Abstract
Bacterial lipopolysaccharide (LPS)-induced exocytosis of granular hemocytes is a key component of the horseshoe crab's innate immunity to infectious microorganisms; stimulation by LPS induces the secretion of various defense molecules from the granular hemocytes. Using a previously uncharacterized assay for exocytosis, we clearly show that hemocytes respond only to LPS and not to other pathogen-associated molecular patterns, such as β-1,3-glucans and peptidoglycans. Furthermore, we show that a granular protein called factor C, an LPS-recognizing serine protease zymogen that initiates the hemolymph coagulation cascade, also exists on the hemocyte surface as a biosensor for LPS. Our data demonstrate that the proteolytic activity of factor C is both necessary and sufficient to trigger exocytosis through a heterotrimeric GTP-binding protein-mediating signaling pathway. Exocytosis of hemocytes was not induced by thrombin, but it was induced by hexapeptides corresponding to the tethered ligands of protease-activated G protein-coupled receptors (PARs). This finding suggested the presence of a PAR-like receptor on the hemocyte surface. We conclude that the serine protease zymogen on the hemocyte surface functions as a pattern-recognition protein for LPS.
The innate immune system is a sensitive non-self-recognizing cascade triggered by microbial cell wall constituents called pathogen-associated molecular patterns (PAMP), such as lipopolysaccharides (LPS) of Gram-negative bacteria, β-1,3-glucans of fungi, and peptidoglycans of Gram-positive bacteria (1–4). These PAMP are recognized via a set of pattern-recognition receptors and proteins that are germ-line-encoded receptors of the innate immune system. Recent studies have revealed that insects and mammals conserve a signaling pathway of the innate immune system through cell-surface receptors called Tolls and through Toll-like receptors (TLRs) (5, 6). In mammals, TLRs on specialized antigen-presenting cells function as signal transducers by way of NF-κB, leading to the production of proinflammatory cytokines and the expression of costimulatory molecules on the cell surface. These cytokines and costimulatory molecules are necessary to activate naive T cells; it may be that TLRs are assembled at the cell membrane as signaling receptor complexes (4, 7, 8).
Although Drosophila Toll controls the host defense to fungal and Gram-positive bacterial infection, it does not function as a pattern-recognition receptor (9). Initially, Drosophila Toll was identified as a transmembrane protein that controls dorsoventral patterning in the Drosophila embryo (10). In the embryo, a proteolytic cascade that includes three proteases (Gastrulation Defective, Snake, and Easter) cleaves a cytokine-like protein, Spaetzle, as a ligand for Drosophila Toll. These three proteases, however, are dispensable in the induction of the Drosophila Toll-mediated immune response (5). During infection, the cleaved form of Spaetzle is produced through another proteolytic cascade, one that includes Persephone, a newly identified serine protease (11, 12). However, no pattern-recognition proteins initiating the proteolytic cascade have been identified. The Drosophila immune system also detects bacteria through peptidoglycan-recognition proteins, and Gram-negative diaminopimelic-acid-type peptidoglycan is the most potent inducer of the Imd pathway (13). Overexpression of peptidoglycan-recognition protein-LE, a receptor for the diaminopimelic-acid-type peptidoglycan, leads to the activation of prophenoloxidase cascade in Drosophila larvae (14).
The presence of circulating hemocytes is essential to invertebrates' innate immunity, such as self-/non-self recognition, phagocytosis, encapsulation, and melanization (15). In horseshoe crabs, granular hemocytes comprise 99% of all hemocytes and are involved in the storage and release of defense molecules, including serine protease zymogens, a clottable protein coagulogen, protease inhibitors, antimicrobial peptides, and lectins (16–18). In response to stimulation by LPS, the defense molecules stored in granules are immediately secreted by exocytosis (16, 19). This response is important for the host defense's ability to engulf and kill invading microbes. The granular components include two PAMP-sensitive zymogens, factor C (20) and factor G (21, 22). These serine protease zymogens are autocatalytically activated by LPS and β-1,3-glucans, respectively. The activated factor C activates factor B, which in turn converts the proclotting enzyme into the clotting enzyme, which converts coagulogen to an insoluble coagulin gel. On the other hand, the activated factor G activates the proclotting enzyme directly.
In horseshoe crabs, however, no specific LPS-recognizing protein has been identified as a pattern-recognition protein involved in signal transduction to trigger exocytosis. Microscopic observation and microfluorometric analysis of intracellular Mg2+ and Ca2+ on LPS-induced exocytosis suggest that the exocytosis is mediated by a heterotrimeric GTP-binding protein (G protein) that stimulates the inositol-1,4,5-triphosphate-signaling pathway leading to the increase of intracellular Mg2+ and Ca2+ (23). The exocytosis is not induced by a phorbol ester that mimics diacylglycerol to activate protein kinase C. Likewise, exocytosis is not affected by ethanol or chelerythrine, both of which inhibit phospholipase D, or by herbimycin, a tyrosine kinase inhibitor (23). In the present study, we have established a quantitative assay for LPS-induced exocytosis. Furthermore, we report that the LPS-sensitive protease zymogen factor C also exists on the hemocyte surface, and that the proteolytic activity triggers a G protein-mediating exocytosis for innate immune responses. We have made an identification and characterization of a pattern-recognition protein that leads to LPS-induced exocytosis.
