Abstract
We previously demonstrated that secreted protein acidic and rich in cysteine (SPARC) increases heat shock protein 27 (HSP27) expression and phosphorylation and promotes glioma cell migration through the p38 mitogen-activated protein kinase (MAPK)/HSP27 signaling pathway. As different regions of the SPARC protein mediate different SPARC functions, elucidating which SPARC domains regulate HSP27 expression, signaling and migration might provide potential therapeutic strategies to target these functions. To investigate the roles of specific domains, we used an SPARC–green fluorescent protein (GFP) fusion protein and constructs of SPARC–GFP with deletions of either the acidic domain (ΔAcidic) or the epidermal growth factor (EGF)-like module (ΔEGF). GFP, SPARC–GFP and the two deletion mutants were expressed in U87MG glioma cells. Characterization of the derived stable clones by confocal imaging and western blotting suggests proper folding, processing and secretion of the deletion constructs. Uptake of the constructs by naive cells suggests enhanced internalization of ΔAcidic and reduced internalization of ΔEGF. Wound and transwell migration assays and western blot analysis confirm our previous results and indicate that ΔAcidic reduces SPARC-induced migration and p38 MAPK/HSP27 signaling and ΔEGF decreases SPARC-induced migration and dramatically decreases the expression and phosphorylation of HSP27 but is poorly internalized. Loss of the EGF-like module suppresses the enhanced HSP27 protein stability conferred by SPARC. In conclusion, deletions of the acidic domain and EGF-like module have differential effects on cell surface binding and HSP27 protein stability; however, both regions regulate SPARC-induced migration and signaling through HSP27. Our data link the domains of SPARC with different functions and suggest one or both of the constructs as potential therapeutic agents to inhibit SPARC-induced migration.
Introduction
Gliomas are the most common primary brain tumors in adults. Even grade II gliomas invade into the normal brain, making these highly malignant tumors. The infiltrative cells are the primary cause of recurrence in glioma patients (1,2). Surgery and radiotherapy are effective in targeting the tumor center but less effective against the invading cells (3). Even with newer therapies, the median survival time of patients with the most malignant, grade IV, gliomas is about 15 months after diagnosis (4), necessitating the development of new therapies.
Secreted protein acidic and rich in cysteine [SPARC (5)], also known as osteonectin (6) or BM-40 (7), is a secreted glycoprotein. SPARC is highly expressed during development, vascular morphogenesis and in tissues undergoing remodeling and repair (8,9). SPARC is secreted into the extracellular matrix (ECM) where it can modulate cell adhesion, motility, proliferation and ECM production (10). We and others have reported that SPARC is highly upregulated early in glioma progression (11) and that SPARC is involved in glioma migration and invasion in vitro (12) and in vivo (13,14).
SPARC is composed of an acidic domain, a follistatin-like domain and an extracellular calcium-binding domain. The acidic domain contains several glutamate residues and binds 5–8 calcium ions with low affinity (kd = 5–10 mM) (8). Data suggest that this domain has a well-defined structure only in the presence of high concentrations of calcium (15). The follistatin-like domain contains an epidermal growth factor (EGF)-like module, which is highly twisted by two disulfide bonds (16) and a copper-binding region that also binds β1 integrin (17). The extracellular calcium-binding domain contains two nearly identical EF-hands and binds two calcium ions cooperatively with high affinity (kd = 170 nM) (18,19). Since extracellular calcium levels are high, the EF-hands are constitutively bound to calcium and so they presumably play a structural role (18).
The functions of SPARC domains have been investigated by adding peptides to cells in culture. A peptide corresponding to a portion of the acidic domain inhibited fibroblast and endothelial cell spreading and an antibody to this peptide blocked the anti-spreading activity of wild-type (wt)-SPARC (20). This peptide also decreased focal adhesions in endothelial cells (21) and, like wt-SPARC, downregulated thrombospondin and fibronectin (FN) and induced the expression of PAI-1 (22). Peptides mimicking the EGF-like module caused focal adhesion disassembly (21), inhibited endothelial cell proliferation (23) and had a biphasic effect on fibroblast proliferation (24). This peptide was also more potent than wt-SPARC in binding to the cell surface of endothelial cells (25) and inhibited angiogenesis induced by neuroblastoma cells in a dose-dependent manner (26). Portions of the extracellular calcium-binding domain are also involved in SPARC function, including de-adhesion, slowing of cell cycle progression and binding to collagens (20,21,27,28). However, as part of this domain is important in the proper folding of SPARC (19), this study focuses on the acidic domain and the EGF-like module.
We have shown previously that SPARC expression increases glioma cell migration on FN through the activation of the p38 mitogen-activated protein kinase (MAPK)/heat shock protein 27 (HSP27) signaling pathway (29). This activation is likely mediated by SPARC binding to a cell surface receptor such as β1 integrin (17,30) to induce signaling through intracellular proteins, such as FAK (31,32) and ILK (33). Indeed, ILK activates p38 MAPK (34). p38 MAPK exists in a complex with unphosphorylated MAPKAPK2, AKT and HSP27. Upon activation, p38 MAPK activates MAPKAPK2, which then phosphorylates HSP27 (35). MAPKAPK2 can also activate AKT (36,37) and AKT can directly phosphorylate HSP27 (38). Once HSP27 is phosphorylated, the complex dissociates (36,38). Unphosphorylated HSP27 participates in actin capping (35). When phosphorylated, HSP27 no longer localizes to the leading edge of the lamellipodia but localizes further back, where it stabilizes the actin filaments to facilitate migration (35,39). We have shown that inhibition of this pathway using HSP27 siRNA results in decreased migration and/or invasion of SPARC-expressing glioma cells in vitro (29).
We have shown that SPARC increases total HSP27 protein and this is accompanied by increased HSP27 transcript abundance (29). As SPARC has also been proposed to be an intracellular protein chaperone (40,41), we further determined whether SPARC mediates HSP27 protein stability and whether deletion of either the acidic domain or the EGF-like module are involved.
The goals of this study were to examine the effects of deleting the acidic domain or the EGF-like module in SPARC-induced migration and signaling. As SPARC is highly overexpressed in the majority of gliomas (11), we expressed the deletion mutants in U87MG glioma cells. Both deletions reduced cell migration and HSP27 expression and signaling but affected internalization of the constructs, cell adhesion and HSP27 protein stability differently. Our results further support our previous conclusions that HSP27 is a good therapeutic target for SPARC-expressing tumors and, in addition, suggest SPARC deletion constructs as potential therapeutic agents.
Materials and methods
Cell maintenance
Cells were maintained in a humidified chamber at 37°C and 5% CO2. U87MG cells and the U87D8 clone were maintained in growth medium: Dulbecco's modified Eagle's medium + 10% fetal bovine serum + 1% penicillin/streptomycin [Pen/Strep (1:1)] + 5 μg/ml gentamicin. The transfected clones were maintained in growth medium + 400 μg/ml geneticin (G418). For experiments, unless otherwise stated, cells were plated in Dulbecco's modified Eagle's medium + 10% fetal bovine serum + 1% penicillin/streptomycin overnight. Then cells were washed twice with phosphate-buffered saline (PBS) and media were changed to serum-free (SF) OptiMEM. Cell culture reagents were purchased from Invitrogen (Grand Island, NY).
Vector constructs, transfection and clone selection
The SPARC–green fluorescent protein (GFP) fusion construct was created previously in our laboratory (29). The deletion mutants were created using QuickChange Site-Directed Mutagenesis (Stratagene, La Jolla, CA). The polyacrylamide gel electrophoresis-purified primers for site-directed mutagenesis for the deletion of the acidic domain (forward 5′-GGGAGGGCCTTGGCAAATCCCTGCCAGAAC-3′ and reverse 5′-GTTCTGGCAGGGATTTGCCAAGGCCCTTCCC-3′) and for the deletion of the EGF-like module (forward 5′-GTGGCGGAAAATCCCGTGTGCCAGGACCCC-3′ and reverse 5′-GGGGTCCTGGCACACGGGATTTTCCGCCAC-3′) were purchased from Invitrogen (Carlsbad, CA). Plasmids were amplified in bacteria and purified by miniprep and/or maxiprep (Qiagen, Valencia, CA). Mutations were verified by enzyme digestion and dye terminator sequencing (Applied Genomics Technology Center, Wayne State University).
