Abstract
N-acetyltransferase 1 (NAT1) catalyzes N-acetylation of arylamines as well as the O-acetylation of N-hydroxylated arylamines. O-acetylation leads to the formation of electrophilic intermediates that result in DNA adducts and mutations. NAT1*10 is the most common variant haplotype and is associated with increased risk for numerous cancers. NAT1 is transcribed from a major promoter, NATb, and an alternative promoter, NATa, resulting in messenger RNAs (mRNAs) with distinct 5′-untranslated regions (UTRs). To best mimic in vivo metabolism and the effect of NAT1*10 polymorphisms on polyadenylation usage, pcDNA5/Flp recombination target plasmid constructs were prepared for transfection of full-length human mRNAs including the 5′-UTR derived from NATb, the open reading frame and 888 nucleotides of the 3′-UTR. Following stable transfection of NAT1*4, NAT1*10 and an additional NAT1*10 variant (termed NAT1*10B) into nucleotide excision repair-deficient Chinese hamster ovary cells, N- and O-acetyltransferase activity (in vitro and in situ), mRNA and protein expression were higher in cells transfected with NAT1*10 and NAT1*10B than in cells transfected with NAT1*4 (P < 0.05). Consistent with NAT1 expression and activity, cytotoxicity and hypoxanthine phosphoribosyl transferase mutants following 4-aminobiphenyl exposures were higher in NAT1*10 than in NAT1*4 transfected cells. Ribonuclease protection assays showed no difference between NAT1*4 and NAT1*10. However, protection of one probe by NAT1*10B was not observed with NAT1*4 or NAT1*10, suggesting additional mechanisms that regulate NAT1*10B. The higher mutants in cells transfected with NAT1*10 and NAT1*10B are consistent with an increased cancer risk for individuals possessing NAT1*10 haplotypes.
Introduction
Human arylamine N-acetyltransferase 1 (NAT1) is a phase II cytosolic isozyme responsible for the biotransformation of many arylamine compounds, including environmental and occupational carcinogens such as 4-aminobiphenyl (ABP) (1). NAT1 has been implicated in several types of cancer due to its role in metabolic activation of arylamine carcinogens, and recent findings report that NAT1 may be important for cell growth and survival of cancer cells (2). NAT1 has been found in nearly all tissues studied (3,4). NAT1 is capable of both N-acetylation and O-acetylation. N-acetylation often results in inactivation followed by urinary excretion. However, following N-hydroxylation, O-acetylation catalyzed by NAT1 generates an unstable N-acetoxyarylamine, which undergoes heterolytic cleavage to yield a highly reactive nitrenium ion. These nitrenium ions are highly electrophilic and can react with DNA that if unrepaired leads to mutations. Therefore, following exposure to arylamine carcinogens, the acetylator phenotype may modulate individual susceptibility to cancer and other diseases.
NAT1*10 is the most common NAT1 variant haplotype and is presently defined (http://n-acetyltransferasenomenclature.louisville.edu) by two single-nucleotide polymorphisms (SNPs) in the 3′-untranslated region (UTR), 1088T > A (rs1057126) and 1095C > A (rs15561). NAT1*10 was associated with elevated NAT1 activity levels in human bladder (5), colon (6), liver (7,8), leukocytes (9) and β-lymphocytes (8) as well as increased N-acetylation capacity in vivo (10). However, other studies did not replicate the increased catalytic activity associated with NAT1*10 (11–13) resulting in a complete lack of consensus regarding NAT1*10 phenotype (14). Further research on SNPs in the 3′-UTR is needed to better understand their functional effects, which may be tissue-specific. There are no amino acid changes due to 3′-UTR polymorphisms, but the 1088T > A causes a change in the second consensus polyadenylation signal (AATAAA—AAAAAA). It has been speculated that this change in polyadenylation signal may give rise to higher acetylation activity (6). The 3′-UTR of a gene contains binding sites for important translational regulatory elements that include microRNAs, proteins or protein complexes, cytoplasmic polyadenylation elements and polyadenylation signals (AAUAAA) (15). It has been shown that SNPs in 3′-UTRs of dihydrofolate reductase, thrombin and resistin genes cause functional effects and alter disease risk (15–17).
In addition to the high allelic frequency, NAT1*10 has been associated with increased risk for many different forms of cancer. NAT1*10 allele or haplotype has been associated with increased risk for non-Hodgkin lymphoma (18,19) as well as for cancers of the urinary bladder (20), lung (21–23), colon/rectum (6,24–26), breast (27–29), prostate (30,31), stomach (32) and pancreas (33). However, other studies have reported no association between NAT1*10 and cancer risk (34–37). Thus, the contribution of NAT1*10 to increased cancer risk is not well understood. It is imperative that the phenotype of NAT1*10 be clearly defined in order to resolve the association of NAT1*10 genotype with increased cancer risk.
The NAT1 gene is located on the small arm of chromosome 8 (38) and spans 53 kb. NAT1 is encoded by a single intronless coding exon containing an open reading frame (ORF) of 870 bp. Several NAT1 transcripts have been identified containing various combinations of the nine non-coding 5′-UTR exons and are known to originate from a major promoter NATb, and an alternative promoter, NATa. NATa originates 51.5 kb upstream of the single NAT1 ORF, whereas NATb originates 11.8 kb upstream of the NAT1 ORF (4,39,40). NATb transcripts are expressed in all tissues studied, whereas NATa transcripts have been identified in kidney, liver, lung and trachea. In addition to polymorphic variation, it may be necessary to consider transcriptional and translational regulation to further understand the variation associated with NAT1 10 acetylation activity and effect on cancer risk. In contrast to previous studies, which included only the NAT1 ORF, the current study employs constructs that mimic the most common transcript originating from the NATb promoter. The constructs contain the ORF, the 3′-UTR and all 5′-non-coding exons found in the most common NAT1 transcript originating at the NATb promoter (39–41). The NATb construct contains exons 4 and 8 (5′-NCEs) and exon 9 (ORF). In addition to the 5′-UTR and the ORF, the NATb constructs also contain 888 nucleotides of the 3′-UTR. The NATb constructs were employed to provide a more comprehensive model of in vivo metabolism and to study any haplotype-specific interactions between the 5′-UTR and NAT1*10 polymorphisms. These constructs were utilized to compare N- and O-acetylation, messenger RNA (mRNA) levels, protein levels, polyadenylation patterns and ABP-induced mutagenesis between cells transfected with NAT1*4 and with variants of NAT1*10.
Materials and methods
Polyadenylation site removal
The bovine growth hormone polyadenylation site from the pcDNA5/Flp recombination target (FRT) (Invitrogen, Carlsbad, CA) vector was removed to allow the endogenous NAT1 polyadenylation sites to be active. This was accomplished by digestion of pcDNA5/FRT at 37°C with restriction endonucleases, ApaI and SphI (New England Biolabs, Ipswich, MA), followed by overhang digestion with T4 DNA polymerase (New England Biolabs) and ligation with T4 Ligase (New England Biolabs).
NATb/NAT1*4 construction
The constructs were created utilizing gene splicing via overlap extension (42) by amplifying the 5′-UTR and the coding region/3′-UTR separately and then fusing the two regions together. Beginning with frequently used transcription start sites (39,40), the 5′-UTRs were amplified from complementary DNA (cDNA) prepared from RNA isolated from homozygous NAT1*4 HepG2 cells. All primer sequences used are shown in Table I. The primers used to amplify the NATb 5′-UTR region were Lkm40P1 and NAT1 (3′) ORF Rev. The coding region and 3′-UTR were amplified as one piece from homozygous NAT1*4 or homozygous NAT1*10 human genomic DNA. The forward primer used to amplify the coding region/3′-UTR was NAT1 (3′) ORF Forward, whereas the reverse primer was pcDNA5distal Reverse. The two sections, the 5′-UTR and the coding region/3′-UTR, were fused together via overlap and amplification of the entire product using nested primers. The forward nested primer for NATb was P1 Fwd Inr NheI. The reverse nested primer was NAT1 Kpn Rev (NAT1*4 and NAT1*10) or NAT1 Kpn Rev 10B (NAT1*10B). Both forward nested primers included the Nhe1 endonuclease restriction site and both reverse nested primers contained the Kpn1 endonuclease restriction site to facilitate cloning. The pcDNA5/FRT vector and NATb/NAT1*4 allelic segments were digested at 37°C with restriction endonucleases KpnI and NheI (New England Biolabs). The NAT1 constructs were then ligated into pcDNA5/FRT using T4 ligase (Invitrogen). All constructs were sequenced to ensure integrity of allelic segments and junction sites.