Materials and Methods
Materials. Factor C and human α-thrombin were purified as described (20, 24). LPS (Salmonella minnesota R595) and lipid A (Escherichia coli K12) were from List Biological Laboratories (Campbell, CA). Laminarin, thapsigargin, pertussis toxin, A23187, l-α-phosphatidylcholine, 1,2-diacyl-sn-glycero-3-phospho-l-serine, l-α-phosphatidylinositol, 1,2-diacyl-snglycero-3-phosphoethanolamine, sphingomyelin, and cholesterol were from Sigma. Peptidoglycan (Staphylococcus aureus) was from Fluka. Curdlan was from Wako Pure Chemical (Osaka). U-73122 and U-73343 were from Calbiochem. Mastoparan, d-Phe-Pro-Arg-chloromethylketone (PPACK), and Ala-Ala-Phe-chloromethylketone were from Bachem. Bovine pancreatic α-chymotr ypsin treated with N-tosyl-l-lysyl chloromethylketone and trypsin treated with N-tosyl-l-phenylalanyl chloromethylketone were from Worthington. Hexapeptides SFLLRN, SLIGRL, and GYPGKF were synthesized at Genenet (Oita, Japan). The COOH-terminal amino acids of these peptides were amidated.
Assay of Exocytosis. Hemolymph (1 ml) was collected into 50 ml of pyrogen-free 10 mM Hepes-NaOH, pH 7.0, containing 0.5 M NaCl. The diluted hemolymph (200 μl each) was applied to pyrogen-free 24-well plates filled with 800 μl of the same buffer, and then incubated at 23°C for 10 min for the attachment of hemocytes. PAMP, proteases, and activators were added in assay buffer (10 mM Hepes-NaOH, pH 7.0, containing 0.5 M NaCl, 50 mM MgCl2, and 10 mM CaCl2). For inhibition studies, hemocytes were pretreated with inhibitors at 23°C for 20 min for U-73122 or 1 h for pertussis toxin. Hemocytes were then stimulated with activators or LPS. After incubation at 23°C for 1 h, each exocytosed fluid was collected by centrifugation at 2,000 × g for 5 min and quantitated by ELISA by using anti-coagulogen or anti-tachylectin-2 (TL-2) antibody.
ELISA. The amount of coagulogen in the exocytosed fluid was determined by using a monoclonal antibody, mAb 14B1. Microtiter plates were coated with 2-fold serial dilutions of the exocytosed supernatant by incubation at 4°C for 2 h. After being washed with 20 mM Tris·HCl, pH 7.5, containing 0.15 M NaCl, the plates were blocked with 2.5% casein in the washing buffer. After mAb 14B1 was added, the plates were incubated at 37°C for 1 h and then washed. Horseradish peroxidase-conjugated goat anti-mouse IgG (Bio-Rad) was added, and the plates were again incubated at 37°C for 1 h. The enzyme activity of horse-radish peroxidase was detected with o-phenylenediamine at 490 nm by using a microplate reader, model 3550 (Bio-Rad). On the other hand, the amount of TL-2 was determined by sandwich ELISA as described (25).
Flow Cytometric Analysis. Hemocytes (150 μl of hemolymph) were fixed, immediately after bleeding, in 10 ml of 1% paraformaldehyde EM (Taab Laboratories Equipment, Berkshire, U.K.) at room temperature for 10 min. After fixation, hemocytes were collected by centrifugation at 100 × g for 5 min and washed twice with 10 mM sodium phosphate, pH 7.0, containing 0.5 M NaCl. The fixed hemocytes (3.0 × 105 cells) were incubated with antibodies in the same buffer containing BSA at 1 mg/ml and were then incubated on ice for 30 min. After being washed with the same buffer containing BSA, the hemocytes were incubated with anti-mouse FITC-conjugated antibody (1:500 dilution, Dako) on ice for 30 min. After another washing, the labeled hemocytes were analyzed by using a FACScan flow cytometer (BD Biosciences). For each sample, 1.0 × 104 cells were analyzed by using cellquest software (BD Biosciences).
Biotinylation of the Surface Proteins on Hemocytes and Immunoprecipitation. The surface proteins of hemocytes were biotinylated as described (26). After biotinylation, the hemocytes were lysed with 50 mM Tris·HCl, pH 7.5, containing 1 mM EDTA, 1% Nonidet P-40, and 0.15 M NaCl at 4°C for 30 min with rocking. The sample was mixed with mAb 2C12 (10 μg/ml) and incubated at 4°C for 1 h. Protein A-Sepharose (Amersham Pharmacia) was then added, and the mixture was further incubated at 4°C for 1 h. The resulting immunocomplex bound to protein A-Sepharose was collected by centrifugation and washed with the same buffer. The pellet was subjected to SDS/PAGE (27) and transferred to nitrocellulose membranes overnight at 20 V by using an electroblot apparatus (Bio-Rad). After blocking, the antigens were visualized by streptavidin-biotinylated horseradish-peroxidase complex (Amersham Pharmacia).
Preparation of Activated Factor C and PPACK-Factor C. The zymogen factor C was activated with α-chymotrypsin as described (28). After activation, 1 μM Ala-Ala-Phe-chloromethylketone was added, and the mixture was incubated at 37°C for 10 min to inactivate the α-chymotrypsin. To prepare the PPACK-factor C, the activated factor C was treated with 10 μM PPACK at 37°C for 10 min.