U87MG cells (ATCC) were transfected by electroporation using the nucleofector program X-01 and solution T (Amaxa, Gaithersburg, MD). Cells were subjected to G418 selection. Cells were diluted, plated in 100-mm dishes and allowed to grow. Fluorescent colonies were visualized using an Olympus IX50 fluorescence microscope and were individually transferred to 24-well plates using cloning discs (Labcor Products, Frederick, MD) soaked in trypsin.
Two clones expressing each of the constructs were chosen based on their having similar expression levels: GFP14 and GFP72 express GFP and SPARCB8 and SPARC83 express SPARC–GFP. The deletion mutant clones include ΔAcidicG3, ΔAcidicE61, ΔEGF1.3 and ΔEGFC1. Throughout this report they will be referred to as GFP, SPARC, ΔAcidic and ΔEGF, respectively.
Selection of the U87D8 clone was also performed using cloning discs. Several clones were analyzed by western blot and U87D8 was selected for the level of endogenous SPARC and the ability to internalize SPARC–GFP.
Western blot analyses
Clones were plated 2 or 4 × 105 on plastic or FN (Millipore, Temecula, CA). Conditioned SF OptiMEM was collected at 24 h and centrifuged. For collection of cell lysates at 3 and 24 h, cells were washed twice with ice-cold PBS and lysed with single detergent lysis buffer (50 mM Tris, 150 mM NaCl and 1% Triton X-100) + 5 mM Na3VO4 + 10 mM NaF + Easy mini protease inhibitor cocktail tablet (Roche, Indianapolis, IN). The lysed cells were scraped, vortexed and centrifuged. For collection of lysates from U87D8 cells exposed to conditioned medium, lysates were collected after 10 and 30 min and 1, 3, 6 and 24 h after exposure.
Lysate protein concentration was determined by BCA protein assay (Thermo, Rockford, IL) and 8–20 μg protein in lysates or 20 μl media were subject to sodium dodecyl sulfate–polyacrylamide gel electrophoresis and western blot as described previously (12). Molecular weights were estimated using the Precision Plus Dual Color Protein Standards (Bio-Rad, Hercules, CA). Primary antibodies include SPARC (1:6000; Haematologic Technologies, Essex Junction, VT), GFP (1:2000; Invitrogen, Eugene, OR), HSP27, actin (1:2000; Santa Cruz Biotechnology, Santa Cruz, CA), p38 MAPK, phospho-p38 MAPK (Thr180/Tyr182), phospho-HSP27 Ser82 (1:1000; Cell Signaling Technology, Danvers, MA), phospho-HSP27 Ser15 and phospho-HSP27 Ser78 (1:1000; Assay Designs, Ann Arbor, MI). Films were scanned using a Hewlett–Packard 8300 series scanner and images captured using Photoshop Software. Densitometry was measured using ImageJ software (National Institutes of Health, Bethesda, MD) and values were normalized to actin. Values represent the average of the clone pairs in three independent experiments and indicate the fold change versus the average of the values for the two GFP control clones.
Immunofluorescence and confocal microscopy
Cells were plated on coverslips coated with 50 μg/ml FN, 1500–2000 cells per well. Clones were exposed to SF OptiMEM for 24 h and U87D8 cells were exposed to conditioned SF OptiMEM for 3 h. Cells were washed with PBS, fixed with 3% paraformaldehyde and permeabilized with 0.05% Triton X-100. Cells were washed twice with PBS, blocked with 1% bovine serum albumin (BSA) and incubated with TGN46 antibody (1:200; AbD Serotec, Oxford, UK) or EEA1 antibody (1:500; BD Biosciences, San Diego, CA). Cells were washed three times with PBS and incubated with Cy3-labeled goat anti-rabbit or Cy3-labeled goat anti-mouse secondary antibody (each 1:1000; Jackson ImmunoResearch Laboratories, West Grove, PA). Cells were washed three times with PBS, once with deionized water and coverslips were mounted using VECTASHIELD Hard-Set mounting medium (Vector Laboratories, Burlingame, CA). Cells were imaged using a Nikon Confocal Microscope C1 System. Images were captured in 0.5-μm sections to generate single-slice images or whole-cell built images using Nikon EZC1 2.30 software.
Adhesion assay
Ninety-six-well plates were coated with FN as indicated, blocked with 1% BSA and washed with PBS. Cells (one clone per construct) were plated in triplicate, 5000 cells per well in SF OptiMEM, incubated on ice for 30 min and 37°C for 24 h. Loose and non-attached cells were removed by shaking the plates at 350 r.p.m. for 6 min. Adherent cells were washed with PBS, fixed with 1% glutaraldehyde, washed three times with PBS, stained with 0.1% crystal violet and washed again. The dye was solubilized in 1% sodium dodecyl sulfate and quantified by reading absorbance at 540 nm. Data represent the average of three independent experiments.
Wound migration assay
Cells were plated in duplicate, 3 × 105 in 60-mm dishes on 50 μg/ml FN and allowed to grow to confluence. Wounds were made with 3/4-inch razor blades, were imaged and distance measured after 20 h as described previously (29). Data represent the average of the clone pairs from three independent experiments.
Growth curve
For each time point, the eight clones were plated in triplicate (10 000 cells per well) on FN (50 μg/ml) in six-well plates. The next day, day 0 cells were trypsinized, stained with 0.4% trypan blue (Invitrogen, Grand Island, NY) and counted using a hemocytometer. For the remaining cells, media were changed to SF OptiMEM and cells were counted at 24 and 48 h.
Transwell migration assay
For migration of cells through transwell filters with 8-μm pores (Fisher Scientific, Pittsburgh, PA), cells (one clone per construct) were plated in triplicate, 5 × 104 cells per transwell insert in SF OptiMEM. Cells settled for 15 min, then lower chambers were filled with OptiMEM containing 10% fetal bovine serum. After 2 h, cells were fixed, stained with hematoxylin and imaged as described previously (29). Nuclei were counted. The data represent the average number of cells per field for three independent experiments.
Inhibition of p38 MAPK
Cells (one clone per construct) were plated 2 × 105 on FN (100 μg/ml) in growth medium overnight. Media were changed to SF OptiMEM + dimethyl sulfoxide (vehicle control) or the p38 MAPK inhibitor SB203580 [20 μM, Calbiochem, La Jolla, CA (42)] for 24 h as described previously (29). Lysates were collected for western blot analysis as described above. Representative images are presented for four independent experiments.
HSP27 stability
Cells (one clone per construct) were plated 2 × 105 on plastic in growth medium overnight. Media were changed to SF OptiMEM + dimethyl sulfoxide (vehicle control) or cycloheximide [50 μg/ml, Sigma–Aldrich, St Louis, MO (43)]. Lysates were collected at 24, 48 and 72 h for western blot analysis as described above. Data represent the average of three independent experiments.
Statistical analyses
For the wound and transwell assays, the means for each construct were compared using generalized linear mixed models. One-way analysis of variance was used for the western blots. The growth curve, adhesion and cycloheximide assays were analyzed using the Student's t-test. All comparisons were considered significant at the <0.05 level.
Results
Creation of the deletion mutant constructs
The deletion mutants were constructed from a previously created SPARC–GFP fusion plasmid (29). The acidic domain deletion mutant (ΔAcidic) was created by deleting base pairs 109–264 corresponding to amino acids 1–52 (Figure 1). This deletion was expected to have little effect on the proper folding of the construct, since it is loosely folded in native SPARC. To delete the EGF-like module (ΔEGF), base pairs 271–336 were deleted corresponding to amino acids 55–76 (Figure 1). This deletion includes four cysteines that form two disulfide bonds (Cys 1–3 and 2–4) (16). With the bonded cysteines deleted, it was expected that this deletion would have little effect on the folding of the construct. The GFP empty vector (GFP), wt-SPARC–GFP (also referred to as SPARC or SPARC–GFP) and the deletion constructs were transfected into U87MG cells, which express very low levels of endogenous SPARC, and stable clones were selected.
Fig. 1.