Table I.
Primers used to construct NAT1*4, NAT1*10 and NAT1*10B in NATb type transcript constructs and for RT–PCR
| Primer | Use | Sequence |
| Lkm40P1 | NATb 5′-UTR forward-specific PCR | 5′-GGCCGCGGCATTCAGTCTAGTTCCTGGTTGCC-3′ |
| P1 Fwd Inr NheI | NATb 5′-UTR forward-specific nested PCR | 5′-TTTAAAGCTAGCATTCAGTCTAGTCTAGTTCCTGGTTGCCGGCT-3′ |
| NAT1 (3′) ORF Reverse | NATb 5′-UTR reverse PCR | 5′-TTCCTCACTCAGAGTCTTGAACTCTATT-3′ |
| NAT1 (3′) ORF Forward | NAT1 coding region forward PCR | 5′-AGACATCTCCATCATCTGTGTTTACTAGT-3′ |
| pcDNA5 FRTdistal Reverse | NAT1 3′-UTR reverse PCR | 5′-CGTGGGGATACCCCCTAGA-3′ |
| NAT1 KPN-Rev | NAT1 3′-UTR reverse nested PCR | 5′-ATAGTAGGTACCTCTGAATTATAGATAAGCAAAGATTCAGATTCT-3′ |
| NAT1 KPN-Rev*10B | NAT1 3′-UTR reverse nested PCR | 5′-ATAGTAGGTACCTCTGAATTATAGATAAGCAAAGATACAGATTCT-3′ |
| NAT1 total spliced Forward | NAT1-specific forward qRT–PCR | 5′-GAATTCAAGCCAGGAAGAAGCA-3′ |
| NAT1 total spliced Reverse | NAT1-specific reverse qRT–PCR | 5′-TCCAAGTCCAATTTGTTCCTAGACT-3′ |
| NAT1 TAQMAN probe | TAQMAN probe for NAT1 total splice | 6FAM-5′-CAATCTGTCTTCTGGATTAA-3′ MGBNFQ |
NATb/NAT1*10 construction
NATb/NAT1*10 constructs were created using the same NATb 5′-UTRs amplified from cDNA prepared from NAT1*4 homozygous RNA isolated from HepG2 cells, whereas the ORF and region 3′ to the ORF were amplified as one piece from NAT1*10/NAT1*10 homozygous human genomic DNA. These two sections, the 5′ UTR and the ORF/region 3′ to the ORF, were fused together using nested primers. Upon sequencing to ensure allelic and junction site integrity, it was discovered that one of the NAT1*10 sources had four additional polymorphisms located in the region 3′ to the ORF including 1571T > C, 1642A > C, 1647 ΔCT and 1716C > T. These NAT1 polymorphisms were verified against NCBI databases and are in linkage disequilibrium (http://egp.gs.washington.edu/). This haplotype is referred to as NAT1*10B and was used to compare N-acetylation activity with that of NAT1*10 and NAT1*4.
Cell culture
UV5-Chinese hamster ovary (CHO) cells, a nuclease excision repair-deficient derivative of AA8 that are hypersensitive to bulky DNA lesions, were obtained from the ATCC (catalog number: CRL-1865). Unless otherwise noted, cells were incubated at 37°C in 5% CO2 in complete alpha-modified minimal essential medium (Lonza, Walkersville, MD) without l-glutamine, ribosides and deoxyribosides supplemented with 10% fetal bovine serum (Hyclone, Logan, UT), 100 U/ml penicillin (Lonza), 100 μg/ml streptomycin (Lonza) and 2 mM l-glutamine (Lonza). The UV5-CHO cells used in this study were previously stably transfected with a single FRT integration site (43). The FRT site allowed stable transfections to utilize the Flp-In System (Invitrogen). When co-transfected with pOG44 (Invitrogen), a Flp recombinase expression plasmid, a site-specific conserved recombination event of pcDNA5/FRT (containing either NATa/NAT1*4 or NATb/NAT1*4) occurs at the FRT site. The FRT site allows recombination to occur immediately downstream of the hygromycin resistance gene, allowing for hygromycin selectivity only after Flp recombinase-mediated integration. The UV5/FRT cells were further modified by stable integration of human CYP1A1 and NADPH-cytochrome P450 reductase gene (43). They are referred to in this manuscript as UV5/1A1 cells.
Removal of the SV40 polyadenylation signal from NATb/NAT1*10B constructs
The SV40 polyadenylation signal was removed from the NAT1*10B pcDNA5/FRT constructs by incubation at 37°C with restriction enzymes, SacII and SapI. The overhangs were filled in or removed using T4 DNA polymerase (New England Biolabs) and then ligated back together using T4 DNA ligase (New England Biolabs).
Transient transfections
UV5/1A1 cells were transiently transfected with pcDNA5/FRT (Invitrogen) or pEF1/V5-His (Invitrogen) containing NAT1*4, NAT1*10 and NAT1*10B constructs using Lipofectamine reagent (Invitrogen) following the manufacturer’s recommendations. The UV5/1A1 cells were co-transfected with pCMV-SPORT-βgal (β-galactosidase transfection control plasmid; Invitrogen). The cells were harvested the next day. Lysate was prepared by centrifuging the cells and resuspending pellet in lysis buffer (0.2% Triton-X-100, 20 mM NaPO4, pH 7.4, 1 mM ethylenediaminetetraacetic acid, 1 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride, 2 μg/ml aprotinin and 2 μM pepstatin A). The resuspended cell pellet was centrifuged at 13 000g for 10 min. The supernatant was used to measure NAT1 and β-galactosidase activities.
Stable transfections
Stable transfections were carried out using the Flp-In System (Invitrogen) into UV5/1A1 cells that were previously stably transfected with an FRT site (as noted above). The pcDNA5/FRT plasmids containing human NAT1*4, NAT1*10 or NAT1*10B were co-transfected with pOG44 (Invitrogen), a Flp recombinase expression plasmid. The UV5/1A1 cells were stably transfected with pcDNA5/FRT containing NATb/NAT1*4, NATb/NAT1*10 and NATb/NAT1*10B constructs using Effectene transfection reagent (Qiagen, Valencia, CA) following the manufacturer’s recommendations. Since the pcDNA5/FRT vector contains a hygromycin resistance cassette, cells were passaged in complete alpha-modified essential medium containing 600 μg/ml hygromycin (Invitrogen) to select for cells containing the pcDNA5/FRT plasmid. Hygromycin-resistant colonies were selected approximately 10 days after transfection and isolated with cloning cylinders.
Determination of in vitro N-acetylation for NAT1 4, NAT1 10 and NAT1 10B
Lysate was prepared as described above. In vitro assays using the NAT1-specific substrate para-aminobenzoic acid (PABA, 300 μM) or ABP (100 μM) were conducted and acetylated products were separated utilizing high-performance liquid chromatography (HPLC) as described previously. N-acetylation activity was determined at a fixed concentration of 1 mM acetyl coenzyme A. Reactions containing substrate, acetyl coenzyme A and enzyme were incubated at 37°C for 10 min. Reactions were terminated by the addition of 1/10 volume of 1 M acetic acid and centrifuged at 15 000g for 10 min. Measurements were adjusted according to baseline measurements using lysates of the UV5/CYP1A1 cell line and normalized by the amount of total protein. Protein concentrations were measured using the method of Bradford (Bio-Rad, Hercules, CA).
In situ N-acetylation by NAT1 4, NAT1 10 and NAT1 10B
In situ N-acetylation activities were determined by a whole cell assay using media spiked with varying concentrations of PABA or ABP between 10 and 300 μM. An initial time course was performed to determine the optimal incubation times for each compound. The cells were incubated at 37°C and media was collected after 1 h (PABA) or 22 min (ABP), 1/10 volume of 1 M acetic acid was added and the mixture was centrifuged at 13 000g for 10 min. The supernatant was injected into the reverse phase HPLC column and N-acetyl-PABA or N-acetyl-ABP was separated and quantitated as described previously (44). Values were normalized to the amount of cells present at time of media removal.
