Abstract
A recent genome-wide association study has identified five new genetic variants for prostate cancer susceptibility in a Japanese population, but it is unknown whether these newly identified variants are associated with prostate cancer risk in other populations, including Chinese men. We genotyped these five variants in a case–control study of 1524 patients diagnosed with prostate cancer and 2169 control subjects from the Chinese Consortium for Prostate Cancer Genetics (ChinaPCa). We found that three of the five genetic variants were associated with prostate cancer risk (P = 4.33 × 10−8 for rs12653946 at 5p15, 4.43 × 10−5 for rs339331 at 6q22 and 8.42 × 10−4 for rs9600079 at 13q22, respectively). A cumulative effect was observed in a dose-dependent manner with increasing numbers of risk variant alleles (Ptrend = 2.58 × 10−13), and men with 5–6 risk alleles had a 2-fold higher risk of prostate cancer than men with 0–2 risk alleles (odds ratio = 2.26, 95% confidence interval = 1.78–2.87). Furthermore, rs339331 T allele was significantly associated with RFX6 and GPRC6A higher messenger RNA expression, compared with the C allele. However, none of the variants was associated with clinical stage, Gleason score or family history. These results provide further evidence that the risk loci identified in Japanese men also contribute to prostate cancer susceptibility in Chinese men.
Introduction
Prostate cancer is the most frequently diagnosed cancer and the second leading cause of cancer deaths in most Western counties, with an estimated 217 730 new cases and 32 050 deaths in 2010 in the USA alone (1). The incidence rates of prostate cancer vary substantially worldwide, with a much higher incidence observed in the Western world than in Asian countries (2). Although prostate cancer incidence in China is still low, it is increasing rapidly in recent years (3).
The three well-established risk factors for prostate cancer are age, race and family history (4), although recent genome-wide association studies (GWAS) mostly in Caucasian populations have identified over 30 risk-conferring loci, providing strong support for a genetic component for this disease (5–7). However, the effect of these variants identified in Caucasians on prostate cancer risk in the Chinese populations is largely unknown.
Recently, Takata et al. (8) conducted a GWAS in a Japanese population (4584 men with prostate cancer and 8801 controls) and identified five new loci for prostate cancer susceptibility at 2p24 (rs13385191 in intron 6 of C2orf43), 5p15 (rs12653946), 6p21 (rs1983891 in FOXP4), 6q22 (rs339331 in intron 4 of RFX6) and 13q22 (rs9600079); none of these regions has been previously associated with prostate carcinogenesis. Meta-analyses and other previous GWAS for prostate cancer did not identify these five loci in European populations (9,10), suggesting that these risk loci may be specific to Asian populations.
In the present study, we assessed the association between the five genetic variants identified by Takata et al. in a large case–control study in a Chinese population. We further evaluated the potential effect of these genetic variants on clinical stage, Gleason score, and family history of prostate cancer.
Materials and methods
Study subjects
This study consisted of 1524 prostate cancer patients and 2169 cancer-free controls, who are part of a part of the Chinese Consortium for Prostate Cancer Genetics (ChinaPCa; http://www.chinapca.org) that was established in July 2009. Most of the subjects included in this study were described previously (11–14). Briefly, all cases in ChinaPCa were histologically confirmed prostate cancer subjects recruited from nine hospitals or universities in the Southern and Eastern parts of China, including Shanghai Cancer Institute, Huashan Hospital, Suzhou Municipal Hospital, Changhai Hospital, Xinhua Hospital, Guangxi Medical University, Ninth People’s Hospital, Sun Yat-sen University, Pudong Gongli Hospital, Fudan University and the First Affiliated Hospital of Nanjing Medical University. According to the international tumor-node-metastasis staging system for prostate cancer, the clinical stage was classified into localized and aggressive stage (localized: T1–2N0M0; aggressive: T3–4NxMx or TxN1Mx or TxNxM1). Pathologic grade was recorded as the Gleason score. Control subjects are men without a history of prostate cancer randomly selected from a community in Shanghai, Taizhou and Nanjing and Jiangsu Province of China. Controls were excluded if they had an abnormal prostate-specific antigen level (≥4.0 ng/ml) or abnormal positive digital rectal examination.