Schematic of the deletion constructs. (A) wt-SPARC–GFP contains the acidic domain (aa 1–52; black), follistatin-like domain (aa 53–130; gray with spots), which includes the EGF-like module (aa 55–76; white) and the β1 integrin-binding site (aa 113–130) and the extracellular calcium-binding domain (aa 131–280; solid gray). ES (horizontal stripes) indicates the export signal, which is not part of the mature protein, and GFP (diagonal stripes) refers to the C-terminal GFP tag. (B) The acidic domain deletion mutant construct has amino acids 1–52 of the mature protein deleted. (C) The EGF-like module deletion mutant construct has amino acids 55–76 deleted. aa, amino acids.
Intracellular localization, expression and secretion of all constructs
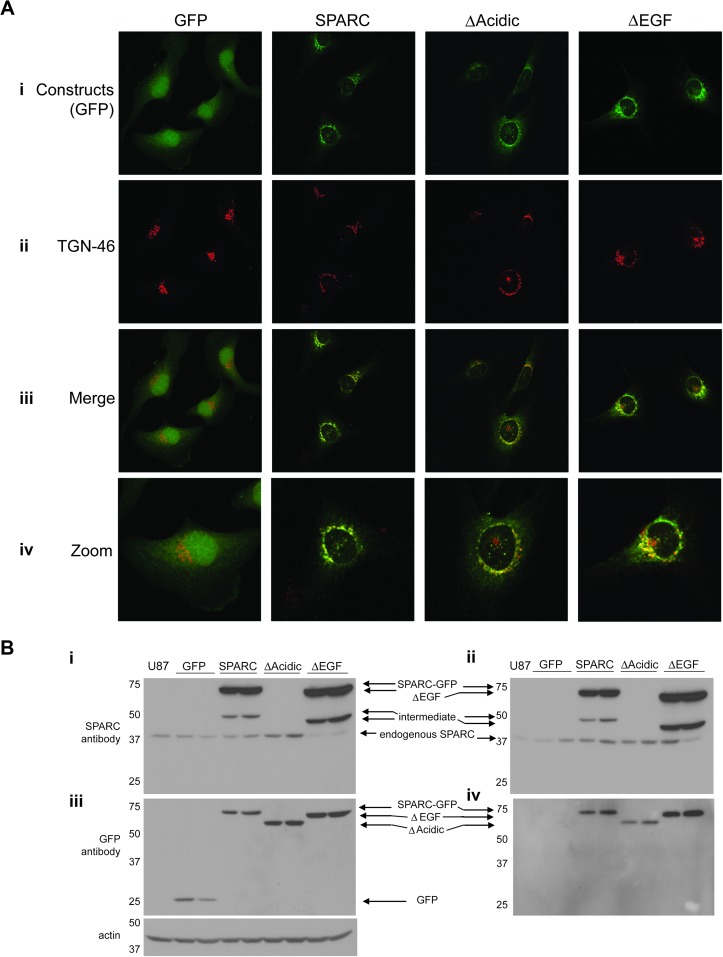
Expression of all constructs was verified by GFP fluorescence confocal imaging (Figure 2A, i). SPARC is secreted through the classical pathway (44) and therefore is processed in the endoplasmic reticulum and the Golgi complex. To ensure that the deletions did not alter protein processing, cells were fixed and immunostained for TGN-46 (Figure 2A, ii), an integral membrane glycoprotein in the trans-Golgi network. Merged GFP and TGN-46 images (Figure 2A, iii and zoom iv) demonstrate that, as expected, GFP is a cytosolic protein and does not co-localize with TGN-46. In contrast, SPARC–GFP and the deletion mutants co-localize with TGN-46, which suggests proper processing of the deletion mutants.
Fig. 2.
Expression and intracellular processing of the constructs. (A) Built confocal images showing one clone expressing each construct. (i) Green fluorescence shows that the control GFP is localized diffusely throughout the cells, whereas SPARC–GFP and the deletion mutants are localized perinuclearly. (ii) TGN-46 immunostaining indicates the trans-Golgi Network. (iii) Merged images indicate co-localization of TGN-46 with SPARC–GFP and both deletion mutants, confirming their localization to the Golgi complex. (iv) Zoomed images better demonstrate co-localization. Images in i–iii were captured at ×60. Zoomed images are ×240. (B) Western blot analyses of cell lysates (i and iii) and conditioned medium (ii and iv) demonstrate the level of intracellular and secreted constructs and endogenous wt-SPARC by the clones. The parental cell line, U87MG, is represented in lane 1 of each blot. (i and ii) Anti-SPARC antibody detects endogenous SPARC, SPARC–GFP and ΔEGF but not ΔAcidic or GFP. The intermediate bands are believed to be due to alternate processing or proteolytic cleavage. (iii and iv) Anti-GFP antibody detects all of the constructs (GFP is not secreted). Actin indicates equal loading of cell lysates. Molecular weights (in kDa) are shown at the left of each blot.
Western blots were used to estimate levels of intracellular and secreted constructs (Figure 2B). Anti-SPARC antibody detects intracellular (Figure 2B, i) and secreted (Figure 2B, ii) SPARC–GFP, ΔEGF and endogenous wt-SPARC. This antibody does not recognize GFP or the ΔAcidic construct. Levels of expression and secretion of all constructs can be seen using an anti-GFP antibody (Figure 2B, iii and iv). GFP is not secreted (Figure 2B, iv). Both deletion mutants are secreted, as expected, since the export signal was not disrupted in either deletion. However, although the expression levels in the lysates of ΔAcidic were similar to SPARC and ΔEGF, the levels in the conditioned medium were consistently lower than that for SPARC and ΔEGF. With the acidic domain being immediately downstream of the export signal, deleting this region may affect secretion.
Effects of the deletions on SPARC uptake by naive U87 cells
SPARC is taken up by normal cells in culture (45–47). SPARC is also taken up by U87MG cells; however, the non-clonal U87MG cell line does not internalize SPARC uniformly (S.A.Rempel, unpublished results) and so we derived a clone from U87MG, termed U87D8. This clone is denoted as naive because it does not express the SPARC–GFP-derived constructs.
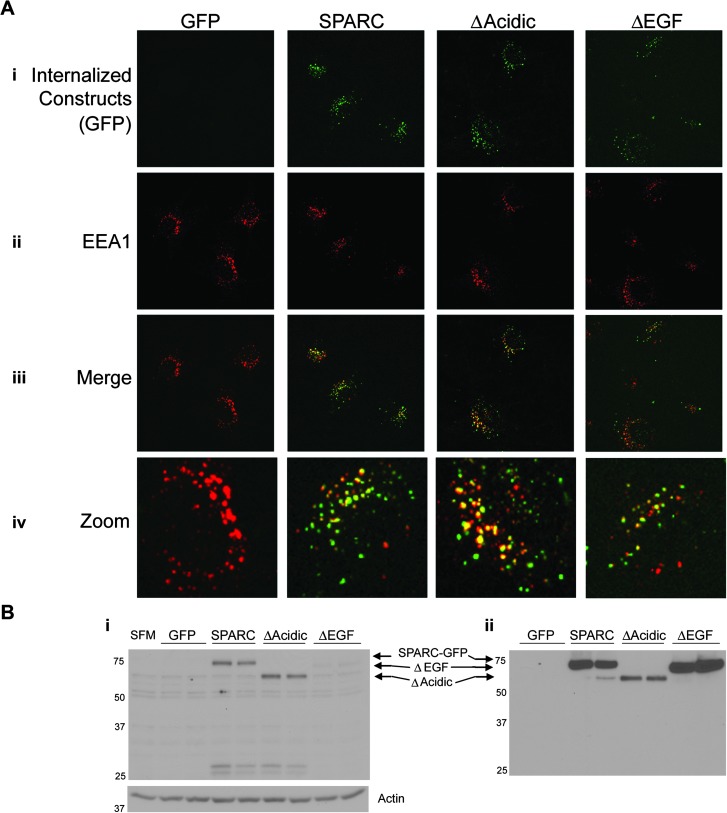
U87D8 cells were exposed to medium conditioned by cells expressing each of the constructs. GFP fluorescence was not detected in cells treated with conditioned medium from GFP-expressing cells (Figure 3A, i) because GFP is not secreted. In contrast, SPARC–GFP, ΔAcidic and ΔEGF were internalized (Figure 3A, i). Cells were immunostained for EEA1, an early endosome marker (Figure 3A, ii). Merged GFP and EEA1 images (Figure 3A, iii and zoom iv) show co-localization, indicating that the constructs are internalized into the early endosome.