Determination of in vitro O-acetylation for NAT1 4 and NAT1 10 and NAT1 10B
N-hydroxy-4-aminobiphenyl (N-OH-ABP) O-acetyltransferase assays were conducted and product was separated and quantified by HPLC as described previously (45). Assays (100 μl) containing 50 μg total protein, N-OH-ABP (100 μM), acetyl coenzyme A (1 mM) and 1 mg/ml deoxyguanosine were incubated at 37°C for 10 min. Reactions were stopped with the addition of 100 μl of water saturated ethyl acetate and centrifuged at 13 000g for 10 min. The organic phase was removed and evaporated to dryness, and the residual was redissolved in 100 μl of 10% acetonitrile and injected onto the HPLC for separation and quantitation of deoxyguanosine-C8-ABP adducts.
Measurement of NAT1 protein
The amount of NAT1 produced in UV5/1A1 cells stably transfected with NATb/NAT1*X was determined by western blot as described previously (45). Cell lysates were isolated as described above. Varying amounts of lysate were mixed 1:1 with 5% β-mercaptoethanol in Laemmli buffer (Bio-Rad), boiled for 5 min and proteins were resolved by 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis. The proteins were then transferred by semi-dry electroblotting to polyvinylidene fluoride membranes. The membranes were probed with G5, a monoclonal mouse anti-NAT1(1:200) Santa Cruz Biotechnology, Santa Cruz, CA) and with horseradish peroxidase-conjugated secondary donkey anti-mouse IgG antibody (1:2000) (Santa Cruz). Supersignal West Pico Chemiluminescent Substrate was used for detection (Pierce). Densitometric analysis was performed using Quantity One Software (Bio-Rad).
Measurement of NAT1 mRNA
Total RNA was isolated from cells using the RNeasy kit (Qiagen) followed by removal of contaminating DNA by treatment with TurboDNase Free (Ambion, Austin, TX). Synthesis of cDNA was performed using qScript cDNA Synthesis Kit (Quanta Biosciences, Gaithersburg, MD) using 1 μg of total RNA in a 20 μl reaction per the manufacturer’s protocol. Quantitative reverse transcription–polymerase chain reaction (qRT–PCR) assays were used to assess the relative amount of NAT1 mRNA in stably transfected cells. The Step One Plus (Applied Biosystems, Foster City, CA) was used to perform qRT–PCR in reactions containing 1× final concentration of qScript One-Step Fast mix (Quanta Biosciences), 300 nm of each primer and 100 nm of probe in a total volume of 20 μl. For qRT–PCR of NAT1 mRNA, a TaqMan probe was used with NAT1 Total Splice Forward and NAT1 Total Splice Reverse primers (Table I) designed using Primer Express 1.5 software (Applied Biosystems). An initial incubation at 50°C was carried out for 2 min and at 94°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. TaqMan® Ribosomal RNA Control Reagents for quantitation of the endogenous control, 18S rRNA (Applied Biosystems), were used to determine ΔCt (NAT1 Ct–18S rRNA Ct). ΔΔCt was determined by subtraction of the smallest ΔCt and relative amounts of NAT1 mRNA were calculated using 2−ΔΔCt as described previously (40).
Measurement of cytotoxicity and mutagenesis
Assays for cell cytotoxicity and mutagenesis were carried as described previously (45). Cells were grown in HAT media (30 μM hypoxanthine, 0.1 μM aminopterin and 30 μM thymidine) for 12 doublings. Cells (1 × 106) were plated, allowed to grow for 24 h and were then treated with 1.56, 3.13, 6.25 or 12.5 μM ABP (Sigma) or vehicle alone (0.5% dimethyl sulfoxide) in media. After 48 h, cells were plated to determine survival and mutagenic response to ABP. To determine cloning efficiency following each dose of ABP, 100 cells were plated in triplicate in six-well plates and allowed to grow for 7 days in non-selective media. Colonies were counted and expressed as percent of vehicle control. To determine mutagenic response following ABP exposure, 5 × 105 cells were plated and subcultured for 7 days and then seeded with 1 × 105 cells/100 mm dish (10 replicates) in complete Dulbecco’s modified Eagle’s medium (Lonza) containing 40 μM 6-thioguanine (Sigma). Mutant hypoxanthine phosphoribosyl transferase (hprt) cells were allowed to grow for 7 days and colonies were counted to determine ABP-induced mutants and corrected by cloning efficiency.
NAT1 mRNA stability assays
Six-well plates containing 1 × 106 stably transfected NAT1*4, NAT1*10 and NAT1*10B cells were treated with complete alpha-modified essential medium media spiked with 5 μg/ml of the transcription inhibitor, Actinomycin D (Sigma, St. Louis, MO). Cells were collected at 0, 1, 2, 4 and 6 h time points and total RNA was isolated as described above. Relative NAT1 mRNA levels were determined from cells transfected with NAT1*4, NAT1*10 and NAT1*10B utilizing qRT–PCR assays as described above. The first-order rate decay constant (slope) of NAT1 mRNA was determined by linear regression.
Ribonuclease protection assay
Biotinylated RNA probes that span the region 3′ to the NAT1 ORF were prepared using the MAXIscript In Vitro Transcription kit (Applied Biosystems/Ambion, Austin, TX). Ribonuclease protection assays (RNAPs) were carried out using RPAIII Kits (Applied Biosystems/Ambion) according to the manufacturer’s protocols. Briefly, total RNA was collected from transiently transfected CHO cells and treated with Turbo DNase Free kit (Applied Biosystems/Ambion). Five μg of total RNA was allowed to hybridize overnight in molar excess of biotinylated RNA probes. The resulting RNA–probe mixture was treated with RNase A/T1 (kit) to degrade any non-hybridized RNA and any remaining probe. The ribonuclease-digested hybridized mixture was then separated on a 12% polyacrylamide gel and transferred to a nitrocellulose membrane. The membrane was probed with Chemiluminescent Nucleic Acid Detection Module (ThermoScientific) and exposed to x-ray film to visualize protected probe fragments.
Statistical analysis
Statistical differences were determined using either an unpaired Student’s t-test or one-way analysis of variance followed by a Bonferroni post-test using Prism Software by Graphpad (La Jolla, CA).
Results
Upon sequencing two human sources of NAT1*10 genomic DNA used to create the NAT1*10 constructs, one source was found to contain four additional polymorphisms in the 3′-UTR. In addition to 1088T > A (rs1057126), 1095C > A (rs15561) and 1191G > T (rs4986993), the second source also possessed 1641A > C (rs8190865), a deletion ΔCT1647 (rs8190866), 1716C > T (rs8190870) and 1735A > T (rs8190871).