Interaction of Factor C with Immobilized Lipids. Sonicated lipids (0.5 mg/ml in 10 mM Hepes-NaOH, pH 7.0, containing 0.15 M NaCl) were immobilized on a sensor chip HPA of the BIAcore 1000 system (BIAcore, Uppsala), according to the manufacturer's specifications. Factor C was injected at a flow rate of 30 μl/min. For the running buffer, 10 mM Hepes-NaOH, pH 7.0, containing 0.15 M NaCl, was used. The change in the mass concentration on the sensor chip was monitored as a resonance signal by using the program supplied by the manufacturer. Sensorgrams of the interactions obtained by using the various concentrations of factor C (10–60 nM) were analyzed by the biaevaluation program, version 3.0.
Results
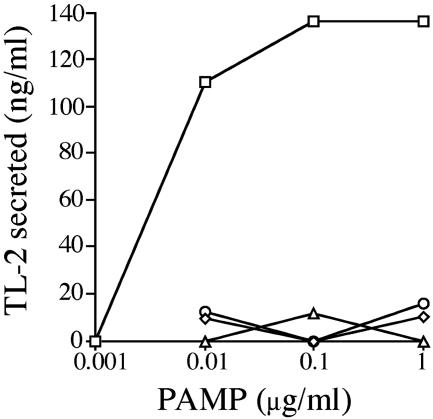
Quantitative Assay of LPS-Induced Exocytosis and the Effects of Activators and Inhibitors of a G Protein-Mediating Signaling Pathway on Exocytosis. Exocytosis of hemocytes was quantitatively assayed by ELISA using an antibody against coagulogen or TL-2. Under the assay conditions used, LPS induced the hemocytes to secrete granular components in a concentration-dependent manner. Maximal secretion was obtained at 0.1 μg/ml with a halfmaximal concentration of 0.005 μg/ml (Fig. 1). In contrast, other PAMP, including β-1,3-glucans (curdlan and laminarin) and peptidoglycans, were unable at 1.0 μg/ml to induce exocytosis of hemocytes, indicating that this assay specifically detects the LPS-induced exocytosis.
Fig. 1.
Effects of PAMP on exocytosis. Hemocytes collected into a sterilized plate were treated with various concentrations of PAMP at 23°C for 1 h. The amount of TL-2 secreted in the supernatant was determined by ELISA. □, LPS; ○, curdlan; ▵, laminarin; ⋄, peptidoglycan.
Hemocytes were efficiently exocytosed in response to LPS stimulation under assay conditions containing 50 mM Mg2+ and 10 mM Ca2+, equivalent to the concentrations of cations in intact hemolymph (29, 30). Cannon et al. (31) reported that the absence of Mg2+ outside of hemocytes causes reversible unresponsiveness of the LPS-induced exocytosis in the American horseshoe crab Limulus polyphemus. As we expected, the absence of divalent cations in the assay buffer resulted in the inability to induce exocytosis even at 10 μg/ml LPS (data not shown).
In L. polyphemus, the LPS-induced exocytosis of hemocytes can be mediated by a G protein that activates phospholipase C, finally leading to intracellular Ca2+ flux (23). The effects of signal transduction activators and inhibitors on exocytosis were examined by the ELISA method (Table 1). A positive control stimulated by LPS at 1 μg/ml was assumed to be a 100% secretion. Pertussis toxin, an inhibitor of G proteins that belong to the Gαi/o family, strongly inhibited exocytosis at 1 μg/ml. Although U-73122, a phospholipase C inhibitor, led to an almost complete inhibition at 10 μM, U-73343, a structural analogue of U-73122, had no effect on exocytosis. The inhibition of LPS-induced exocytosis by U-73122 was reversed by adding A23187, a Ca2+ ionophore. Also inducing exocytosis in the absence of LPS was thapsigargin, an inhibitor of endomembranous Ca2+-ATPase that increases the cytoplasmic concentration of Ca2+.In addition, a synthetic mastoparan effectively induced exocytosis of hemocytes. Mastoparan, an amphiphilic tetradecapeptide from wasp venom, mimics receptors by activating G proteins directly (32). These data indicate that the LPS-induced exocytosis of hemocytes occurs through a G protein-mediating signaling pathway.
Table 1. Effects of activators and inhibitors on exocytosis of the horseshoe crab hemocytes.