At recruitment, study participants were interviewed to confirm age and ascertain information on family history of cancer (defined as cancer in first-degree relatives of parents, siblings or children) and epidemiologic risk factors. After interview, venous blood sample was collected from each subject, from which genomic DNAs were extracted using the QIAamp DNA mini kit (Qiagen). This study was approved by the Institutional Review Boards at Fudan University, Nanjing Medical University, Shanghai Cancer Institute, US National Cancer Institute and each participating hospital.
Single-nucleotide polymorphism selection and genotyping
We selected five genetic variants that have been previously been identified to be associated with prostate cancer in Japanese men (8) for genotyping. Two assay platforms were used for genotyping. For 825 cases and 1379 controls included in the study, genotyping was done on the MassARRAY iPLEX (Sequenom, San Diego, CA) platform through the use of an allele-specific matrix-assisted laser desorption/ionization time-of-flight mass spectrometry assay. For quality control, duplicates from two subjects (as positive controls and for calculating concordance) and two water samples (as negative controls) were included in each 96-well plate; the overall concordance rate was 99% for these five variants among the 58 duplicate samples. The remaining 699 cases and 790 controls (all from Nanjing Medical University) were genotyped using the TaqMan allelic discrimination assay on the 7900HT Real-time PCR System (Applied Biosystems, Foster City, CA); the sequences of primer and probe for each SNP are available on request. For quality control, ∼5% of the samples were randomly selected for duplicate assays; there was 100% concordant. For both assays platforms, technicians who performed the genotyping were blinded to the case–control status of all subjects. About 5% samples were cross-validated by our two platforms, and the concordant was 100%. Furthermore, the results of these SNPs in additive model did not show significant difference in these two genotyping platforms.
Real-time reverse transcription–polymerase chain reaction
Because rs339331 at 6q22 was located in the regions of RFX6 and GPRC6A, we further examined the correlation between rs339331 and the expressions of these two genes. Thirty-four prostate cancer tissues were obtained from patients after prostatectomy. RNA from the tissues with different genotypes was extracted by using the Trizol Reagent (Invitrogen, USA).Total RNA was measured by quantitative real-time reverse transcription–polymerase chain reaction. The primers used for amplification are shown in Supplementary Table I, available at Carcinogenesis Online. GAPDH gene was used as an internal quantitative control, and each assay was done in triplicate.
Statistical methods
Hardy–Weinberg equilibrium of the genotype distribution among controls was tested by a goodness-of-fit chi-square test. Unconditional logistic regression analysis was done to estimate odds ratios (ORs) and their respective 95% confidence intervals (CIs) for risk variants in relation to risk of prostate cancer; variants were assessed in both categorical (heterozygous and homozygous minor allele in relation to homozygous major allele) and additive models (linearly according to 0, 1 or 2 risk alleles). Bonferroni correction for multiple testing was also applied. Linkage disequilibrium was calculated using the Haploview 4.2 software (15), by determining D′ and r2 values. For SNPs that were significant in the main effects analysis, we assessed the cumulative effects of the significant risk loci on prostate cancer, by first summarizing the three SNPs by counting the number of risk alleles in each subject and modeling the summary variable categorically (0–2 as reference, 3, 4 and 5–6) in unconditional logistic regression analysis. Because the cumulative effects on individual SNP could induce a positive bias (16), a permutation method with 10 000 times was used to correct the potential bias. In addition, for cases, we assessed the association of the significant risk variants with pathological characteristics including clinical stage (localized and aggressive tumors), Gleason score (<7 and ≥7) and family history (no and yes). A linear regression model adjusted for age was used to assess the correlation between genotypes (independent variable, coded as 0, 1 or 2) and gene messenger RNA expression levels. All analyses were performed using the Statistical Analysis System (SAS) software (version 9.2; SAS Institute, Cary, NC). All statistical tests were two sided.
Results
A total of 1524 prostate cancer patients and 2169 controls were included in this study. As shown in Table I, the mean age was 71.7 years old for patients and 68.6 years old for controls. The median prostate-specific antigen level among cases was 31.9 ng/ml. Cases were more likely to have family members with cancer than controls (20.9 versus 12.9%, P < 0.001). Among cases, 68.5% of the cases had Gleason score >7 (n = 835). The allele frequencies of five genetic variants among cases and controls and their association with prostate cancer risk are shown in Table II. The observed genotype frequency among the control subjects was in agreement with the Hardy–Weinberg equilibrium, except for one SNP (rs13385191). Significant differences of allele distributions between cases and controls were observed for three of the five SNPs (rs12653946, rs339331 and rs9600079, P = 4.33 × 10−8, 4.43 × 10−5 and 8.42 × 10−4, respectively), which remained significant after Bonferroni correction (P = 2.17 × 10−7, 2.21 × 10−4 and 4.21 × 10−3, respectively). No significant association was found for rs13385191 or rs1983891 (P = 0.481 and 0.842, respectively). Furthermore, results were similar after additional adjustment for age and family history of cancer.
