal coimmunoprecipitation analysis in a wide variety of cell types. Genetic analysis indicates that a specific tryptophancontaining motif on T antigen is required for its interaction with Bub1. Interaction with Bub1 is not required for immortalization by T antigen but may be necessary for transformation. T antigen expression results in a compromise of the spindle assembly checkpoint, such that cell arrest mediated by the presence of a microtubule-depolymerizing drug is attenuated. A T antigen mutant that fails to interact with Bub1 cannot compromise the spindle checkpoint. This finding raises the intriguing possibility that perturbations of the spindle checkpoint resulting in aneuploidy and genetic instability may be a critical component of the mechanism by which T antigen transforms cells.
The binding site for Bub1 on T antigen is distinct from the binding sites for previously reported T antigen interactors. Our genetic analysis of Bub1 binding to T antigen indicates that determinants in the region of helix 4, between amino acids 89 and 97, are critical for the interaction. This segment of T antigen is located between the DnaJ domain (residues 1–70) and the LXCXE motif (residues 103–107) required for binding to pRB family members. The dl89–97 mutant was the most defective mutant for binding Bub1, although each of the conserved tryptophans W91, W94, and W95 in the WEXWW motif were important for efficient binding. The deletion in mutant dl89–97 is unlikely to be grossly perturbing T antigen structure, because dl89–97 T antigen has been shown to possess the ability to alter the phosphorylation state of p130 and target it for degradation, indicating that the DnaJ domain and pocket protein binding functions are intact (13). Moreover, mutant dl89–97, as well as each of the tryptophan substitution mutants, was still capable of immortalizing REFs as well as or better than WT T antigen (Fig. 3c and data not shown). Dl89–97 T antigen can also bind to Hsc70, pRB, and p53, activate an E2F transcriptional reporter like WT T antigen, and successfully overcome a p53-dependent cell cycle arrest (ref. 35 and data not shown). Thus, the overall structural integrity of dl89–97 T antigen is likely to be preserved. The WEXWW motif is conserved between SV40, BK, JC, and bovine polyomavirus T antigens. The degree of conservation between the viral T antigens suggests an important function for this motif, perhaps in a similar manner to the conserved HPD(K/R) motif of DnaJ domains. We believe that Bub1 binding constitutes one important function that depends on this conserved sequence motif; however, we cannot rule out the existence of another binding partner that shares the WEXWW binding motif.
An important question arises regarding the functional consequences of T antigen binding to Bub1. We find that T antigen causes an override of the spindle checkpoint in a Bub1 binding-dependent manner. Thus, the mitotic index of T antigen-expressing cells is consistently lower than that of dl89–97-expressing or parental cells in response to nocodazole treatment. This bypass of the checkpoint is accompanied by enhanced rates of endoreduplication. Although the checkpoint is clearly compromised, it is by no means completely abrogated, hence explaining why our phenotype is not as severe as a knockout of a spindle checkpoint gene such as mad2 (36). We believe that T antigen only causes a limited perturbation of the checkpoint, that nevertheless has the potential to result in chromosome segregation defects and concomitant aneuploidy.
The interaction of Bub1 and T antigen may explain several previous reports that demonstrated that T antigen is able to induce aneuploidy and genetic instability, giving rise to both structural and numerical chromosome aberrations (14–17, 37). It had been proposed that this instability was most likely caused by the ability of T antigen to interact with and inactivate the p53 protein. However, it was subsequently shown that an amino-terminal 147-aa fragment of T antigen that is unable to interact with p53 can still efficiently induce instability, and that interaction with pRB was also not strictly required for the induction of genomic instability (15). Moreover, it was shown that loss of p53 in somatic cells did not result in aneuploidy, although a slight tendency toward tetraploidization was observed (38). Thus, the mechanism by which T antigen induces genetic instability is not clear. Nevertheless, a potential correlation between the ability of T antigen to deregulate mitotic checkpoints including the spindle assembly checkpoint and to induce chromosomal abnormalities has been proposed (17). Our data presented here, demonstrating that T antigen interacts with Bub1 within the amino terminus, suggests one potential mechanism for how T antigen, especially the 147-aa N-terminal fragment, may trigger genomic instability. The interaction of T antigen with Bub1 is especially significant in this context because Bub1 has been found to be occasionally mutated in human cancers (19, 21, 22). One such Bub1 mutant is capable of compromising the normal spindle checkpoint in a dominant negative manner in the presence of WT protein, presumably causing errors in chromosome segregation and aneuploidy (19). Other reports also support the idea that mutations in mitotic checkpoint proteins can cause genomic instability and in rare cases give rise to cancer (36, 39). Because tetraploidy is frequently an intermediate step for progression to aneuploidy, it is particularly interesting that T antigen has already been shown to induce tetraploid DNA content in both permissive and nonpermissive cells (37, 40). We found that dl89–97 T antigen, which does not interact with Bub1, was defective for induction of endoreduplication in the presence of spindle damage. Future experiments should be aimed at determining whether the interaction of T antigen with Bub1 gives rise to structural aberrations such as chromatid exchange, dicentric chromosomes, or alterations in the number of chromosomes.