Fig. 3.
Internalization of the constructs by naive cells. (A) Built confocal images showing U87D8 exposed to conditioned medium from one clone per construct for 3 h. (i) Internalized constructs. Note that GFP is not secreted and so there is no GFP in the conditioned medium to be taken up by the cells. (ii) EEA1 immunostaining indicates the early endosome. (iii) Merged images show co-localization of EEA1 with SPARC–GFP, ΔAcidic and ΔEGF, indicating that they are internalized into the endosomes. Images i–iii were captured at ×60. (iv) Zoomed images (×240) better demonstrate co-localization. (B) Western blot analysis. (i) Lysates collected from U87D8 after 6 h exposure to conditioned media or SF OptiMEM (SFM) shows that SPARC–GFP and both deletion mutants are present in cell lysates of the naive cells. The blot is labeled according to the conditioned media to which the U87D8 cells were exposed. Actin indicates equal loading of cell lysates. (ii) The level of SPARC–GFP or deletion mutant constructs present in the conditioned media. Molecular weights (in kDa) are indicated at the left of each blot.
The uptake of ΔEGF by U87D8 cells appeared less than SPARC and ΔAcidic (Figure 3A). This was surprising as the amount of ΔEGF in the conditioned media was as high as that observed for SPARC–GFP (Figure 2B). Therefore, we assessed the levels of uptake relative to the amount present in the conditioned media by western blot analysis (Figure 3B). Low levels of internalized constructs were observed within 10 min (data not shown); however, uptake was maximally observed at 6 h (Figure 3B, i) or later (data not shown). Comparing the levels of the constructs in the conditioned medium (Figure 3B, ii) relative to the levels that were internalized by the U87D8 cells (Figure 3B, i), ΔEGF was taken up to a lesser extent, whereas ΔAcidic was taken up to a greater extent than SPARC. Interestingly, degradation products of the SPARC and ΔAcidic constructs, which are presumed to be the GFP tag since it is ∼29 kDa and is detectable with the GFP antibody, were detectable as early as 3 h (also at 6 h as in Figure 3B). However, there was no degradation product observed in the U87D8 cells exposed to ΔEGF conditioned medium at any time point examined and ΔEGF did not accumulate within the cells. These data suggest domain-specific interactions with a cell surface receptor, which could alter SPARC function in adhesion, migration and signaling.
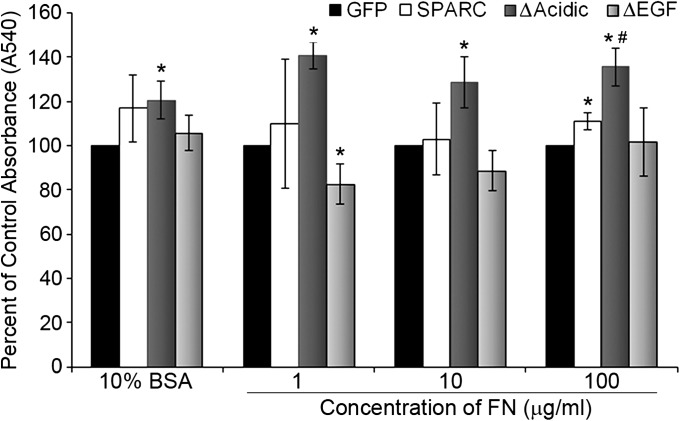
ΔAcidic increases cell adhesion on FN
To determine the effects of the deletions on cell adhesion, clones were plated on 10% BSA (control) or on increasing concentrations of FN. Adhesion at 24 h was studied to correlate the level of adhesion with distance of migration on FN (see below). All cells were more adherent on FN than on 10% BSA (data not shown). The values illustrated in Figure 4 reflect the percent absorbance relative to the control cells for each matrix. Compared with GFP, SPARC did not affect adhesion when cells were plated on 10% BSA or the lower FN concentrations but increased adhesion on 100 μg/ml FN. When plated on 10% BSA, ΔAcidic cells were more adherent than GFP control cells but were not significantly different from SPARC or ΔEGF. ΔAcidic increased adhesion on all levels of FN compared with GFP or ΔEGF. These cells were also more adherent than SPARC on the highest levels of FN. ΔEGF cells were less adherent than GFP cells on the lowest levels of FN but were not significantly different than SPARC on any FN concentration. The opposing effects of the deleted regions may balance out and explain why cells expressing SPARC have similar adhesion to control cells under most conditions.
Fig. 4.
Deletion of the acidic domain increases cell adhesion. Adhesion was measured at 24 h. SPARC-expressing cells were more adherent than GFP cells on the highest level of FN. Expression of ΔAcidic increased adhesion compared with control on all levels of FN and on BSA. ΔAcidic increased adhesion on 100 μg/ml FN compared with SPARC. Expression of ΔEGF decreased adhesion compared with control cells on 1 μg/ml FN. ΔEGF cells were not significantly different from SPARC but were less adherent than ΔAcidic on all concentrations of FN (P ≤ 0.03). Concentration of FN shown is in μg/ml. * Significantly different from GFP (P ≤ 0.036), #Significantly greater than SPARC (P = 0.0095).
Deletion of the acidic domain or EGF-like module reduces SPARC-induced migration
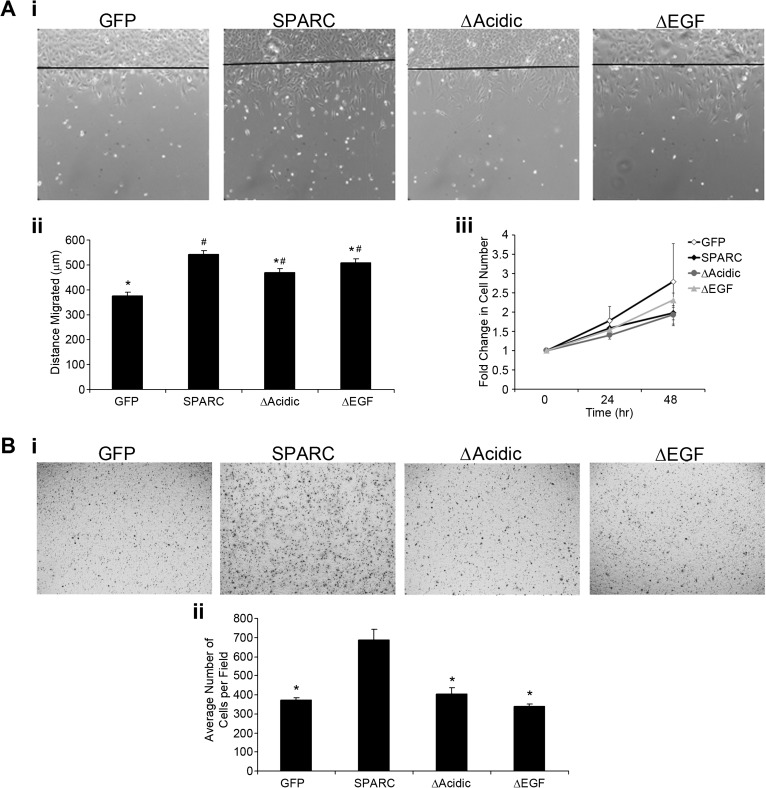
Wound assays were used to measure distance migrated on FN over 20 h. Figure 5A, i shows representative images of one clone for each construct (a line indicates the start of migration) and Figure 5A, ii illustrates the average distance migrated. Consistent with our previous data (29), SPARC-expressing cells migrated farther than GFP-expressing cells. Either deletion resulted in decreased migration relative to that of SPARC-expressing cells; however, migration was greater than that observed for control cells. ΔAcidic had a greater effect on reducing migration than ΔEGF. To show that the cells that are present in the wound area were a result of migration and not due to increased proliferation, cell proliferation was measured over 48 h. Approximately, twice the amount of time allowed for migration was used to amplify subtle changes in proliferation. Figure 5A, iii indicates that there was no difference in proliferation over this period of time.
Fig. 5.