| Concentrations | Exocytosed coagulogen or TL-2, % | |
|---|---|---|
| Activators | ||
| LPS | 1 μg/ml | 100 |
| Thapsigargin | 0.01 μM | 0 |
| 0.1 μM | 78 | |
| 1 μM | 83 | |
| Ca2+ ionophore | 10 μM | 76 |
| Mastoparan | 1 μM | 48 |
| 2 μM | 77 | |
| 5 μM | 78 | |
| Inhibitors + LPS (1 μg/ml) | ||
| Pertussis toxin | 0.01 μg/ml | 69 |
| 0.1 μg/ml | 81 | |
| 1 μg/ml | 18 | |
| U-73122 | 0.1 μM | 97 |
| 1 μM | 56 | |
| 10 μM | 8 |
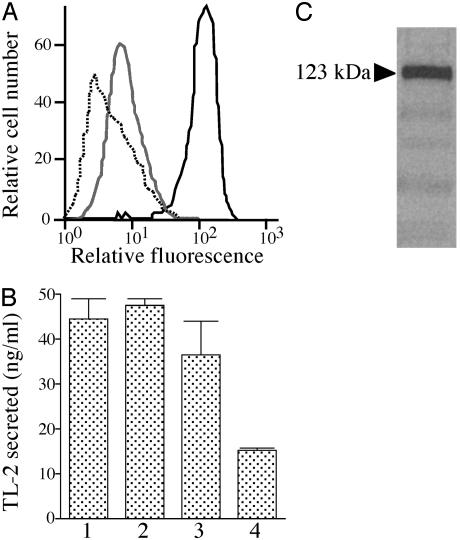
Flow Cytometry of Hemocytes by Using Antibodies Against the Granular Components. Two PAMP-binding proteins, factor C for LPS and factor G for β-1,3-glucans, have been identified from the large granules of the horseshoe crab hemocytes (16). Flow cytometric analysis was performed to determine whether or not these PAMP-binding proteins exist on the hemocyte surface. A polyclonal antibody against factor C strongly reacted with the hemocytes (Fig. 2A). In contrast, a polyclonal antibody against factor G did not react with hemocytes. Preincubation of the anti-factor C antibody with an excess amount of purified factor C completely inhibited binding to hemocytes. In addition, six kinds of mAbs against factor C all reacted with hemocytes (data not shown): five mAbs (2C12, 1B6, 2F6, 3D2, and 4H1) are directed against the H-chain of the two-chain form of factor C, and one mAb (2A7) is directed against the L-chain containing the serine protease domain (33). In contrast, a polyclonal antibody against TL-2, a major component of the large granules of hemocytes, did not specifically react with hemocytes, indicating the specific expression of the factor C antigen on hemocytes (data not shown).
Fig. 2.
Factor C as a surface protein on hemocytes. (A) The fixed hemocytes were stained with 1 μg/ml of a polyclonal antibody against factor C followed by FITC-labeled secondary antibody (solid line). The gray line shows cytometric analysis with the first polyclonal antibody pretreated with an excess amount of the purified factor C, and the dotted line shows the negative control without the first antibody. (B) Inhibition of the LPS-induced exocytosis by mAb 2C12. Hemocytes were preincubated with mAb 2C12 at 0.1 μg/ml (bar 2), 0.2 μg/ml (bar 3), and 0.5 μg/ml (bar 4) or without mAb 2C12 (bar 1) at 23°C for 1 h. Then, exocytosis was induced by LPS at 1.0 μg/ml. (C) Biotinylated surface antigens were immunoprecipitated by mAb 2C12 and subjected to SDS/PAGE under reducing conditions. The factor C-like cell surface antigen was visualized by streptavidin-biotinylated horseradish peroxidase. The arrowhead shows the apparent molecular mass.
The binding of mAb 2C12 to hemocytes dose-dependently inhibited exocytosis elicited by LPS (Fig. 2B). However, 6C1-1F, a mAb against proxin, which is a recently identified surface protein on hemocytes (26), had no effect on the LPS-induced exocytosis (data not shown). To confirm the presence of the factor C antigen on hemocytes, the surface proteins of hemocytes were biotinylated, and the labeled proteins were immunoprecipitated by mAb 2C12. The precipitated antigen was subjected to SDS/PAGE, and the streptavidin-biotinylated horseradish peroxidase complex detected a major (123-kDa) protein band corresponding to the single-chain form of factor C (Fig. 2C). Washing hemocytes with buffer containing 10 mM EDTA before fixing them had no effect on the detection of the factor C antigen by flow cytometric analysis, suggesting that the localization of the factor C antigen on the hemocyte surface is not dependent on divalent cations (data not shown).
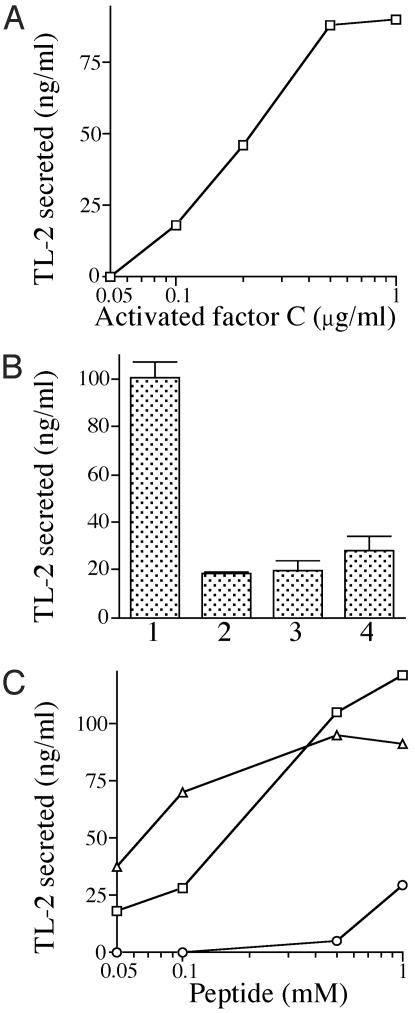
Proteolytic Activity of the Activated Factor C on Hemocytes Is Required for LPS-Induced Exocytosis. Factor C is activated autocatalytically by forming a complex with LPS through the lipid A portion (34). On the other hand, chymotrypsin converts factor C to the active protease through the hydrolysis of a specific Phe-Ile bond (35). Therefore, purified factor C was activated by chymotrypsin and then added to hemocytes. This chymotrypsin-activated factor C was also competent for exocytosis in the absence of LPS (Fig. 3A), and exocytosis was inhibited by the treatment of hemocytes with U73122 (Fig. 3B, bar 2). Furthermore, alkylating the His residues of the catalytic site of the activated factor C with d-Phe-Pro-Arg-chloromethylketone (PPACK-factor C) caused the complete loss of exocytotic ability (Fig. 3B, bar 3). The substrate specificity of the activated factor C for synthetic peptide substrates is quite similar to that of thrombin, a mammalian clotting factor (20, 28). However, human α-thrombin did not efficiently induce exocytosis (Fig. 3B, bar 4), suggesting that a specific protein substrate for the activated factor C exists on the hemocyte surface.