The most straightforward way to assemble all of the data presented here is that T antigen uses Bub1 in the transformation process by destabilizing the host genome. Interestingly, inhibition of Bub1 in human fibroblasts was recently demonstrated to produce at low frequency aneuploid clones capable of anchorage-independent growth, thus supporting a potential role for Bub1 in T antigen-mediated oncogenic transformation (41). The fact that most cells expressing T antigen fail to grow as foci suggests that secondary genetic or epigenetic changes may play a role in T antigen-mediated transformation. Also consistent with this idea is the fact that some cells transformed by a temperature-sensitive T antigen allele remain transformed at the nonpermissive temperature (42, 43). Clearly, these concepts are not mutually exclusive, nor do the rule out other mechanisms.
In summary, the yeast two-hybrid analysis presented here has enabled us to identify an interaction of T antigen with Bub1, a member of the spindle checkpoint protein complex. We have further shown that this interaction causes cells to arrest less efficiently in response to spindle damage and is not required for the ability of T antigen to immortalize cells, but it may be required for transformation. These results provide further insight into the potential mechanism by which T antigen transforms cells.
Supplementary Material
Acknowledgments
We thank P. Sorger, H. Yu, J. van Deursen, M. Kirschner, R. Agami, and A. Chestukhin for providing valuable reagents; R. Subramanian, I. Serebriiskii, D. Davies, K. Adams, J. Hyams, J. Pines, K. Hardwick, M. O'Hare, and D. Pellman for advice and helpful discussions; and B. Schaffhausen for critical reading of the manuscript. This work was supported by National Institutes of Health Grants PO1-CA50661 and CA30002 (to T.M.R.). M.C. was supported by a Medical Research Council Ph.D. studentship. R.L.L. is supported by a Biotechnology and Biological Sciences Research Council Ph.D. studentship.
Abbreviations: SV40, simian virus 40; REF, rat embryo fibroblast; pRB, retinoblastoma protein; siRNA, small interfering RNA.
References
- 1.Sullivan, C. S. & Pipas, J. M. (2002) Microbiol. Mol. Biol. Rev. 66, 179–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fanning, E. & Knippers, R. (1992) Annu. Rev. Biochem. 61, 55–85. [DOI] [PubMed] [Google Scholar]
- 3.Manfredi, J. J. & Prives, C. (1994) Biochim. Biophys. Acta 1198, 65–83. [DOI] [PubMed] [Google Scholar]
- 4.Petit, C. A., Gardes, M. & Feunteun, J. (1983) Virology 127, 74–82. [DOI] [PubMed] [Google Scholar]
- 5.Jat, P. S. & Sharp, P. A. (1986) J. Virol. 59, 746–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, M., McCormack, M., Zinn, K. G., Farrell, M. P., Bikel, I. & Livingston, D. M. (1986) J. Virol. 60, 290–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linzer, D. I. & Levine, A. J. (1979) Cell 17, 43–52. [DOI] [PubMed] [Google Scholar]
- 8.Lane, D. P. & Crawford, L. V. (1979) Nature 278, 261–263. [DOI] [PubMed] [Google Scholar]
- 9.DeCaprio, J. A., Ludlow, J. W., Figge, J., Shew, J. Y., Huang, C. M., Lee, W. H., Marsilio, E., Paucha, E. & Livingston, D. M. (1988) Cell 54, 275–283. [DOI] [PubMed] [Google Scholar]
- 10.Zalvide, J. & DeCaprio, J. A. (1995) Mol. Cell. Biol. 15, 5800–5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell, K. S., Mullane, K. P., Aksoy, I. A., Stubdal, H., Zalvide, J., Pipas, J. M., Silver, P. A., Roberts, T. M., Schaffhausen, B. S. & DeCaprio, J. A. (1997) Genes Dev. 11, 1098–1110. [DOI] [PubMed] [Google Scholar]
- 12.Srinivasan, A., McClellan, A. J., Vartikar, J., Marks, I., Cantalupo, P., Li, Y., Whyte, P., Rundell, K., Brodsky, J. L. & Pipas, J. M. (1997) Mol. Cell. Biol. 17, 4761–4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stubdal, H., Zalvide, J., Campbell, K. S., Schweitzer, C., Roberts, T. M. & DeCaprio, J. A. (1997) Mol. Cell. Biol. 17, 4979–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ray, F. A., Peabody, D. S., Cooper, J. L., Cram, L. S. & Kraemer, P. M. (1990) J. Cell Biochem. 42, 13–31. [DOI] [PubMed] [Google Scholar]