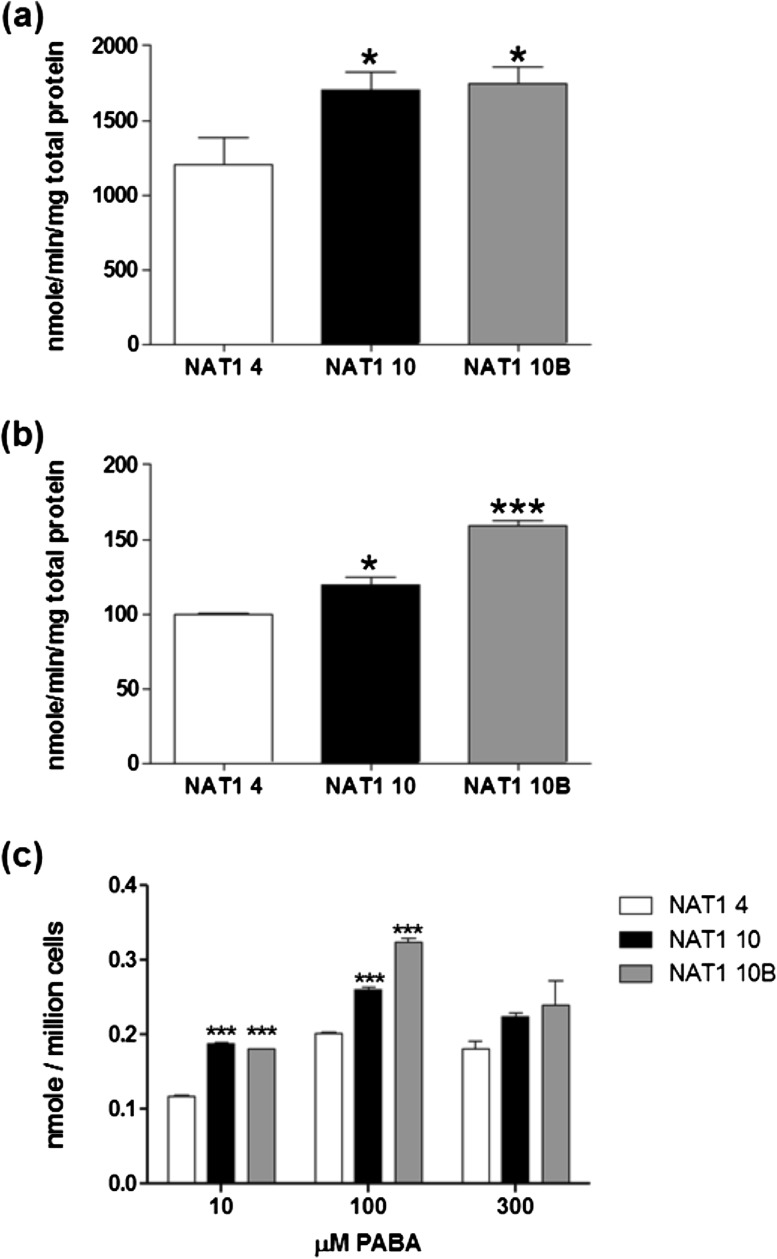
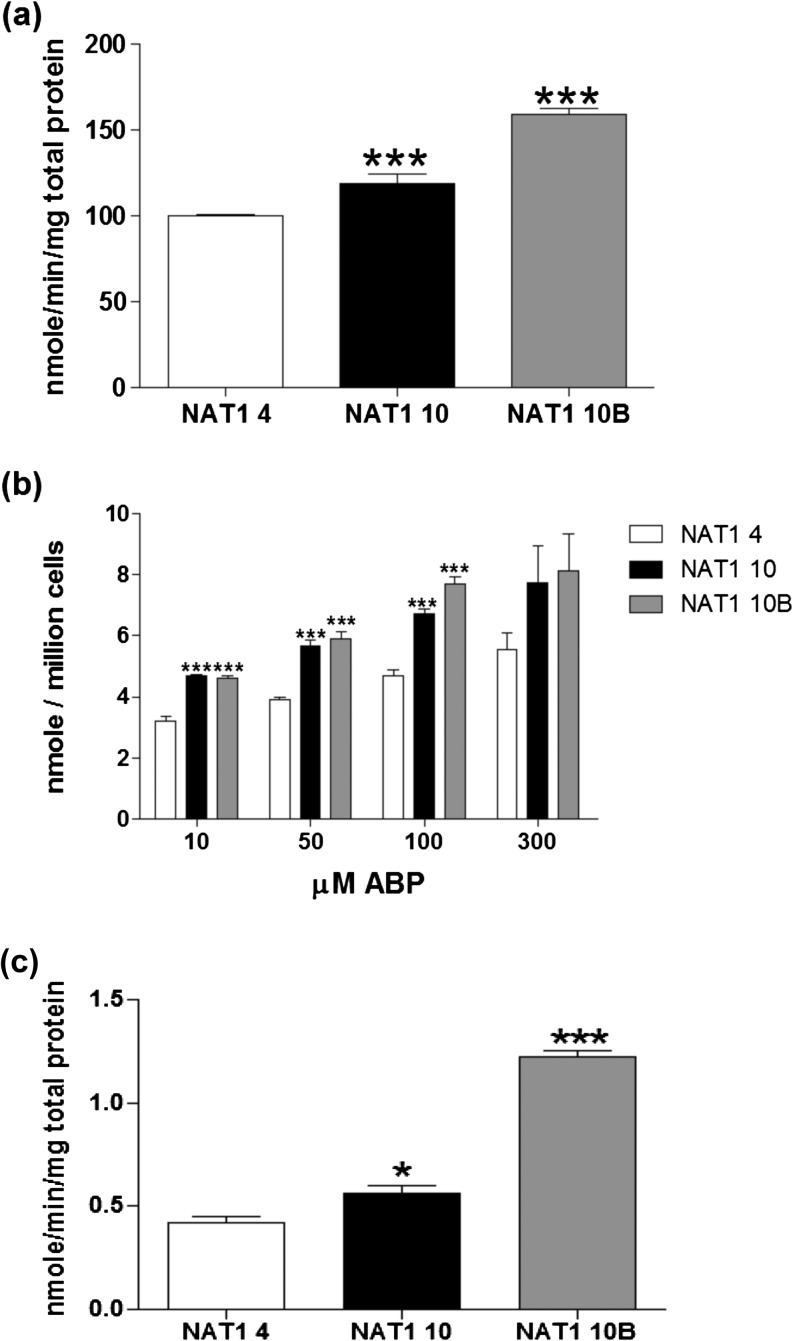
NAT1 activity was examined using PABA, ABP or N-OH-ABP as substrates. Significantly more N-acetylation of PABA (Figure 1) and ABP (Figure 2) was detected in NATb/NAT1*10 and NATb/NAT1*10B than in NATb/NAT1*4 (P < 0.05) in both transiently and stably transfected UV5/1A1 cells. Significantly more O-acetylation of N-OH-ABP was also detected in NAT1*10 and NAT1*10B than in NAT1*4 (P<0.05) in stably transfected UV5/1A1 cells (Figure 2).
Fig. 1.
PABA N-acetylation by NAT1 4 (white bars), NAT1 10 (black bars) and NAT1 10B (gray bars). (a) PABA N-acetylation (in vitro) following transient transfection with pcDNA5/FRT; (b) PABA N-acetylation (in vitro) following stable transfection with pcDNA5/FRT; (c) PABA N-acetylation (in situ) following stable transfection with pcDNA5/FRT. Each bar represents mean ± SEM for three transient transfections (a) or three separate collections performed in triplicate (b and c). Significantly higher than NAT1 4 denoted by *P < 0.05 and ***P < 0.0001 following analysis with one-way analysis of variance and Bonferroni post-test.
Fig. 2.
ABP N-acetylation and N-OH-ABP O-acetylation by NAT1 4 (white bars), NAT1 10 (black bars) and NAT1 10B (gray bars) following stable transfection in UV5/1A1 cells. (a) ABP N-acetylation (in vitro); (b) ABP N-acetylation (in situ); (c) O-acetylation of N-OH-ABP (in vitro). Each bar represents mean ± SEM for three separate collections performed in triplicate. Significantly higher than NAT1 4 denoted by *P < 0.05 and ***P < 0.0001 following analysis with one-way analysis of variance.