Fig. 3.
Proteolytic activity of the activated factor C is involved in exocytosis. (A) Hemocytes were treated with various concentrations of the activated factor C in the absence of LPS at 23°C for 1 h. After the treatments, the amounts of TL-2 exocytosed were determined by ELISA. (B) Hemocytes were mixed with the following proteases at 1 μg/ml to examine whether or not exocytosis would occur. Bar 1, the activated factor C; bar 2, the activated factor C in the presence of U-73122 at 10 μM; bar 3, PPACK-factor C; bar 4, human α-thrombin. (C) Effects of agonist peptides for PAR1, PAR2, and PAR4 on the exocytosis of hemocytes were examined. Hemocytes were treated with each of various concentrations of the agonist peptides SFLLRN (PAR-1), SLIGRL (PAR-2), and GYPGKF (PAR-4) in the absence of LPS at 23°C for 1 h. After the treatments, the amounts of TL-2 exocytosed were determined by ELISA. □, SFLLRN; ▵, SLIGRL; ○, GYPGKF.
Extracellular proteases trigger cellular responses in part via protease-activated G protein-coupled receptors (PARs) (36). Four kinds of PARs have been identified; whereas PAR2 is activated by trypsin or trypsin-like proteases but not by thrombin, PAR1, PAR3, and PAR4 can be activated by thrombin (37). Thrombin activates PAR1 by binding to and cleaving its NH2-terminal exodomain to unmask a new NH2-terminal region of the receptor that serves as a tethered peptide ligand, binding intramolecularly to the body of the receptor to lead intracellular signaling. The synthetic peptides, which mimic the first six amino acid residues of the NH2-terminal portions newly exposed by receptor cleavage, function as agonists for PARs. The hexapeptide agonists are known to induce platelet aggregation at concentrations of 0.1–1.0 mM (36, 38–40). We therefore evaluated whether or not these synthetic peptides could induce exocytosis of horseshoe crab hemocytes. The hexapeptides SFLLRN and SLIGRL, corresponding respectively to the tethered ligands of human PAR1 and mouse PAR2, readily induced exocytosis in a dose-dependent manner. On the other hand, GYPGKF, corresponding to the tethered ligand of mouse PAR4, had little effect at 1.0 mM (Fig. 3C). These data suggest that a G protein-coupled receptor belonging to the PAR family is involved in the exocytosis of horseshoe crab hemocytes.
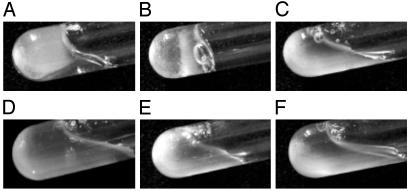
Next, the effects of PAMP and of the proteolytic activity of factor C on intact hemolymph were examined. Each of these substances was mixed with freshly prepared hemolymph under sterile conditions. The chymotrypsin-activated factor C triggered hemolymph coagulation in a few minutes, as did LPS (Fig. 4 A and B). In contrast, PPACK-factor C and other PAMP, including β-1,3-glucans and peptidoglycans, had no effect on hemolymph coagulation (Fig. 4 C–E). These data indicate that the proteolytic activity of factor C is both necessary and sufficient for the exocytosis of hemocytes, which leads to coagulation.
Fig. 4.
Effects of PAMP and proteases on hemolymph coagulation. Hemolymph (1 ml) was collected into sterilized glass tubes and incubated with PAMP or proteases at 1 μg/ml at 23°C for 30 min. (A) LPS. (B) Activated factor C. (C) PPACK-factor C. (D) Curdlan. (E) Peptidoglycan. (F) Negative control (no stimulators).
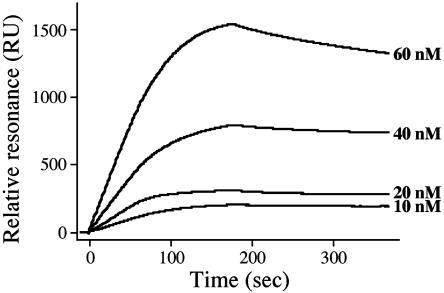
Binding Parameters of Factor C for Lipid A and Other Lipids. The binding parameters of factor C with lipid A from E. coli K12 were determined by surface plasmon resonance analysis (Fig. 5). The passage of factor C at various concentrations over lipid A immobilized on a sensor chip yielded an association rate constant of ka = 6.3 × 105 M–1·s–1 and a dissociation rate constant of kd = 4.8 × 10–4·s–1, for a consequent dissociation constant of Kd (kd/ka) = 7.6 × 10–10 M. This high binding affinity between factor C and lipid A suggests that the factor C antigen on hemocytes effectively functions as a pattern-recognition protein for LPS.
Fig. 5.
Association and dissociation of factor C with the immobilized lipid A. The binding parameters of factor C with lipid A were determined by surface plasmon resonance analysis. Sensorgrams for the binding of factor C to the lipid A immobilized on a sensor chip were overlaid at various concentrations of factor C.