Deletion of the acidic domain or the EGF-like module reduces SPARC-induced migration. (A) Wound migration assay. (i) Representative ×10 images of one clone expressing each construct indicate cell migration from the start of the wound after 20 h. (ii) Average distance migrated for both clones expressing each construct. SPARC significantly increased migration over control. Deletion of the acidic domain or the EGF-like module reduced migration compared with SPARC. The deletion mutants migrated farther than control cells. (iii) Fold change in cell number at 24 and 48 h relative to 0 h indicates that increased migration is not due to increased proliferation. (B) Transwell migration assay. (i) Representative ×10 images of one clone expressing each construct indicate migration through transwell filters after 2 h. (ii) Average number of cells per field. Expression of SPARC–GFP increases cell migration compared with control cells. Both deletions reduced migration to control levels. *Significantly less than SPARC (P ≤ 0.033), #Significantly greater than GFP (P < 0.01).
To confirm the effects of the deletions on cell migration, the transwell assay was used to assess migration as the number of cells that migrated through 8-μm pores. Representative ×10 images are shown (Figure 5B, i). Figure 5B, ii indicates the average number of cells per field. As in the wound assay, SPARC significantly increased migration compared with GFP control cells and both deletion mutants reduced migration. However, in this assay, both deletions reduced migration to control levels. The differences observed between wound and transwell migration assays indicate that the ECM may influence the activity of SPARC and the deletion mutants to affect migration.
ΔAcidic and ΔEGF reduce SPARC-induced activation of the p38 MAPK/HSP27 pathway
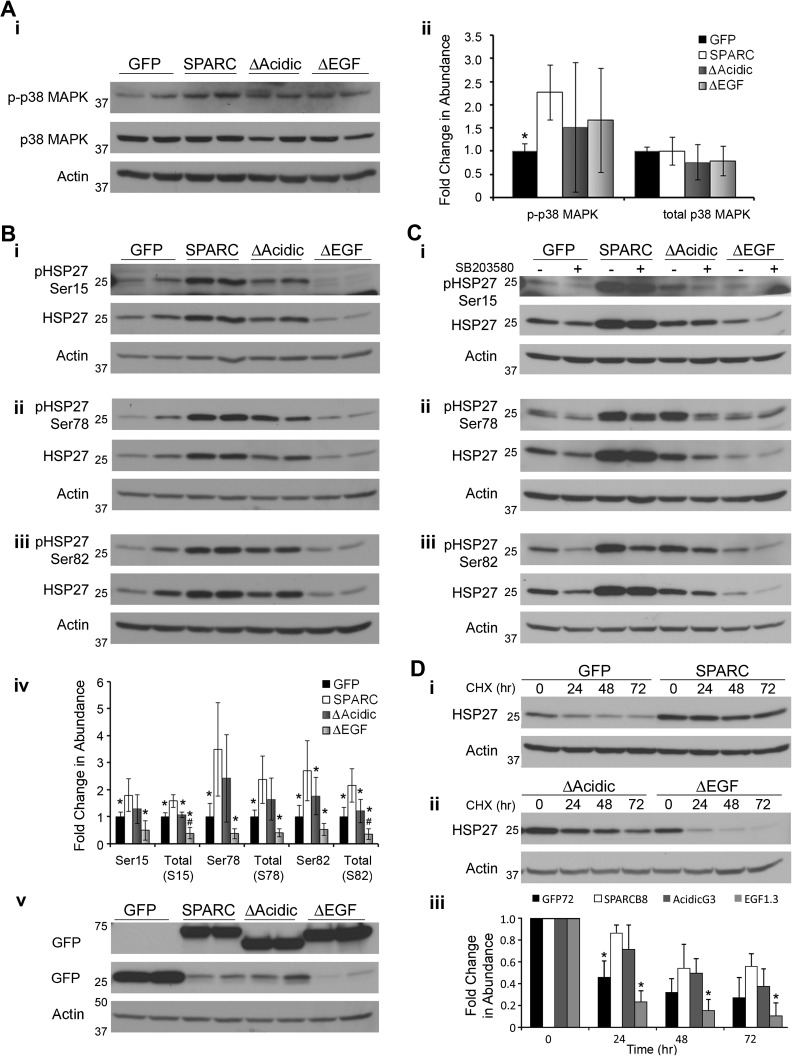
We have previously demonstrated that p38 MAPK and HSP27 mediate SPARC-induced migration (29). We used western blot analysis to determine the effects of the deletions on this signaling pathway (Figure 6). As previously reported, SPARC expression increases p38 MAPK phosphorylation (Figure 6A). p38 MAPK activation was maximal at 3 h when the cells were plated on 100 μg/ml FN. As previously reported, SPARC increases expression of HSP27 and increases phosphorylation of HSP27 at all three serine residues (Figure 6B, i–iii and quantitated in iv). ΔAcidic and ΔEGF showed a trend for intermediate activation of p38 MAPK but were not significantly different from SPARC or GFP cells (Figure 6A). Deletion of the acidic domain resulted in a slight reduction in expression of HSP27 compared with SPARC; however, phosphorylation of HSP27 was only significantly reduced at serine 82. HSP27 expression and phosphorylation were not significantly different between ΔAcidic and GFP cells. Deletion of the EGF-like module reduced expression of HSP27 compared with SPARC, ΔAcidic and GFP cells. Phosphorylation of HSP27 in the ΔEGF cells was significantly less than SPARC and ΔAcidic cells but was not significantly less than phosphorylation of HSP27 in GFP cells. That phosphorylation of HSP27 was through p38 MAPK signaling was confirmed using the inhibitor SB203580 (Figure 6C, i–iii), as previously reported (29).
Fig. 6.
Deletion of the acidic domain or EGF-like module reduces SPARC-mediated signaling through the p38 MAPK/HSP27 pathway. (A) (i) Western blot analysis of expression and activation of p38 MAPK and (ii) densitometric analysis indicate that SPARC increases phosphorylation of p38 MAPK. The deletion mutants were not significantly different from GFP- or SPARC-expressing cells. (B) Western blot analysis of HSP27 expression and phosphorylation at (i) serine 15, (ii) serine 78 and (iii) serine 82 and (iv) densitometric analysis show that SPARC increases HSP27 expression and phosphorylation at all three serines. Deletion of the acidic domain reduces HSP27 expression and phosphorylation at serine 82. ΔAcidic was not significantly different from control cells. Deletion of the EGF-like module decreased HSP27 expression and phosphorylation to or below control levels. Equal expression of the constructs is confirmed using anti-GFP antibody in v. (C) Cells were treated with 20 μM SB203580 for 24 h. Western blot analysis of HSP27 expression and phosphorylation at (i) serine 15, (ii) serine 78 and (iii) serine 82 indicates that HSP27 phosphorylation is mediated by p38 MAPK. (D) Cells were exposed to 50 μg/ml cycloheximide for the times indicated. (i, ii) Western blot analysis and (iii) densitometric analysis indicate that the increased levels of HSP27 in the SPARC-expressing clones is due at least in part to increased protein stability and this increased stability is lost with the deletion of the EGF-like module. * Significantly less than SPARC (P < 0.05), #Significantly less than GFP (P < 0.016). Molecular weights (in kDa) are indicated at the left of each blot.
Because the level of HSP27 in the ΔEGF cells did not correlate with the level of migration in the wound assay, we examined other signaling molecules involved in migration. There was no change in β1 integrin expression (data not shown). Under the conditions and time points tested, we did not see consistent effects on FAK expression or activation by SPARC or the deletion mutants (data not shown). There was also no change in the intracellular localization of phosphorylated FAK as determined by confocal imaging (data not shown). Western blot analysis also indicated no change in the expression or activation of extracellular signal-regulated kinase or myosin light chain (data not shown).
As signaling can also be regulated by the level of total HSP27 available for phosphorylation, we determined whether the increase in total HSP27 induced by SPARC was in part due to increased protein stability. We found that SPARC stabilized HSP27 protein, and the half-life was not reached within 72 h post-cycloheximide treatment (Figure 6D, i). Deletion of the acidic domain had no significant effect, whereas deletion of the EGF-like module shortened the half-life to control levels (Figure 6D, ii). The quantitation in Figure 6D, iii indicates levels of HSP27 in each clone relative to the 0 h time point.