Table I.
Clinical and demographic characteristics of subjects
| Variables | Cases (n = 1524) |
Controls (n = 2169) |
||
| N | % | N | % | |
| Age (mean years ± SD) | 71.7 ± 7.6 | 68.6 ± 6.6 | ||
| Family history | ||||
| No | 797 | 79.1 | 1804 | 87.1 |
| Yes | 210 | 20.9 | 267 | 12.9 |
| PSA (ng/ml; mean ± SD) | 90.9 ± 331.0 | 1.5 ± 2.1 | ||
| Missing data | 179 | 737 | ||
| T-stage | ||||
| No. of subjects | 1254 | NA | ||
| T0 | 3 | 0.2 | ||
| T1 | 253 | 20.2 | ||
| T2 | 498 | 39.7 | ||
| T3 | 367 | 29.3 | ||
| T4 | 83 | 6.6 | ||
| Tx | 50 | 4.0 | ||
| Missing data | 270 | |||
| N-stage | ||||
| No. of subjects | 1253 | NA | ||
| N0 | 810 | 64.6 | ||
| N1 | 297 | 23.7 | ||
| Nx | 146 | 11.7 | ||
| Missing data | 271 | |||
| M-stage | ||||
| No. of subjects | 1240 | NA | ||
| M0 | 794 | 64.0 | ||
| M1 | 426 | 34.4 | ||
| Mx | 20 | 1.6 | ||
| Missing data | 284 | |||
| Gleason score | ||||
| No. of subjects | 1219 | NA | ||
| <7 | 384 | 31.5 | ||
| ≥7 | 835 | 68.5 | ||
| Missing data | 305 | |||
PSA, prostate-specific antigen.
Table II.
Association of five previously identified SNPs with prostate cancer in a Chinese population
| SNP ID | Regiona | Locationb | Allele (major/minor) | Risk allele | Minor allele frequency |
PHWEc | OR (95% CI)d |
Pe | Pf | |||
| Cases | Controls | Heterozygous | Homozygous | Additive model | ||||||||
| rs13385191 | 2p24 | 20751746 | G/A | A | 0.442 | 0.433 | 0.003 | 1.08 (0.93–1.25) | 1.02 (0.85–1.23) | 1.03 (0.94–1.13) | 0.48 | |
| rs12653946 | 5p15 | 1948829 | C/T | T | 0.481 | 0.354 | 0.321 | 1.30 (1.12–1.50) | 1.70 (1.39–2.07) | 1.30 (1.19–1.43) | 4.33 × 10−8 | 2.17 × 10−7 |
| rs1983891 | 6p21 | 41644405 | C/T | C | 0.323 | 0.325 | 0.459 | 1.00 (0.87–1.15) | 0.88 (0.70–1.11) | 0.99 (0.90–1.09) | 0.84 | |
| rs339331 | 6q22 | 117316745 | T/C | T | 0.311 | 0.358 | 0.385 | 0.88 (0.77–1.01) | 0.68 (0.54–0.85) | 0.81 (0.74–0.90) | 4.43 × 10−5 | 2.21 × 10−4 |
| rs9600079 | 13q22 | 72626140 | G/T | T | 0.485 | 0.445 | 0.778 | 1.25 (1.08–1.46) | 1.31 (1.08–1.58) | 1.18 (1.07–1.30) | 8.42 × 10−4 | 4.21 × 10−3 |
Relative to SNP position.
Based on the NCBI database, build 36.
Hardy–Weinberg equilibrium (HWE) among control subjects.
Reference genotype or allele is major homozygote or allele.
Additive model.
After Bonferroni correction.