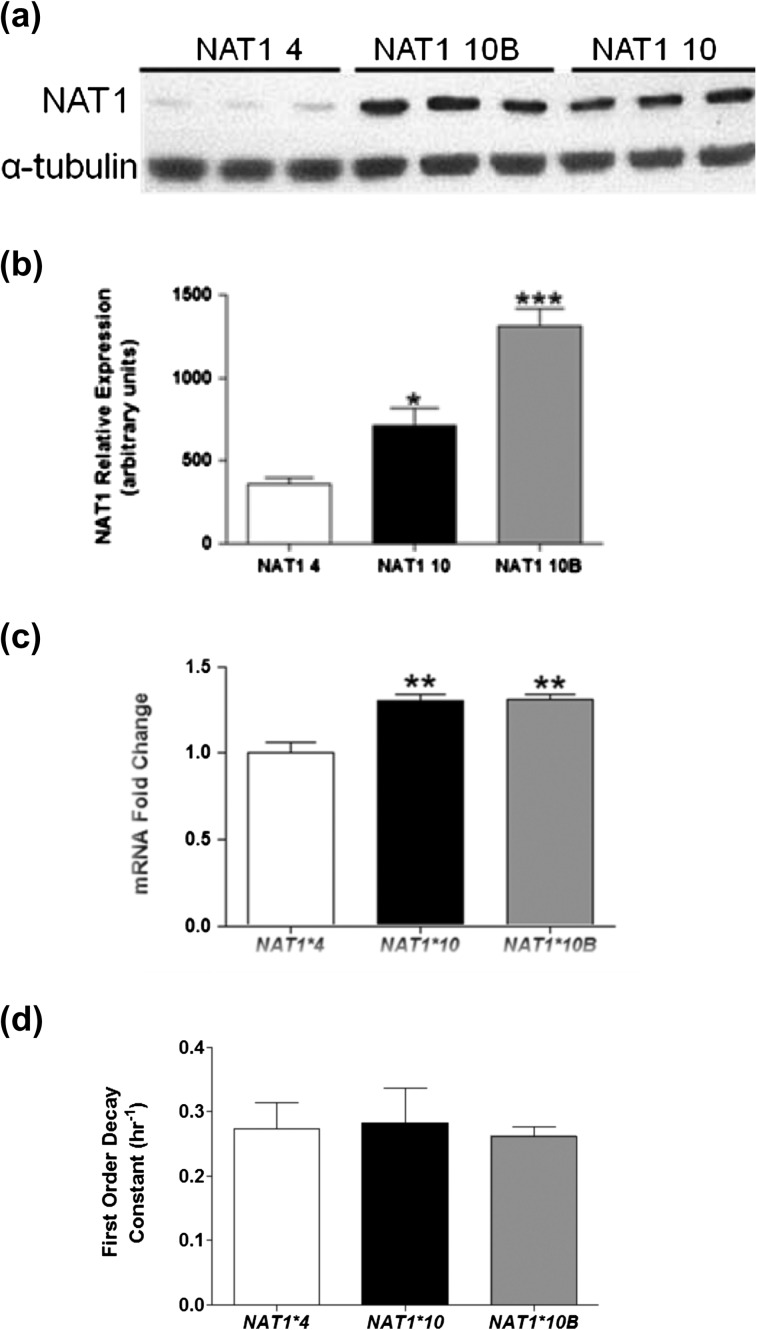
Western blots were performed to examine NAT1 expression in stably transfected UV5/1A1 cells (Figure 3). Significantly more NAT1 (P < 0.05) was detected in NATb/NAT1*10 and NAT1*10B than in NAT1*4 transfected cells (Figure 3b). Significantly (P < 0.001) more NAT1 mRNA was also observed in NAT1*10 and NAT1*10B than NAT1*4 transfected cells (Figure 3c). However, no differences were observed in stability between NAT1*4, NAT1*10 and NAT1*10B mRNA (Figure 3d).
Fig. 3.
Representative western blot of NAT1 4, NAT1 10 and NAT1 10B stable expression (a) and densitometric analysis (b). NAT1 mRNA expression levels in stably transfected NATb constructs (c) and NAT1 mRNA stability (d). Each bar represents mean ± SEM of two western blots or three collections of mRNA each performed in triplicate. Analysis done with Quantity One software (BioRad). Significantly higher than NAT1 4 denoted by *P < 0.05, **P < 0.001 and ***P < 0.0001 following analysis with one-way analysis of variance.
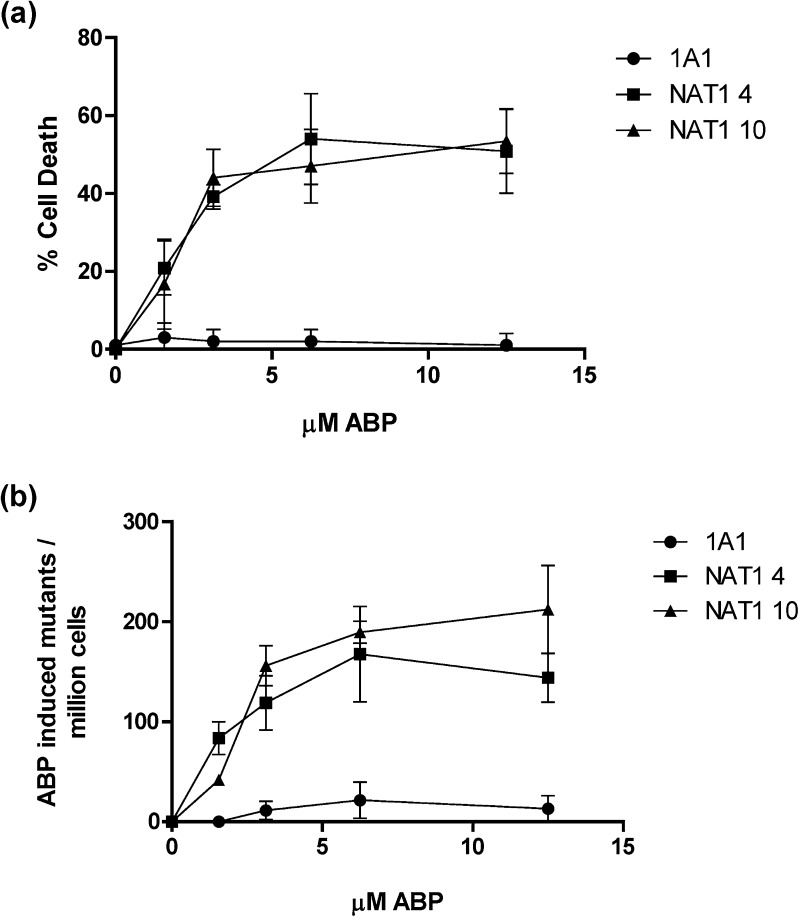
Cytotoxicity and hprt mutants were examined in cells stably transfected with NAT1*4 and NAT1*10 following exposure to ABP. Increased ABP-induced cytotoxicity (Figure 4a) and hprt mutants (Figure 4b) were observed in all NAT1 transfected cells compared with non-transfected cells. Cells transfected with NAT1*10 resulted in higher ABP-induced hprt mutants than cells transfected with NAT1*4 following exposure to 3.2, 6.3 and 12.5 μM ABP.
Fig. 4.
(a) ABP-induced cytotoxicity and (b) ABP-induced hprt mutants per million cells in UV5/1A1 cells that were stably transfected. CYP1A1 only (circle), CYP1A1/NAT1 4 (quare) and CYP1A1/NAT1 10 (triangle). Each data point represents mean ± SEM for three determinations.
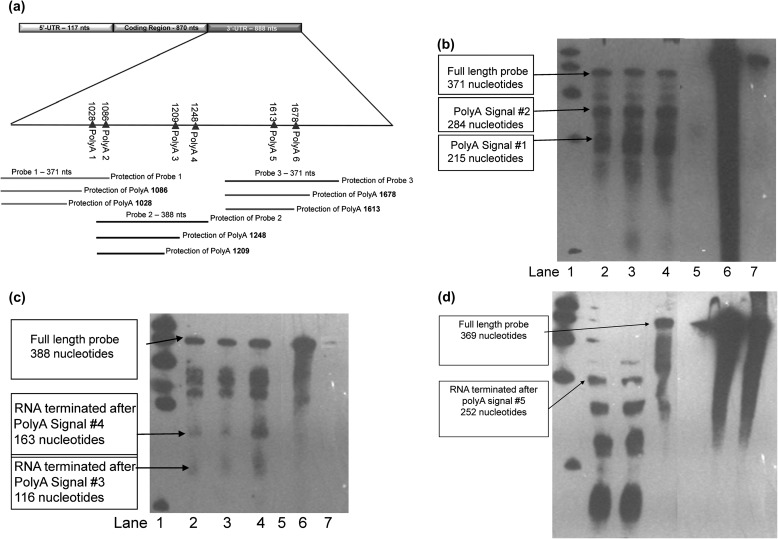
NAT1 mRNAs with intact polyadenylation tails contained in the public sequence databases including UCSC genomic browser, Unigene and EST ended 10–35 nucleotides beyond four different polyadenylation signals located at positions 1088, 1209, 1248 and 1613. Three biotinylated RNA probes were used to determine the polyadenylation pattern of NAT1*4, NAT1*10 and NAT1*10B in transiently transfected UV5/1A1 cells via an RNAP (Figure 5). RNAPs detected no difference in polyadenylation site usage between mRNA isolated from cells transfected with NATb/NAT1*4 or NATb/NAT1*10. Bands corresponding to polyadenylation signal 1 located at position 1028 (215 nucleotides), polyadenylation signal 2 located at position 1088 (284 nucleotides), polyadenylation signal 3 located at position 1209 (118 nucleotides), polyadenylation signal 4 located at position 1248 (163 nucleotides) and polyadenylation signal 5 located at position 1613 (252 nucleotides) were observed for all constructs. Full-length probe 1 (371 nucleotides) and full-length probe 2 (388 nucleotides) were observed for NAT1*4, NAT1*10 and NAT1*10B (Figure 5b and c). Full-length protection of probe 3 was observed only in NAT1*10B transfected cells (369 nucleotides) (Figure 5d). No band was observed in the lane with yeast RNA (negative control) for any probe.