Discussion
SPARC is highly expressed in gliomas and we have demonstrated that it increases migration in vitro (12) and in vivo (13). We have further shown that SPARC increases migration at least in part through the p38 MAPK/HSP27 signaling pathway (29). We have also demonstrated that SPARC suppresses proliferation in vitro (48) and in vivo (13). Thus, therapeutically, it might be useful to inhibit SPARC-induced migration and invasion but retain the negative effects on proliferation. The evidence that different domains of SPARC may be involved in different SPARC functions, prompted us to question whether deletion of specific domains would alter adhesion and/or signaling so as to reduce or eliminate SPARC-induced migration. To this end, we evaluated the effects of deleting the acidic domain and the EGF-like module. We found that deletion of the acidic domain resulted in increased uptake, increased adhesion, decreased p38 MAPK/HSP27 signaling and decreased migration. Deletion of the EGF-like module dramatically reduced the expression of HSP27, due in part to a decrease in the HSP27 protein stability conferred by wt-SPARC. This deletion also resulted in decreased uptake and migration. The decreased uptake suggests that this mutant either cannot bind as efficiently, and then probably would not serve as a therapeutic agent to interfere with wt-SPARC function, or it binds, but cannot transduce the signal, and then would be a good therapeutic agent. Further investigation is necessary to clarify the effects of ΔEGF. In contrast, the acidic domain deletion mutant seems capable of interacting at the cell surface and the increased uptake appears to attenuate signaling and migration. These data suggest that the ΔAcidic construct could interfere with wt-SPARC function to suppress signaling through HSP27 and suppress migration. These studies help to further define the function of these specific domains and suggest potential therapeutic strategies to inhibit SPARC-induced migration.
We first evaluated the effects of the deletions on intracellular protein processing in U87MG cells expressing the constructs. Both deletion mutants were localized to the trans-Golgi network (Figure 2A) and were secreted (Figure 2B), suggesting that the deletions did not significantly alter protein processing. Therefore, the constructs could be used to examine the biological effects of the deletions on SPARC function in internalization, adhesion, migration and signaling.
SPARC is internalized by binding to α5β1 integrin on adipose stromal cells (30). Although the mechanism for uptake of SPARC in glioma cells is unknown, α5β1 integrin is a candidate receptor as U87MG cells express α5β1 [(49); S.A.Rempel, unpublished results]. Both deletion mutants have the β1 integrin-binding region (Figure 1). We investigated the uptake of the constructs and, as expected, both deletion mutants were internalized; however, uptake of ΔEGF was reduced, whereas ΔAcidic was enhanced compared with SPARC (Figure 3). The data suggest that the EGF-like module may increase the affinity of SPARC for β1 integrin. This agrees with an observation by Yost and Sage (25), who demonstrated that a peptide mimicking the EGF-like module was a more potent competitor for cell surface binding than wt-SPARC. Deletion of the acidic domain places the EGF-like module at the N-terminus of the mature protein (Figure 1) and resulted in enhanced uptake compared with SPARC, which further suggests that the EGF-like module promotes the binding of SPARC to β1 integrin. However, it is possible that SPARC binding to the cell surface and internalization are mediated by different receptors, requiring further investigation into these mechanisms.
Previous studies of SPARC peptides involving cell spreading and adhesion prompted us to investigate the effects of the deletion mutants on adhesion, migration and signaling. Cell adhesion and migration are dynamic processes and can be affected by the type and concentration of ECM. We assessed adhesion on increasing levels of FN at 24 h to correlate status of adhesion and distance of migration. We chose to use FN because gliomas tend to migrate along blood vessels (1,14), the site of FN in the normal brain (50). Gliomas can secrete FN (51) and express FN receptors, including the SPARC-binding α5β1 integrin (1,51). Additionally, tumor cells may induce host cells to produce ECM proteins including FN (50).
Although SPARC is a de-adhesive protein when added to normal cells in culture (20,27), these studies did not indicate whether the cells were plated on a matrix. We found that SPARC had no effect on glioma cell adhesion on lower concentrations of FN and increased at the highest concentration of FN. Our results reflect a difference in tumor versus normal cells and/or matrix-specific effects of SPARC. Additionally, our results differ only slightly from previous studies in our laboratory, which showed that SPARC expression did not alter attachment on FN (48), and differences observed may be an effect of the different time points examined. The adhesion results show that the loss of the acidic domain increases adhesion, suggesting that the acidic domain has de-adhesive properties, which is consistent with other reports (20,21). The acidic domain is also involved in the suppressive effect of SPARC on FN production (22). Deletion of this domain would result in greater deposition of FN compared with SPARC and ΔEGF, resulting in increased adhesion. In contrast, deletion of the EGF-like module had a moderately suppressive effect on adhesion. By deleting the EGF-like module, the acidic domain is positioned closer to the β1 integrin-binding site on SPARC and so it may have a greater ability to disrupt the interaction between β1 and FN, resulting in decreased cell adhesion. As with SPARC, this effect was overcome by increasing the concentration of FN. The opposing effects of the two regions on adhesion may balance out and explain why cells expressing SPARC are similarly adherent to control cells on FN.
Both deletion mutants decreased migration when compared with SPARC–GFP. However, the mutant-expressing cells migrated significantly more than the control cells in the wound assay (Figure 5B) and similarly to control cells in the transwell assay (Figure 5E). In the wound assay, cells were plated on FN and migration was along a flat surface, without spatial constraints (Figure 5A). In the transwell assay, ECM was not added to the inserts, but the cells had to move through pores (Figure 5D), which would provide spatial constraint. The differences between the two assays indicate that the actions of the deletion mutants are dependent on the matrix or that they are involved in SPARC signaling to myosin II. Myosin II is required for glioma migration through the pores of transwell filters (52). Others have demonstrated that SPARC activates myosin through ILK (33); however, there were no changes in phosphorylated myosin light chain in our study under the conditions tested. Additionally, we have previously demonstrated that SPARC-induced migration, in the wound assay as well as invasion through Matrigel-coated transwell filters, is inhibited by HSP27 siRNA (29), indicating that SPARC-induced migration, with or without spatial constraints, is mediated by HSP27. Therefore, it seems more likely that the differences observed in migration between the two assays were due to the presence or absence of FN.
We have shown previously that SPARC-induced migration is mediated through the p38 MAPK/HSP27 signaling pathway (29). Using inhibitor SB203580, we demonstrate that the deletion constructs also signal though p38 MAPK to affect HSP27 phosphorylation. In cells expressing ΔAcidic, the decreased migration (Figure 5) correlated with decreased expression of HSP27 and phosphorylation at serine 82 (Figure 6B) compared with SPARC-expressing cells. The ΔAcidic cells were not significantly different from the GFP cells in the level of HSP27 expression and phosphorylation (Figure 6), which correlates well with the similar level of migration as measured by the transwell assay (Figure 5). Since the uptake data suggest that this construct interacts with a cell surface receptor, the decreased signaling indicates that this domain is important in SPARC-induced activation of this pathway.
Despite the low levels of HSP27, clones expressing ΔEGF migrated a greater distance than ΔAcidic and GFP cells in the wound assay (Figure 5). Deletion of the EGF-like module resulted in a reduction in the expression of HSP27 to less than GFP control levels; however, the levels of phosphorylated HSP27 in ΔEGF cells were not significantly less than in GFP cells. The relative levels of phosphorylated and unphosphorylated HSP27 may be critical in the function of this protein. Unphosphorylated HSP27 participates in actin capping (35), whereas phosphorylated HSP27 stabilizes actin filaments at the base of the lamellipodia facilitating migration (53). The high degree of phosphorylation relative to the low level of total HSP27 present in ΔEGF cells may be able to promote migration due to a low availability of unphosphorylated HSP27 to contribute to actin capping. The mechanism by which SPARC upregulates HSP27 is unknown; however, we have previously demonstrated that SPARC upregulates HSP27 transcript abundance (29) and now show that SPARC promotes HSP27 protein stability. The latter function may be due to its proposed intracellular chaperone function (40,41). It is clear from this study that the EGF-like module is essential for SPARC-induced HSP27 protein stability. Future studies of this construct may yield further insight into the SPARC–HSP27 relationship.