The linkage disequilibrium analysis revealed that these five SNPs were in weak linkage disequilibrium (all r2 = 0.00 for each pair of the loci). We further investigated the cumulative effects of the three significant loci (rs12653946, rs339331 and rs9600079). As shown in Table III, individuals with multiple risk alleles had a higher risk of prostate cancer, compared with those with 0–2 risk alleles of the three variants, with a dose-dependent manner with increasing numbers of risk variant alleles conferring increasing risk (Ptrend = 2.58 × 10−13). Specifically, compared with patients with 0–2 risk alleles, patients carrying 3, 4 or 5–6 risk alleles had ORs of 1.43 (95% CI = 1.21–1.69), 1.62 (95% CI = 1.35–1.94) or 2.26 (95% CI = 1.78–2.87), respectively. These permutation-adjusted OR showed the similar results. In addition, we assessed the cumulative effects of all the five loci (rs13385191, rs12653946, rs1983891, rs339331 and rs9600079) and found that individuals multiple risk alleles also had an increased risk of prostate cancer (Ptrend = 5.51 × 10−10).
Table III.
Cumulative effect of the three risk loci on the risk of prostate cancer
| No. of risk allelesa | Cases |
Controls |
OR (95% CI) | P | OR (95% CI)b | ||
| N | % | N | % | ||||
| 0–2 | 404 | 27.9 | 787 | 38.4 | 1.00 (reference) | 1.00 (reference) | |
| 3 | 459 | 31.7 | 626 | 30.5 | 1.43 (1.21–1.69) | 3.95 × 10−5 | 1.42 (1.21–1.68) |
| 4 | 390 | 26.9 | 469 | 22.9 | 1.62 (1.35–1.94) | 1.52 × 10−7 | 1.62 (1.35–1.94) |
| 5–6 | 196 | 13.5 | 169 | 8.2 | 2.26 (1.78–2.87) | 1.98 × 10−11 | 2.26 (1.78–2.87) |
| Ptrend | 2.58 × 10−13 | ||||||
Risk alleles included rs12653946 T, rs339331 T and rs9600079 T alleles.
Based on 10 000 permutations.
The association between the three risk variants (rs12653946, rs339331 and rs9600079) and prostate cancer stratified by clinical stage (aggressive versus localized), Gleason score (≥7 versus <7) and family history of cancer (yes versus no) was further evaluated among cases. As shown in Table IV, none of the three genetic variants were significantly associated with the clinical stage, Gleason score and family of history of cancer among cases. Furthermore, no association with these clinical variables was found when multiple variants associated with prostate cancer were considered simultaneously (data not shown).
Table IV.
Association of the three genetic variants with pathologic characteristics of prostate cancer and family history of cancer among cases
| Variable | No. of subjects | rs12653946 |
No. of subjects | rs339331 |
No. of subjects | rs9600079 |
|||||||||
| CC | CT | TT | OR (95% CI)a | TT | CT | CC | OR (95% CI)a | GG | GT | TT | OR (95% CI)a | ||||
| Clinical stage | |||||||||||||||
| Localized | 537 | 117 | 261 | 99 | 1.00 (reference) | 532 | 242 | 235 | 55 | 1.00 (reference) | 521 | 128 | 289 | 104 | 1.00 (reference) |
| Aggressive | 692 | 240 | 327 | 125 | 0.96 (0.82–1.13) | 690 | 332 | 295 | 63 | 0.91 (0.77–1.09) | 675 | 180 | 333 | 162 | 1.04 (0.88–1.23) |
| P | 0.61 | 0.31 | 0.63 | ||||||||||||
| Gleason score | |||||||||||||||
| <7 | 383 | 135 | 174 | 74 | 1.00 (reference) | 378 | 168 | 173 | 37 | 1.00 (reference) | 368 | 94 | 200 | 74 | 1.00 (reference) |
| ≥7 | 828 | 279 | 407 | 142 | 0.99 (0.83–1.17) | 824 | 392 | 355 | 77 | 0.92 (0.76–1.10) | 809 | 208 | 410 | 191 | 1.07 (0.90–1.28) |
| P | 0.89 | 0.38 | 0.45 | ||||||||||||
| Family history | |||||||||||||||
| No | 793 | 270 | 380 | 143 | 1.00 (reference) | 791 | 375 | 338 | 78 | 1.00 (reference) | 775 | 192 | 416 | 167 | 1.00 (reference) |
| Yes | 209 | 77 | 91 | 41 | 0.98 (0.79–1.21) | 204 | 99 | 91 | 14 | 0.91 (0.71–1.15) | 201 | 66 | 89 | 46 | 0.87 (0.69–1.09) |
| P | 0.83 | 0.42 | 0.22 | ||||||||||||
Additive model.