Fig. 5.
RNAPs examining pattern of polyadenylation site usage. (a) Schematic representation of NAT1 3′-UTR and probes. (b) The first and second polyadenylation sites mapped with probe 1. (c) The third and fourth polyadenylation sites mapped with probe 2. (d) The fifth and sixth polyadenylation sites mapped with probe 3. Lane 1: biotinylated marker; lane 2: RNA isolated from transiently transfected NAT1*4; lane 3: NAT1*10; lane 4: NAT1*10B. lanes 5–7 are control lanes; lane 5: yeast (no target) RNA; lane 6: no ribonuclease; lane 7: probe alone. Lanes 2–5 were hybridized to probe and treated with ribonuclease.
The pcDNA5/FRT expression vector utilized in these experiments contained an SV40 polyadenylation signal for the hygromycin cassette. It was removed to ensure that the presence of NAT1*10B transcripts beyond the third probe were not vector-induced. Following removal of the SV40 polyadenylation signal from the pcDNA5/FRT, no significant (P > 0.05) difference was observed in PABA N-acetylation in UV5/1A1 cells transiently transfected with NAT1*10 or NAT1*10B.
Discussion
NAT1*10 has been associated with higher risk for many different forms of cancer, including breast, colorectal, lung, pancreatic, prostate and urinary bladder cancers, gastric adenocarcinoma and non-Hodgkin’s lymphoma. Several studies suggest that NAT1*10 has higher acetylation capacity than the referent haplotype, NAT1*4, whereas others have reported no difference. Because the allelic frequency of NAT1*10 is high in many diverse populations (46–48), particularly among those of African descent (49,50), it is important to identify mechanisms responsible for the increased cancer risk associated with NAT1*10. To better understand the association of NAT1*10 and cancer, NAT1 10 acetylation activity was studied (in vitro and in situ) using complete NATb mRNA constructs to better mimic in vivo gene expression.
Differences in expression and activity of the referent protein, NAT1 4, and the variants, NAT1 10 and NAT1 10B, were studied in UV5/1A1 CHO cells transiently and stably transfected with NATb-type mRNA. Increased N- and O-acetylation as well as increased mRNA and protein expression were observed in cells transfected with NAT1*10 and NAT1*10B when compared with cells transfected with NAT1*4. This difference was observed in both transiently and stably transfected cells. In addition to 1088T > A (rs1057126), 1095C > A (rs15561) and 1191G > T (rs4986993), we identified a NAT1*10 variant haplotype (termed NAT1*10B) that also has 1641A > C (rs8190865), a deletion ΔCT1647 (rs8190866), 1716C > T (rs8190870) and 1735A > T (rs8190871). The dbSNP Genotype and Allele Frequency Report (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_gf.cgi) database reports an allele frequency of 0.106 for 1641A > C, 1735A > T and ΔCT1647 and an allelic frequency of 0.156 for 1716C > T. Cells transfected with NAT1*10B resulted in higher N- and O-acetylation activity and protein expression compared with NAT1*10. Since these additional polymorphisms found in NAT1*10B are not assessed in association studies and they are present with high allelic frequency, it is possible that some of the discrepancies concerning NAT1*10 phenotype could be attributed to misclassification of NAT1*10B as NAT1*10.
Alternative polyadenylation plays a role in regulation of gene expression, and it is known that ≥50% of human genes encode multiple transcripts derived from alternative polyadenylation sites (51). Differential processing at multiple polyadenylation sites can be influenced by physiological conditions such as cell growth, differentiation and development or by pathological events such as cancer; however, these mechanisms are largely unknown (52). There are six potential polyadenylation signals located in the region 3′ to the NAT1 ORF. Previous studies have described the use of two consensus polyadenylation signals located at positions 1088 and 1209 in the NAT1 3′-UTR (4) or most recently, the use of 3 NAT1 polyadenylation signals including the use of an additional polyadenylation signal at position 1613 (8). Our study included a survey of NCBI databases, which identified NAT1 transcripts that utilize four of the six potential polyadenylation signals located at positions 1088, 1209, 1248 and 1613. The 1088T > A SNP present in NAT1*10 alters the second polyadenylation signal (AATAAA—AAAAAA). It has been suggested that this change in polyadenylation signal may increase the stability of the NAT1*10 mRNA which could then result in differences between NAT1*10 and NAT1*4 in acetylation capacity (6). However, we did not observe differences in NAT1 mRNA stability between NAT1*4, NAT1*10 or NAT1*10B haplotypes. RNAPs were performed to map the polyadenylation pattern cells transfected with NAT1*4, NAT1*10 and NAT1*10B. Bands were observed corresponding to the first five potential polyadenylation signals and no qualitative differences were observed between NAT1*4 and NAT1*10. However, full-length protection of probe 3 was observed for NAT1*10B but not for NAT1*10 or NAT1*4, suggesting the presence of NAT1*10B transcripts that extend beyond probe 3. Other bands present may be due to either RNA cruciform structures or probe–probe interactions. Because NAT1 transcripts have been identified which utilize multiple polyadenylation signals both in this study and in NCBI databases, alternative polyadenylation usage may be important in NAT1 regulation.
NAT1*10 has been associated with increased risk for many cancers. A recent study reported an association between higher NAT1 10 activity and increased translation efficiency in HepG2 cells (8). However, our study demonstrated higher NAT1 10 and NAT1 10B acetylation associated with higher mRNA expression. The higher acetylation activity observed in NAT1 10 compared with NAT1 4 in this study could be partly responsible for the increased risk associated with individuals possessing NAT1*10. Butcher et al. (53) suggested cell type-specific expression of an RNA-binding protein that would allow increased mRNA stability in some cell types. Previously published studies have reported allele-specific effects of RNA-binding proteins associated with increased cancer risk. For example, SF2 has been identified as a critical, allele-selective RNA binding protein that contributes to prostate tumor growth (54). Additionally, RNA-binding proteins such as ESRP-1 and ESRP-2 have been reported to be cell type specific (55). Therefore, it is possible that an allele-specific RNA-binding protein could bind to NAT1*4, NAT1*10and NAT1*10B mRNA differently in certain cell types and therefore modify NAT1 differently in those cell-types. NAT1*10 expression appears to be under the control of multiple types of regulation which could be cell type dependent.