- 15.Woods, C., LeFeuvre, C., Stewart, N. & Bacchetti, S. (1994) Oncogene 9, 2943–2950. [PubMed] [Google Scholar]
- 16.Stewart, N. & Bacchetti, S. (1991) Virology 180, 49–57. [DOI] [PubMed] [Google Scholar]
- 17.Chang, T. H., Ray, F. A., Thompson, D. A. & Schlegel, R. (1997) Oncogene 14, 2383–2393. [DOI] [PubMed] [Google Scholar]
- 18.Roberts, B. T., Farr, K. A. & Hoyt, M. A. (1994) Mol. Cell. Biol. 14, 8282–8291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cahill, D. P., Lengauer, C., Yu, J., Riggins, G. J., Willson, J. K., Markowitz, S. D., Kinzler, K. W. & Vogelstein, B. (1998) Nature 392, 300–303. [DOI] [PubMed] [Google Scholar]
- 20.Taylor, S. S. & McKeon, F. (1997) Cell 89, 727–735. [DOI] [PubMed] [Google Scholar]
- 21.Ru, H. Y., Chen, R. L., Lu, W. C. & Chen, J. H. (2002) Oncogene 21, 4673–4679. [DOI] [PubMed] [Google Scholar]
- 22.Shichiri, M., Yoshinaga, K., Hisatomi, H., Sugihara, K. & Hirata, Y. (2002) Cancer Res. 62, 13–17. [PubMed] [Google Scholar]
- 23.Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 24.Harlow, E. & Lane, D. (1990) Antibodies: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 25.Tang, Z., Bharadwaj, R., Li, B. & Yu, H. (2001) Dev. Cell 1, 227–237. [DOI] [PubMed] [Google Scholar]
- 26.Martinez-Exposito, M. J., Kaplan, K. B., Copeland, J. & Sorger, P. K. (1999) Proc. Natl. Acad. Sci. USA 96, 8493–8498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jat, P. S. & Sharp, P. A. (1989) Mol. Cell. Biol. 9, 1672–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoyt, M. A., Totis, L. & Roberts, B. T. (1991) Cell 66, 507–517. [DOI] [PubMed] [Google Scholar]
- 29.Taylor, S. S., Ha, E. & McKeon, F. (1998) J. Cell Biol. 142, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalderon, D. & Smith, A. E. (1984) Virology 139, 109–137. [DOI] [PubMed] [Google Scholar]
- 31.Li, Y. & Benezra, R. (1996) Science 274, 246–248. [DOI] [PubMed] [Google Scholar]
- 32.Thompson, D. A., Belinsky, G., Chang, T. H., Jones, D. L., Schlegel, R. & Munger, K. (1997) Oncogene 15, 3025–3035. [DOI] [PubMed] [Google Scholar]
- 33.Thomas, J. T. & Laimins, L. A. (1998) J. Virol. 72, 1131–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin, D. Y., Spencer, F. & Jeang, K. T. (1998) Cell 93, 81–91. [DOI] [PubMed] [Google Scholar]
- 35.Gjoerup, O., Chao, H., DeCaprio, J. A. & Roberts, T. M. (2000) J. Virol. 74, 864–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michel, L. S., Liberal, V., Chatterjee, A., Kirchwegger, R., Pasche, B., Gerald, W., Dobles, M., Sorger, P. K., Murty, V. V. & Benezra, R. (2001) Nature 409, 355–359. [DOI] [PubMed] [Google Scholar]
- 37.Levine, D. S., Sanchez, C. A., Rabinovitch, P. S. & Reid, B. J. (1991) Proc. Natl. Acad. Sci. USA 88, 6427–6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bunz, F., Fauth, C., Speicher, M. R., Dutriaux, A., Sedivy, J. M., Kinzler, K. W., Vogelstein, B. & Lengauer, C. (2002) Cancer Res. 62, 1129–1133. [PubMed] [Google Scholar]
- 39.Lee, H., Trainer, A. H., Friedman, L. S., Thistlethwaite, F. C., Evans, M. J., Ponder, B. A. & Venkitaraman, A. R. (1999) Mol. Cell 4, 1–10. [DOI] [PubMed] [Google Scholar]
- 40.Friedrich, T. D., Laffin, J. & Lehman, J. M. (1992) J. Virol. 66, 4576–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Musio, A., Montagna, C., Zambroni, D., Indino, E., Barbieri, O., Citti, L., Villa, A., Ried, T. & Vezzoni, P. (2003) Cancer Res. 63, 2855–2863. [PubMed] [Google Scholar]
- 42.Seif, R. & Martin, R. G. (1979) J. Virol. 31, 350–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rassoulzadegan, M., Perbal, B. & Cuzin, F. (1978) J. Virol. 28, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.