Studies have implicated SPARC or SPARC peptides as a therapy for cancers in which SPARC proves a positive prognostic marker (54,55). For example, others have shown that peptides that mimic all or part of the EGF-like module were effective in blocking angiogenesis associated with neuroblastoma (26,55). Our study does not indicate whether the ΔEGF construct would be a good therapeutic agent in gliomas. However, although some of the effects of the ΔAcidic construct must be examined in more detail, expression of this mutant increased glioma cell adhesion and reduced SPARC-induced migration and signaling through HSP27. ΔAcidic has an intact EGF-like module and the absence of the acidic domain places the EGF-like module at the N-terminus of the protein, resulting in a protein that is internalized more efficiently and may compete for wt-SPARC binding but with limited induction of signaling. Therefore, this construct may reduce cell migration and retain anti-angiogenic activity. Further analysis of these constructs as therapeutic agents is warranted.
Funding
National Institutes of Health/National Cancer Institute (R01CA86997 and R01CA138401 to S.A.R.). The authors are grateful to the Barbara Jane Levy family for their continued support.
Acknowledgments
We thank Shelly Yoshida for assistance in generating the EGF-like module deletion construct and Meghan M.Hinds for assistance with western blots and uptake studies. Authors are grateful to Dr Xiao-Ping Yang for assistance with the transwell assay microscopy and Dr Chaya Brodie for use of the confocal microscope.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- BSA
bovine serum albumin
- ECM
extracellular matrix
- EGF
epidermal growth factor
- FN
fibronectin
- GFP
green fluorescent protein
- HSP27
heat shock protein 27
- MAPK
mitogen-activated protein kinase
- PBS
phosphate-buffered saline
- SF
serum-free
- SPARC
secreted protein acidic and rich in cysteine
- wt
wild-type
References
- 1.Rempel SA, et al. Tumor invasiveness and anti-invasion strategies. In: Newton HB, editor. Handbook of Brain Tumor Chemotherapy. 2006. Elsevier Inc., San Diego, CA, pp. 193–218. [Google Scholar]
- 2.Lefranc F, et al. Possible future issues in the treatment of glioblastomas: special emphasis on cell migration and the resistance of migrating glioblastoma cells to apoptosis. J. Clin. Oncol. 2005;23:2411–2422. doi: 10.1200/JCO.2005.03.089. [DOI] [PubMed] [Google Scholar]
- 3.Louis DN. Molecular pathology of malignant gliomas. Annu. Rev. Pathol. 2006;1:97–117. doi: 10.1146/annurev.pathol.1.110304.100043. [DOI] [PubMed] [Google Scholar]
- 4.Demuth T, et al. Molecular mechanisms of glioma cell migration and invasion. J. Neurooncol. 2004;70:217–228. doi: 10.1007/s11060-004-2751-6. [DOI] [PubMed] [Google Scholar]
- 5.Sage H, et al. Characterization of a novel serum albumin-binding glycoprotein secreted by endothelial cells in culture. J. Biol. Chem. 1984;259:3993–4007. [PubMed] [Google Scholar]
- 6.Termine JD, et al. Osteonectin, a bone-specific protein linking mineral to collagen. Cell. 1981;26:99–105. doi: 10.1016/0092-8674(81)90037-4. [DOI] [PubMed] [Google Scholar]
- 7.Mann K, et al. Solubilization of protein BM-40 from a basement membrane tumor with chelating agents and evidence for its identity with osteonectin and SPARC. FEBS. Lett. 1987;218:167–172. doi: 10.1016/0014-5793(87)81040-2. [DOI] [PubMed] [Google Scholar]
- 8.Lane TF, et al. The biology of SPARC, a protein that modulates cell-matrix interactions. FASEB J. 1994;8:163–173. [PubMed] [Google Scholar]
- 9.Sage EH. Terms of attachment: SPARC and tumorigenesis. Nat. Med. 1997;3:144–146. doi: 10.1038/nm0297-144. [DOI] [PubMed] [Google Scholar]
- 10.Framson PE, et al. SPARC and tumor growth: where the seed meets the soil? J. Cell. Biochem. 2004;92:679–690. doi: 10.1002/jcb.20091. [DOI] [PubMed] [Google Scholar]
- 11.Rempel SA, et al. SPARC: a signal of astrocytic neoplastic transformation and reactive response in human primary and xenograft gliomas. J. Neuropathol. Exp. Neurol. 1998;57:1112–1121. doi: 10.1097/00005072-199812000-00002. [DOI] [PubMed] [Google Scholar]
- 12.Golembieski WA, et al. Increased SPARC expression promotes U87 glioblastoma invasion in vitro. Int. J. Dev. Neurosci. 1999;17:463–472. doi: 10.1016/s0736-5748(99)00009-x. [DOI] [PubMed] [Google Scholar]
- 13.Schultz C, et al. Secreted protein acidic and rich in cysteine promotes glioma invasion and delays tumor growth in vivo. Cancer Res. 2002;62:6270–6277. [PubMed] [Google Scholar]
- 14.Rich JN, et al. Bone-related genes expressed in advanced malignancies induce invasion and metastasis in a genetically defined human cancer model. J. Biol. Chem. 2003;278:15951–15957. doi: 10.1074/jbc.M211498200. [DOI] [PubMed] [Google Scholar]
- 15.Maurer P, et al. High-affinity and low-affinity calcium binding and stability of the multidomain extracellular 40-kDa basement membrane glycoprotein (BM-40/SPARC/osteonectin) Eur. J. Biochem. 1992;205:233–240. doi: 10.1111/j.1432-1033.1992.tb16773.x. [DOI] [PubMed] [Google Scholar]
- 16.Hohenester E, et al. Crystal structure of a pair of follistatin-like and EF-hand calcium-binding domains in BM-40. EMBO J. 1997;16:3778–3786. doi: 10.1093/emboj/16.13.3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weaver MS, et al. The copper binding domain of SPARC mediates cell survival in vitro via interaction with integrin beta1 and activation of integrin-linked kinase. J. Biol. Chem. 2008;283:22826–22837. doi: 10.1074/jbc.M706563200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hohenester E, et al. Structure of a novel extracellular Ca(2+)-binding module in BM-40. Nat. Struct. Biol. 1996;3:67–73. doi: 10.1038/nsb0196-67. [DOI] [PubMed] [Google Scholar]
- 19.Busch E, et al. Calcium affinity, cooperativity, and domain interactions of extracellular EF-hands present in BM-40. J. Biol. Chem. 2000;275:25508–25515. doi: 10.1074/jbc.M001770200. [DOI] [PubMed] [Google Scholar]
- 20.Lane TF, et al. Functional mapping of SPARC: peptides from two distinct Ca+(+)-binding sites modulate cell shape. J. Cell. Biol. 1990;111:3065–3076. doi: 10.1083/jcb.111.6.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy-Ullrich JE, et al. SPARC mediates focal adhesion disassembly in endothelial cells through a follistatin-like region and the Ca(2+)-binding EF-hand. J. Cell. Biochem. 1995;57:341–350. doi: 10.1002/jcb.240570218. [DOI] [PubMed] [Google Scholar]
- 22.Lane TF, et al. Regulation of gene expression by SPARC during angiogenesis in vitro. Changes in fibronectin, thrombospondin-1, and plasminogen activator inhibitor-1. J. Biol. Chem. 1992;267:16736–16745. [PubMed] [Google Scholar]
- 23.Funk SE, et al. The Ca2(+)-binding glycoprotein SPARC modulates cell cycle progression in bovine aortic endothelial cells. Proc. Natl. Acad. Sci. USA. 1991;88:2648–2652. doi: 10.1073/pnas.88.7.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Funk SE, et al. Differential effects of SPARC and cationic SPARC peptides on DNA synthesis by endothelial cells and fibroblasts. J. Cell. Physiol. 1993;154:53–63. doi: 10.1002/jcp.1041540108. [DOI] [PubMed] [Google Scholar]
- 25.Yost JC, et al. Specific interaction of SPARC with endothelial cells is mediated through a carboxyl-terminal sequence containing a calcium-binding EF hand. J. Biol. Chem. 1993;268:25790–25796. [PubMed] [Google Scholar]