Because the allelic frequency of NAT1*10 haplotype is high, clearly defining the NAT1*10 phenotype would allow cancer risk and other toxicities related to environmental arylamine exposure to be better understood. This study has shown that cells transfected with NAT1*10 and NAT1*10B have higher N- and O-acetylation activity, NAT1 mRNA and protein expression compared with cells transfected with NAT1*4. Additionally, higher ABP-induced DNA adducts and mutants were observed in cells transfected with NAT1*10 compared with NAT1*4. However, no differences between NAT1*4 and NAT1*10 polyadenylation pattern were observed. Additionally, no differences in mRNA stability were observed between NAT1*4, NAT1*10 and NAT1*10B in stably transfected UV5/1A1 cells (Figure 3d) or in NAT1 endogenously expressed in HepG2 cells (Zhang,X., Doll,M.A. and Hein,D.W, unpublished data). Other mechanisms such as RNA-binding proteins may be responsible for the higher amount of NAT1*10 mRNA, protein, acetylation activity, ABP-induced DNA adducts and mutants compared with NAT1*4. A difference was observed in the NAT1*10B polyadenylation pattern compared with NAT1*10 and NAT1*4, suggesting that additional mechanisms are involved in regulation of NAT1*10B compared with NAT1*10. Because the NAT1*10B polymorphisms are associated with a high allele frequency (>0.106) and current genotyping methods do not differentiate between NAT1*10 and NAT1*10B, misclassification of NAT1*10B could contribute to divergent findings regarding the role of NAT1*10 in cancer risk. Further studies should be conducted to determine the responsible mechanisms for regulation of NAT1*10 and NAT1*10B. The higher acetylation and ABP-induced mutants observed in NAT1 10 compared with NAT1 4 is consistent with increased risk for cancers associated with arylamine exposure in individuals possessing NAT1*10 haplotypes.
Funding
The National Cancer Institute (R01-CA034627); The National Institute of Environmental Health Sciences (T32-ES011564 and P30-ES014443); The Department of Defense Breast Cancer Research Program (BC083107).
Acknowledgments
Portions of this work constitute partial fulfillment for the PhD in pharmacology and toxicology at the University of Louisville to Lori Millner.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- ABP
4-aminobiphenyl
- cDNA
complementary DNA
- CHO
Chinese hamster ovary
- FRT
Flp recombination target
- HPLC
high performance liquid chromatography
- hprt
hypoxanthine phosphoribosyl transferase
- mRNA
messenger RNA
- NAT1
N-acetyltransferase 1
- N-OH-ABP
N-hydroxy-4-aminobiphenyl
- ORF
open reading frame
- PABA
para-aminobenzoic acid
- qRT–PCR
quantitative reverse transcription–polymerase chain reaction
- RNAP
Ribonuclease protection assay
- SNP
single-nucleotide polymorphism
- UTR
untranslated region
References
- 1.Hein DW, et al. Molecular genetics and epidemiology of the NAT1 and NAT2 acetylation polymorphisms. Cancer Epidemiol. Biomarkers Prev. 2000;9:29–42. [PubMed] [Google Scholar]
- 2.Tiang JM, et al. RNAi-mediated knock-down of arylamine N-acetyltransferase-1 expression induces E-cadherin up-regulation and cell-cell contact growth inhibition. PLoS One. 2011;6:e17031. doi: 10.1371/journal.pone.0017031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pacifici GM, et al. Acetyltransferase in humans: development and tissue distribution. Pharmacology. 1986;32:283–291. doi: 10.1159/000138181. [DOI] [PubMed] [Google Scholar]
- 4.Boukouvala S, et al. Structural analysis of the genes for human arylamine N-acetyltransferases and characterisation of alternative transcripts. Basic Clin. Pharmacol. Toxicol. 2005;96:343–351. doi: 10.1111/j.1742-7843.2005.pto_02.x. [DOI] [PubMed] [Google Scholar]
- 5.Badawi AF, et al. Role of aromatic amine acetyltransferases, NAT1 and NAT2, in carcinogen-DNA adduct formation in the human urinary bladder. Cancer Res. 1995;55:5230–5237. [PubMed] [Google Scholar]
- 6.Bell DA, et al. Polymorphism in the N-acetyltransferase 1 (NAT1) polyadenylation signal: association of NAT1*10 allele with higher N-acetylation activity in bladder and colon tissue. Cancer Res. 1995;55:5226–5229. [PubMed] [Google Scholar]
- 7.Zenser TV, et al. Human N-acetylation of benzidine: role of NAT1 and NAT2. Cancer Res. 1996;56:3941–3947. [PubMed] [Google Scholar]
- 8.Wang D, et al. Human N-acetyltransferase 1 *10 and *11 alleles increase protein expression through distinct mechanisms and associate with sulfamethoxazole-induced hypersensitivity. Pharmacogenet. Genomics. 2011;21:652–664. doi: 10.1097/FPC.0b013e3283498ee9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhangwei X, et al. N-Acetyltransferase-1 gene polymorphisms and correlation between genotype and its activity in a central Chinese Han population. Clin. Chim. Acta. 2006;371:85–91. doi: 10.1016/j.cca.2006.02.025. [DOI] [PubMed] [Google Scholar]
- 10.Hein DW, et al. Pharmacogenetics of the arylamine N-acetyltransferases: a symposium in honor of Wendell W. Weber. Drug Metab. Dispos. 2000;28:1425–1432. [PubMed] [Google Scholar]
- 11.Payton MA, et al. Genotyping human arylamine N-acetyltransferase type 1 (NAT1): the identification of two novel allelic variants. Biochem. Pharmacol. 1998;55:361–366. doi: 10.1016/s0006-2952(97)00478-4. [DOI] [PubMed] [Google Scholar]
- 12.Bruhn C, et al. Correlation between genotype and phenotype of the human arylamine N-acetyltransferase type 1 (NAT1) Biochem. Pharmacol. 1999;58:1759–1764. doi: 10.1016/s0006-2952(99)00269-5. [DOI] [PubMed] [Google Scholar]
- 13.de Leon JH, et al. Characterization of naturally occurring and recombinant human N-acetyltransferase variants encoded by NAT1. Mol. Pharmacol. 2000;58:288–299. doi: 10.1124/mol.58.2.288. [DOI] [PubMed] [Google Scholar]
- 14.Hein DW. N-acetyltransferase SNPs: emerging concepts serve as a paradigm for understanding complexities of personalized medicine. Expert Opin. Drug Metab. Toxicol. 2009;5:353–366. doi: 10.1517/17425250902877698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mishra PJ, et al. MiRSNPs or MiR-polymorphisms, new players in microRNA mediated regulation of the cell: introducing microRNA pharmacogenomics. Cell Cycle. 2008;7:853–858. doi: 10.4161/cc.7.7.5666. [DOI] [PubMed] [Google Scholar]
- 16.Pizzuti A, et al. An ATG repeat in the 3′-untranslated region of the human resistin gene is associated with a decreased risk of insulin resistance. J. Clin. Endocrinol. Metab. 2002;87:4403–4406. doi: 10.1210/jc.2002-020096. [DOI] [PubMed] [Google Scholar]
- 17.Gehring NH, et al. Increased efficiency of mRNA 3′ end formation: a new genetic mechanism contributing to hereditary thrombophilia. Nat. Genet. 2001;28:389–392. doi: 10.1038/ng578. [DOI] [PubMed] [Google Scholar]
- 18.Morton LM, et al. Hair dye use, genetic variation in N-acetyltransferase 1 (NAT1) and 2 (NAT2), and risk of non-Hodgkin lymphoma. Carcinogenesis. 2007;28:1759–1764. doi: 10.1093/carcin/bgm121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morton LM, et al. Genetic variation in N-acetyltransferase 1 (NAT1) and 2 (NAT2) and risk of non-Hodgkin lymphoma. Pharmacogenet. Genomics. 2006;16:537–545. doi: 10.1097/01.fpc.0000215071.59836.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor JA, et al. The role of N-acetylation polymorphisms in smoking-associated bladder cancer: evidence of a gene-gene-exposure three-way interaction. Cancer Res. 1998;58:3603–3610. [PubMed] [Google Scholar]
- 21.Abdel-Rahman SZ, et al. Association of the NAT1*10 genotype with increased chromosome aberrations and higher lung cancer risk in cigarette smokers. Mutat. Res. 1998;398:43–54. doi: 10.1016/s0027-5107(97)00238-8. [DOI] [PubMed] [Google Scholar]