Nakamura et al. (34) reported that factor C interacts with not only LPS but also acidic phospholipids, such as phosphatidylserine and phosphatidylinositol. Therefore, we hypothesize that the factor C antigen is localized on the hemocyte surface by interacting with specific lipids on the cell membrane. Surface plasmon resonance analysis demonstrated that factor C specifically interacts with phosphatidylserine and phosphatidylinositol with Kd of 2.5 × 10–9 and 4.7 × 10–9 M, respectively (Table 2). Factor C also showed tight interaction with cholesterol with Kd of 1.4 × 10–9 M. In contrast, the specific binding was not detectable between factor C and phosphatidylcholine, phosphatidylethanolamine, or sphingomyelin. The interaction of factor C with lipid A was completely inhibited by the addition of 50 μg/ml lipid A, phosphatidylserine, or phosphatidylinositol to the running buffer. Interestingly, cholesterol did not inhibit the interaction of factor C with lipid A, suggesting that factor C interacts with cholesterol through a binding site different from that for lipid A.
Table 2. Binding parameters of factor C for lipid A and other lipids.
| ka, M-1·s-1 | kd, s-1 | Kd, M | |
|---|---|---|---|
| Lipid A | 6.30 × 105 ± 3.82 × 105 | 4.76 × 10-4 ± 1.74 × 10-4 | 7.56 × 10-10 |
| Phosphatidylserine | 5.73 × 105 ± 4.28 × 105 | 1.45 × 10-3 ± 3.01 × 10-4 | 2.53 × 10-9 |
| Phosphatidylinositol | 8.15 × 105 ± 2.25 × 105 | 3.82 × 10-3 ± 1.10 × 10-4 | 4.69 × 10-9 |
| Cholesterol | 2.01 × 106 ± 1.19 × 106 | 2.87 × 10-3 ± 6.01 × 10-4 | 1.43 × 10-9 |
Discussion
The pattern-recognition protein on granular hemocytes of horseshoe crabs is an essential protein that is key to the innate immune responses, such as hemolymph coagulation and microbial killing, which are initiated by the granular components secreted via the LPS-induced exocytosis of hemocytes (16). Using an assay for exocytosis, we clearly showed that hemocytes respond only to LPS and not to other PAMP, such as β-1,3-glucans and peptidoglycans, although the β-1,3-glucan-sensitive protease zymogen factor G is stored in granules of hemocytes (Fig. 1). This assay enables the quantitative detection of LPS-induced exocytosis, ranging from ≈0.005 to 0.1 μg/ml LPS. Under the assay conditions, the formation of coagulin fiber was not observed because the protein concentration of coagulin was too low to initiate self-polymerization (41). LPS-induced exocytosis in the American horseshoe crab L. polyphemus has been reported to occur through a G protein-mediating pathway that activates a phospholipase C, leading to intracellular Ca2+ fluxes (23). A similar signaling pathway through a G protein was also observed in hemocytes of the Japanese horseshoe crab Tachypleus tridentatus by the assay method (Table 1).
Here, we focused on identifying and characterizing a pattern-recognition protein for LPS on hemocytes. By subjecting the cell surface proteins of hemocytes to flow cytometric analysis and biotinylation, we identified a factor C antigen with an apparent molecular mass of 123 kDa, corresponding to the single-chain zymogen of factor C. This zymogen was recognized by the polyclonal antibody and also by the six kinds of mAbs with different epitopes against factor C (Fig. 2 A). In addition, mAb 2C12 specifically inhibited LPS-induced exocytosis (Fig. 2B). These data indicate that the factor C antigen on the hemocyte surface is either identical with or closely related to the factor C that is stored in secretory granules. The granular factor C purified from hemocytes has mainly a two-chain form, including an H-chain of 80 kDa and an L-chain of 43 kDa (35). Determination of the nucleotide sequence of factor C clarified that factor C is synthesized as a single-chain form of zymogen (42). Therefore, the single-chain factor C is possibly cleaved to the two-chain form during purification.
Factor C is a mosaic protein, consisting of one Cys-rich domain, one EGF-like domain, one C-type lectin domain, five β2-glycoprotein I-like (Sushi-like) domains, and a serine protease domain. Although no obvious transmembrane domain is found in the primary sequence, factor C binds to acidic phospholipids, such as phosphatidylserine and phosphatidylinositol (34). Here, we used surface plasmon resonance analysis to observe the specific interactions of factor C with these acidic phospholipids, and with cholesterol (Table 2). Microdomains or lipid rafts on the cell surface, which are enriched with glycosphingolipids and cholesterol, have been accepted to regulate the signal transduction of cell surface receptors (43). Therefore, the factor C antigen may localize on lipid rafts of hemocytes through an interaction with the specific lipid components.
Factor C forms a complex with the lipid A portion of LPS through the H-chain region of factor C, generating proteolytic activity to initiate the coagulation cascade (34, 44). During LPS's activation of factor C, a cleavage of the L-chain occurs, resulting in the activation of factor C with three chains (80 kDa, 34 kDa, and 8.5 kDa); the same activation process is triggered by chymotrypsin (35). As detected by gel filtration, the activated factor C remains closely associated with LPS as a high-molecular-weight complex (44). Furthermore, part of the H-chain region has been identified as an LPS binding site, as judged by the recombinant proteins (45, 46). Interestingly, our data show that the chymotrypsin-activated factor C, but not the active site-alkylated factor C by PPACK, is also competent for exocytosis in the absence of LPS (Fig. 3 A and B). Therefore, proteolytic activity is both necessary and sufficient to trigger exocytosis.