- 26.Chlenski A, et al. Neuroblastoma angiogenesis is inhibited with a folded synthetic molecule corresponding to the epidermal growth factor-like module of the follistatin domain of SPARC. Cancer Res. 2004;64:7420–7425. doi: 10.1158/0008-5472.CAN-04-2141. [DOI] [PubMed] [Google Scholar]
- 27.Motamed K, et al. SPARC inhibits endothelial cell adhesion but not proliferation through a tyrosine phosphorylation-dependent pathway. J. Cell. Biochem. 1998;70:543–552. doi: 10.1002/(sici)1097-4644(19980915)70:4<543::aid-jcb10>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 28.Mayer U, et al. Calcium-dependent binding of basement membrane protein BM-40 (osteonectin, SPARC) to basement membrane collagen type IV. Eur. J. Biochem. 1991;198:141–150. doi: 10.1111/j.1432-1033.1991.tb15996.x. [DOI] [PubMed] [Google Scholar]
- 29.Golembieski WA, et al. HSP27 mediates SPARC-induced changes in glioma morphology, migration, and invasion. Glia. 2008;56:1061–1075. doi: 10.1002/glia.20679. [DOI] [PubMed] [Google Scholar]
- 30.Nie J, et al. IFATS collection: combinatorial peptides identify alpha5beta1 integrin as a receptor for the matricellular protein SPARC on adipose stromal cells. Stem Cells. 2008;26:2735–2745. doi: 10.1634/stemcells.2008-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas SL, et al. PTEN augments SPARC suppression of proliferation and inhibits SPARC-induced migration by suppressing SHC-RAF-ERK and AKT signaling. Neuro Oncol. 2010;12:941–955. doi: 10.1093/neuonc/noq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi Q, et al. Targeting SPARC expression decreases glioma cellular survival and invasion associated with reduced activities of FAK and ILK kinases. Oncogene. 2007;26:4084–4094. doi: 10.1038/sj.onc.1210181. [DOI] [PubMed] [Google Scholar]
- 33.Barker TH, et al. SPARC regulates extracellular matrix organization through its modulation of integrin-linked kinase activity. J. Biol. Chem. 2005;280:36483–36493. doi: 10.1074/jbc.M504663200. [DOI] [PubMed] [Google Scholar]
- 34.Esfandiarei M, et al. Integrin-linked kinase functions as a downstream signal of platelet-derived growth factor to regulate actin polymerization and vascular smooth muscle cell migration. BMC Cell Biol. 2010;11:16. doi: 10.1186/1471-2121-11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guay J, et al. Regulation of actin filament dynamics by p38 map kinase-mediated phosphorylation of heat shock protein 27. J. Cell Sci. 1997;110(Pt 3):357–368. doi: 10.1242/jcs.110.3.357. [DOI] [PubMed] [Google Scholar]
- 36.Rane MJ, et al. p38 Kinase-dependent MAPKAPK-2 activation functions as 3-phosphoinositide-dependent kinase-2 for Akt in human neutrophils. J. Biol. Chem. 2001;276:3517–3523. doi: 10.1074/jbc.M005953200. [DOI] [PubMed] [Google Scholar]
- 37.Wu R, et al. Hsp27 regulates Akt activation and polymorphonuclear leukocyte apoptosis by scaffolding MK2 to Akt signal complex. J. Biol. Chem. 2007;282:21598–21608. doi: 10.1074/jbc.M611316200. [DOI] [PubMed] [Google Scholar]
- 38.Zheng C, et al. MAPK-activated protein kinase-2 (MK2)-mediated formation and phosphorylation-regulated dissociation of the signal complex consisting of p38, MK2, Akt, and Hsp27. J. Biol. Chem. 2006;281:37215–37226. doi: 10.1074/jbc.M603622200. [DOI] [PubMed] [Google Scholar]
- 39.Schafer C, et al. HSP27 expression regulates CCK-induced changes of the actin cytoskeleton in CHO-CCK-A cells. Am. J. Physiol. 1999;277:C1032–C1043. doi: 10.1152/ajpcell.1999.277.6.C1032. [DOI] [PubMed] [Google Scholar]
- 40.Emerson RO, et al. Chaperone-like activity revealed in the matricellular protein SPARC. J. Cell. Biochem. 2006;98:701–705. doi: 10.1002/jcb.20867. [DOI] [PubMed] [Google Scholar]
- 41.Martinek N, et al. Is SPARC an evolutionarily conserved collagen chaperone? J. Dent. Res. 2007;86:296–305. doi: 10.1177/154405910708600402. [DOI] [PubMed] [Google Scholar]
- 42.Nomura N, et al. Phorbol 12-myristate 13-acetate (PMA)-induced cell migration of glioblastoma cells is mediated via p38 MAPK/HSP27 pathway. Biochem. Pharmacol. 2007;74:690–701. doi: 10.1016/j.bcp.2007.06.018. [DOI] [PubMed] [Google Scholar]
- 43.Mimnaugh EG, et al. Simultaneous inhibition of hsp 90 and the proteasome promotes protein ubiquitination, causes endoplasmic reticulum-derived cytosolic vacuolization, and enhances antitumor activity. Mol. Cancer Ther. 2004;3:551–566. [PubMed] [Google Scholar]
- 44.Mason IJ, et al. Evidence from molecular cloning that SPARC, a major product of mouse embryo parietal endoderm, is related to an endothelial cell ‘culture shock’ glycoprotein of Mr 43,000. EMBO J. 1986;5:1465–1472. doi: 10.1002/j.1460-2075.1986.tb04383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gooden MD, et al. Cell cycle-dependent nuclear location of the matricellular protein SPARC: association with the nuclear matrix. J. Cell. Biochem. 1999;74:152–167. [PubMed] [Google Scholar]
- 46.Kzhyshkowska J, et al. Alternatively activated macrophages regulate extracellular levels of the hormone placental lactogen via receptor-mediated uptake and transcytosis. J. Immunol. 2008;180:3028–3037. doi: 10.4049/jimmunol.180.5.3028. [DOI] [PubMed] [Google Scholar]
- 47.Zhang J, et al. A novel GGA-binding site is required for intracellular sorting mediated by stabilin-1. Mol. Cell. Biol. 2009;29:6097–6105. doi: 10.1128/MCB.00505-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rempel SA, et al. SPARC modulates cell growth, attachment and migration of U87 glioma cells on brain extracellular matrix proteins. J. Neurooncol. 2001;53:149–160. doi: 10.1023/a:1012201300188. [DOI] [PubMed] [Google Scholar]
- 49.Maglott A, et al. The small alpha5beta1 integrin antagonist, SJ749, reduces proliferation and clonogenicity of human astrocytoma cells. Cancer Res. 2006;66:6002–6007. doi: 10.1158/0008-5472.CAN-05-4105. [DOI] [PubMed] [Google Scholar]
- 50.Mahesparan R, et al. Expression of extracellular matrix components in a highly infiltrative in vivo glioma model. Acta. Neuropathol. 2003;105:49–57. doi: 10.1007/s00401-002-0610-0. [DOI] [PubMed] [Google Scholar]
- 51.Ohnishi T, et al. Role of fibronectin-stimulated tumor cell migration in glioma invasion in vivo: clinical significance of fibronectin and fibronectin receptor expressed in human glioma tissues. Clin. Exp. Metastasis. 1998;16:729–741. doi: 10.1023/a:1006532812408. [DOI] [PubMed] [Google Scholar]
- 52.Beadle C, et al. The role of myosin II in glioma invasion of the brain. Mol. Biol. Cell. 2008;19:3357–3368. doi: 10.1091/mbc.E08-03-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pichon S, et al. Control of actin dynamics by p38 MAP kinase—Hsp27 distribution in the lamellipodium of smooth muscle cells. J. Cell Sci. 2004;117:2569–2577. doi: 10.1242/jcs.01110. [DOI] [PubMed] [Google Scholar]
- 54.Atorrasagasti C, et al. Overexpression of SPARC obliterates the in vivo tumorigenicity of human hepatocellular carcinoma cells. Int. J. Cancer. 2010;126:2726–2740. doi: 10.1002/ijc.24966. [DOI] [PubMed] [Google Scholar]
- 55.Chlenski A, et al. Modulation of matrix remodeling by SPARC in neoplastic progression. Semin. Cell Dev. Biol. 2010;21:55–65. doi: 10.1016/j.semcdb.2009.11.018. [DOI] [PubMed] [Google Scholar]