- 22.Wikman H, et al. Relevance of N-acetyltransferase 1 and 2 (NAT1, NAT2) genetic polymorphisms in non-small cell lung cancer susceptibility. Pharmacogenetics. 2001;11:157–168. doi: 10.1097/00008571-200103000-00006. [DOI] [PubMed] [Google Scholar]
- 23.Gemignani F, et al. Development of lung cancer before the age of 50: the role of xenobiotic metabolizing genes. Carcinogenesis. 2007;28:1287–1293. doi: 10.1093/carcin/bgm021. [DOI] [PubMed] [Google Scholar]
- 24.Chen J, et al. A prospective study of N-acetyltransferase genotype, red meat intake, and risk of colorectal cancer. Cancer Res. 1998;58:3307–3311. [PubMed] [Google Scholar]
- 25.Ishibe N, et al. Genetic polymorphisms in heterocyclic amine metabolism and risk of colorectal adenomas. Pharmacogenetics. 2002;12:145–150. doi: 10.1097/00008571-200203000-00008. [DOI] [PubMed] [Google Scholar]
- 26.Lilla C, et al. Effect of NAT1 and NAT2 genetic polymorphisms on colorectal cancer risk associated with exposure to tobacco smoke and meat consumption. Cancer Epidemiol. Biomarkers Prev. 2006;15:99–107. doi: 10.1158/1055-9965.EPI-05-0618. [DOI] [PubMed] [Google Scholar]
- 27.Ambrosone CB, et al. Hair dye use, meat intake, and tobacco exposure and presence of carcinogen-DNA adducts in exfoliated breast ductal epithelial cells. Arch. Biochem. Biophys. 2007;464:169–175. doi: 10.1016/j.abb.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 28.Millikan RC, et al. Cigarette smoking, N-acetyltransferases 1 and 2, and breast cancer risk. Cancer Epidemiol. Biomarkers Prev. 1998;7:371–378. [PubMed] [Google Scholar]
- 29.Zheng W, et al. N-acetyltransferase 1 genetic polymorphism, cigarette smoking, well-done meat intake, and breast cancer risk. Cancer Epidemiol. Biomarkers Prev. 1999;8:233–239. [PubMed] [Google Scholar]
- 30.Hein DW, et al. Association of prostate cancer with rapid N-acetyltransferase 1 (NAT1*10) in combination with slow N-acetyltransferase 2 acetylator genotypes in a pilot case-control study. Environ. Mol. Mutagen. 2002;40:161–167. doi: 10.1002/em.10103. [DOI] [PubMed] [Google Scholar]
- 31.Rovito PM, Jr, et al. Heterocyclic amines and genotype of N-acetyltransferases as risk factors for prostate cancer. Prostate Cancer Prostatic Dis. 2005;8:69–74. doi: 10.1038/sj.pcan.4500780. [DOI] [PubMed] [Google Scholar]
- 32.Boissy RJ, et al. A pilot study investigating the role of NAT1 and NAT2 polymorphisms in gastric adenocarcinoma. Int. J. Cancer. 2000;87:507–511. [PubMed] [Google Scholar]
- 33.Li D, et al. Polymorphisms of cytochrome P4501A2 and N-acetyltransferase genes, smoking, and risk of pancreatic cancer. Carcinogenesis. 2006;27:103–111. doi: 10.1093/carcin/bgi171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brockton N, et al. N-acetyltransferase polymorphisms and colorectal cancer: a HuGE review. Am. J. Epidemiol. 2000;151:846–861. doi: 10.1093/oxfordjournals.aje.a010289. [DOI] [PubMed] [Google Scholar]
- 35.Gu J, et al. Effects of N-acetyl transferase 1 and 2 polymorphisms on bladder cancer risk in Caucasians. Mutat. Res. 2005;581:97–104. doi: 10.1016/j.mrgentox.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 36.Sanderson S, et al. Joint effects of the N-acetyltransferase 1 and 2 (NAT1 and NAT2) genes and smoking on bladder carcinogenesis: a literature-based systematic HuGE review and evidence synthesis. Am. J. Epidemiol. 2007;166:741–751. doi: 10.1093/aje/kwm167. [DOI] [PubMed] [Google Scholar]
- 37.Gong C, et al. A meta-analysis of the NAT1 and NAT2 polymorphisms and prostate cancer: a huge review. Med. Oncol. 2011;28:365–376. doi: 10.1007/s12032-010-9423-5. [DOI] [PubMed] [Google Scholar]
- 38.Blum M, et al. Human arylamine N-acetyltransferase genes: isolation, chromosomal localization, and functional expression. DNA Cell Biol. 1990;9:193–203. doi: 10.1089/dna.1990.9.193. [DOI] [PubMed] [Google Scholar]
- 39.Husain A, et al. Identification of the major promoter and non-coding exons of the human arylamine N-acetyltransferase 1 gene (NAT1) Pharmacogenetics. 2004;14:397–406. doi: 10.1097/01.fpc.0000114755.08559.6e. [DOI] [PubMed] [Google Scholar]
- 40.Barker DF, et al. Functional properties of an alternative, tissue-specific promoter for human arylamine N-acetyltransferase 1. Pharmacogenet. Genomics. 2006;16:515–525. doi: 10.1097/01.fpc.0000215066.29342.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Husain A, et al. Functional analysis of the human N-acetyltransferase 1 major promoter: quantitation of tissue expression and identification of critical sequence elements. Drug Metab. Dispos. 2007;35:1649–1656. doi: 10.1124/dmd.107.016485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horton RM, et al. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 43.Bendaly J, et al. 2-Amino-3,8-dimethylimidazo-[4,5-f]quinoxaline-induced DNA adduct formation and mutagenesis in DNA repair-deficient Chinese hamster ovary cells expressing human cytochrome P4501A1 and rapid or slow acetylator N-acetyltransferase 2. Cancer Epidemiol. Biomarkers Prev. 2007;16:1503–1509. doi: 10.1158/1055-9965.EPI-07-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hein DW, et al. Tissue distribution of N-acetyltransferase 1 and 2 catalyzing the N-acetylation of 4-aminobiphenyl and O-acetylation of N-hydroxy-4-aminobiphenyl in the congenic rapid and slow acetylator Syrian hamster. Mol. Carcinog. 2006;45:230–238. doi: 10.1002/mc.20164. [DOI] [PubMed] [Google Scholar]
- 45.Millner LM, et al. NATb/NAT1*4 promotes greater arylamine N-acetyltransferase 1 mediated DNA adducts and mutations than NATa/NAT1*4 following exposure to 4-aminobiphenyl. Mol. Carcinog. 2011 doi: 10.1002/mc.20836. doi: 10.1002/mc.20836. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lo-Guidice JM, et al. Molecular analysis of the N-acetyltransferase 1 gene (NAT1*) using polymerase chain reaction-restriction fragment-single strand conformation polymorphism assay. Pharmacogenetics. 2000;10:293–300. doi: 10.1097/00008571-200006000-00003. [DOI] [PubMed] [Google Scholar]
- 47.Vaziri SA, et al. Variation in enzymes of arylamine procarcinogen biotransformation among bladder cancer patients and control subjects. Pharmacogenetics. 2001;11:7–20. doi: 10.1097/00008571-200102000-00002. [DOI] [PubMed] [Google Scholar]
- 48.Cascorbi I, et al. Association of NAT1 and NAT2 polymorphisms to urinary bladder cancer: significantly reduced risk in subjects with NAT1*10. Cancer Res. 2001;61:5051–5056. [PubMed] [Google Scholar]
- 49.Loktionov A, et al. Differences in N-acetylation genotypes between Caucasians and Black South Africans: implications for cancer prevention. Cancer Detect. Prev. 2002;26:15–22. doi: 10.1016/s0361-090x(02)00010-7. [DOI] [PubMed] [Google Scholar]
- 50.Kidd LC, et al. No association between variant N-acetyltransferase genes, cigarette smoking and prostate cancer susceptibility among men of African descent. Biomark. Cancer. 2011;3:1–13. doi: 10.4137/BIC.S6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tian B, et al. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Di Giammartino DC, et al. Mechanisms and consequences of alternative polyadenylation. Mol. Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Butcher NJ, et al. Regulation of arylamine N-acetyltransferases. Curr. Drug Metab. 2008;9:498–504. doi: 10.2174/138920008784892128. [DOI] [PubMed] [Google Scholar]
- 54.Olshavsky NA, et al. Identification of ASF/SF2 as a critical, allele-specific effector of the cyclin D1b oncogene. Cancer Res. 2010;70:3975–3984. doi: 10.1158/0008-5472.CAN-09-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Warzecha CC, et al. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol. Cell. 2009;33:591–601. doi: 10.1016/j.molcel.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]