A diverse collection of stimuli have been detected through a broad family of seven-transmembrane G protein-coupled receptors (47, 48). In addition, thrombin activates platelets in part via PARs (36, 37). Although human thrombin did not induce exocytosis of horseshoe crab hemocytes, the hexapeptides corresponding to the tethered ligands of human PAR1 and mouse PAR2 both induced exocytosis of hemocytes (Fig. 3C), suggesting the presence of a PAR-like receptor on the hemocyte surface. Our data did not reveal whether or not the factor C activity can directly activate the PAR-like receptor. It is possible that a new protease cascade initiated by the cell surface factor C is localized on hemocytes, leading to the LPS-induced exocytosis. PAMP-induced exocytosis is not specific to hemocytes of the horseshoe crab. For example, mouse Paneth cells in small-intestinal crypts secrete antimicrobial α-defensins in response to the stimulation by PAMP, such as LPS, lipoteichoic acid, and muramyl dipeptide (49). It is not yet known whether TLRs trigger the secretory response of Paneth cells or not. A PAMP-induced protease system may be conserved in the vertebrate's innate immunity.
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research on Priority Area 839 (The Molecular Basis of the Recognition of Non-Self Substances in Innate Immunity) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to S.K.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: LPS, lipopolysaccharide; PAR, protease-activated G protein-coupled receptor; PAMP, pathogen-associated molecular patterns; TLR, Toll-like receptor; PPACK, d-Phe-Pro-Arg-chloromethylketone; TL-2, tachylectin-2.
References
- 1.Janeway, C. A., Jr. (1989) Cold Spring Harbor Symp. Quant. Biol. 54, 1–13. [DOI] [PubMed] [Google Scholar]
- 2.Söderhäll, K. & Cerenius, L. (1998) Curr. Opin. Immunol. 10, 23–28. [DOI] [PubMed] [Google Scholar]
- 3.Ashida, M. & Brey, P. T. (1998) in Molecular Mechanisms of Immune Responses in Insects, eds. Brey, P. T. & Hultmark, D. (Chapman & Hall, London), pp. 135–172.
- 4.Hoffmann, J. A., Kafatos, F. C., Janeway, C. A. & Ezekowitz, R. A. B. (1999) Science 284, 1313–1318. [DOI] [PubMed] [Google Scholar]
- 5.Lemaitre, B., Nicolas, E., Michaut, L., Reichhart, J. M. & Hoffmann, J. A. (1996) Cell 86, 973–983. [DOI] [PubMed] [Google Scholar]
- 6.Medzhitov, R., Preston-Hurlburt, P. & Janeway, C. A., Jr. (1997) Nature 388, 394–397. [DOI] [PubMed] [Google Scholar]
- 7.Akira, S., Takeda, K. & Kaisho, T. (2001) Nat. Immunol. 2, 675–680. [DOI] [PubMed] [Google Scholar]
- 8.Medzhitov, R. & Janeway, C. A. (2002) Science 296, 298–300. [DOI] [PubMed] [Google Scholar]
- 9.Levashina, E. A., Langley, E., Green, C., Gubb, D., Ashburner, M., Hoffmann, J. A. & Reichhart, J. M. (1999) Science 285, 1917–1919. [DOI] [PubMed] [Google Scholar]
- 10.Anderson, K. V. & Nüsslein-Volhard, C. (1984) Nature 311, 223–227. [DOI] [PubMed] [Google Scholar]
- 11.Ligoxygakis, P., Pelte, N., Hoffmann, J. A. & Reichhart, J. M. (2002) Science 297, 114–116. [DOI] [PubMed] [Google Scholar]
- 12.Hoffmann, J. A. & Reichhart, J. M. (2002) Nat. Immunol. 3, 121–126. [DOI] [PubMed] [Google Scholar]
- 13.Leulier, F., Parquet, C., Pili-Floury, S., Ryu, J.-H., Caroff, M., Lee, W.-J., Mengin-Lecreulx, D. & Lemaitre, B. (2003) Nat. Immunol. 4, 478–484. [DOI] [PubMed] [Google Scholar]
- 14.Takehana, A., Katsuyama, T., Yano, T., Oshima, Y., Takada, H., Aigaki, T. & Kurata, S. (2002) Proc. Natl. Acad. Sci. USA 99, 13705–13710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rowley, A. F. & Ratcliffe, N. A. (1981) in Invertebrate Blood Cells 2, eds. Ratcliffe, N. A. & Rowley, A. F. (Academic, London), pp. 421–488.
- 16.Iwanaga, S., Kawabata, S. & Muta, T. (1998) J. Biochem. 123, 1–15. [DOI] [PubMed] [Google Scholar]
- 17.Iwanaga, S. (2002) Curr. Opin. Immunol. 14, 87–95. [DOI] [PubMed] [Google Scholar]
- 18.Kawabata, S. & Tsuda, R. (2002) Biochim. Biophys. Acta 1572, 414–421. [DOI] [PubMed] [Google Scholar]
- 19.Armstrong, P. B. (1985) in Blood Cells of Marine Invertebrates: Experimental Systems in Cell Biology and Comparative Physiology, ed. Cohen, W. D. (Liss, New York), pp. 77–124.
- 20.Nakamura, T., Morita, T. & Iwanaga, S. (1986) Eur. J. Biochem. 154, 511–521. [DOI] [PubMed] [Google Scholar]
- 21.Morita, T., Tanaka, S., Nakamura, T. & Iwanaga, S. (1981) FEBS Lett. 129, 318–321. [Google Scholar]
- 22.Muta, T., Seki, N., Takaki, Y., Hashimoto, R., Oda, T., Iwanaga, A., Tokunaga, F. & Iwanaga, S. (1995) J. Biol. Chem. 270, 892–897. [PubMed] [Google Scholar]
- 23.Solon, E., Gupta, A. P. & Gaugler, R. (1996) Dev. Comp. Immunol. 20, 307–321. [DOI] [PubMed] [Google Scholar]
- 24.Kawabata, S., Morita, T., Iwanaga, S. & Igarashi, H. (1985) J. Biochem. 97, 1073–1078. [DOI] [PubMed] [Google Scholar]
- 25.Inamori, K., Saito, T., Iwaki, D., Nagira, T., Iwanaga, S., Arisaka, F. & Kawabata, S. (1999) J. Biol. Chem. 274, 3272–3278. [DOI] [PubMed] [Google Scholar]
- 26.Osaki, T., Okino, N., Tokunaga, F., Iwanaga, S. & Kawabata, S. (2002) J. Biol. Chem. 277, 40084–40090. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli, U. K. (1970) Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 28.Tokunaga, F., Nakajima, H. & Iwanaga, S. (1991) J. Biochem. 109, 150–157. [DOI] [PubMed] [Google Scholar]
- 29.Robertson, J. D. (1970) Biol. Bull. 138, 157–183. [Google Scholar]
- 30.Levin, J. (1985) in Blood Cells of Marine Invertebrates: Experimental Systems in Cell Biology and Comparative Physiology, ed. Cohen, W. D. (Liss, New York), pp. 145–163.
- 31.Cannon, G. W., Tsuchiya, M., Rittschof, D. & Bonaventura, J. (1986) Biol. Bull. 171, 330–337. [Google Scholar]
- 32.Higashijima, T., Uzu, S., Nakajima, T. & Ross, E. M. (1988) J. Biol. Chem. 263, 6491–6494. [PubMed] [Google Scholar]
- 33.Miura, Y., Tokunaga, F., Miyata, T., Moriyasu, M., Yoshikawa, K. & Iwanaga, S. (1992) J. Biochem. 112, 476–481. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura, T., Tokunaga, F., Morita, T., Iwanaga, S., Kusumoto, S., Shiba, T., Kobayashi, T. & Inoue, K. (1988) Eur. J. Biochem. 176, 89–94. [DOI] [PubMed] [Google Scholar]
- 35.Tokunaga, F., Miyata, T., Nakamura, T., Morita, T., Kuma, K., Miyata, T. & Iwanaga, S. (1987) Eur. J. Biochem. 167, 405–416. [DOI] [PubMed] [Google Scholar]
- 36.Vu, T. K., Hung, D. T., Wheaton, V. I. & Coughlin S. R. (1991) Cell 64, 1057–1068. [DOI] [PubMed] [Google Scholar]
- 37.Coughlin, S. R. (1999) Proc. Natl. Acad. Sci. USA 96, 11023–11027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kahn, M. L., Nakanishi-Matsui, M., Shapiro, M. J., Ishihara, H. & Coughlin, S. R. (1999) J. Clin. Invest. 103, 879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andersen, H., Greenberg, D. L., Fujikawa, K., Xu, W., Chung, D. W. & Davie, E. W. (1999) Proc. Natl. Acad. Sci. USA 96, 11189–11193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Camerer, E., Huang, W. & Coughlin S. R. (2000) Proc. Natl. Acad. Sci. USA 97, 5255–5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawasaki, H., Nose, T., Muta, T., Iwanaga, S., Shimohigashi, Y. & Kawabata, S. (2000) J. Biol. Chem. 275, 35297–35301. [DOI] [PubMed] [Google Scholar]
- 42.Muta, T., Miyata, T., Misumi, Y., Tokunaga, F., Nakamura, T., Toh, Y., Ikehara, Y. & Iwanaga, S. (1991) J. Biol. Chem. 266, 6554–6561. [PubMed] [Google Scholar]
- 43.Simons, K. & Ikonen, K. (1997) Nature 387, 569–572. [DOI] [PubMed] [Google Scholar]
- 44.Nakamura, T., Tokunaga, F., Morita, T. & Iwanaga, S. (1988) J. Biochem. 103, 370–374. [DOI] [PubMed] [Google Scholar]
- 45.Tan, N. S., Ng, M. L. P., Yau, Y. H., Chong, P. K. W., Ho, B. & Ding, J. L. (2000) FASEB J. 14, 1801–1813. [DOI] [PubMed] [Google Scholar]
- 46.Wang, J., Tan, N. S., Ho, B. & Ding, J. L. (2002) J. Biol. Chem. 277, 36363–36372. [DOI] [PubMed] [Google Scholar]
- 47.Wess, J. (1998) Pharmacol. Ther. 80, 231–264. [DOI] [PubMed] [Google Scholar]
- 48.Schöneberg, T., Schultz, G. & Gudermann, T. (1999) Mol. Cell. Endocrinol. 151, 181–193. [DOI] [PubMed] [Google Scholar]
- 49.Ayabe, T., Satchell, D. P., Wilson, C. L., Parks, W. C., Selsted, M. E. & Quellette, A. J. (2000) Nat. Immunol. 1, 113–118. [DOI] [PubMed] [Google Scholar]