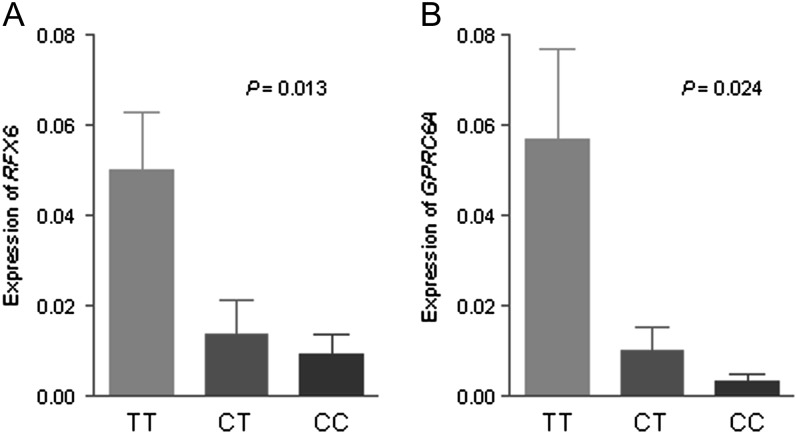
We assessed rs339331 in RFX6 and GPRC6A genes expression in 34 prostate cancer tissues using quantitative real-time reverse transcription–polymerase chain reaction. A significant association between rs339331 and RFX6 expression was observed (P = 0.013). Similarly, rs339331 was significantly associated with the expression of GPRC6A in this study (P = 0.024) (Figure 1).
Fig. 1.
Association between rs339331 and messenger RNA expression of (A) RFX6 and (B) GPRC6A. The transcript in prostate cancer tissues was detected by quantitative real-time reverse transcription–polymerase chain reaction. The frequency distributions of the TT, CT and CC genotypes were 16, 14 and 4, respectively. The fold change was normalized against GAPDH using the ΔΔCt method. The P-value was calculated with a linear regression model adjusted for age.
Discussion
In this large study among Chinese men, we found that three (rs12653946, rs339331 and rs9600079) of the five genetic variants reported in Japanese men were also associated with prostate cancer risk in Chinese men. Furthermore, there seems to be a cumulative effect of the three significant genetic variants with increasing risk in a dose-dependent fashion.
In this study, rs12653946 was the most significant prostate cancer susceptibility locus for Chinese men, which is consistent with the GWAS findings in Japanese men (8). Both this variant and rs9600079 on 13q22 are located in regions with no known genes, similar to the genetic variants reported in other GWAS of Caucasian populations (17).
A third locus found to be significantly associated with prostate cancer risk in our Chinese population was rs339331 at 6q22, which was mapped in the region including two genes, RFX6 and GPRC6A. RFX6 is a member of the regulatory factor X family of transcription factors, which has been shown to be primarily expressed in pancreas and with a lower expression in the liver, prostate and other tissues (18). The function of the RFX6 in the prostate is unknown, although it has been shown that mutations of this gene have a deleterious effect on the development of the pancreas (19). GPRC6A is a member of the G protein-coupled receptors family C, which is broadly expressed in many tissues and organs, including the testis (20–22). Pi et al. (23) showed that Gprc6a-null mice had altered levels of circulating testosterone and estrogen, which led to feminization of male mice, hyperglycemia among other phenotypes. We have also provided in vivo evidence that rs339331 was associated with RFX6 and GPRC6A messenger RNA expression, suggesting that a plausible relationship between rs339331 and prostate cancer.
In the present study, we found no evidence for an association of genetic variants with the aggressive stage, high Gleason score and family history of cancer. In the GWAS in Japanese men, none of the loci was found to be associated with the risk of aggressive prostate cancer either (8). In previous GWAS of European populations, none of the nearly 30 reported loci was consistently associated with aggressive prostate cancer, though some studies have reported stronger effects in more aggressive as compared with less aggressive tumors (24–26). Therefore, another approach or more genetic variants are needed to search for genetic markers in clinical utility (7,27).
In our study, we found that three of significant variants (rs12653946, rs339331 and rs9600079) had risk estimates in the same direction as that reported for Japanese men, and three significant variants were associated with a range of ORs of 1.03–1.30, which had a strong cumulative association with prostate cancer risk with the increasing risk alleles. In the study by Takata et al. (8), the effect sizes of each variant were in the range of ORs of 1.11–1.31, which showed a moderate association with prostate cancer risk. Men who carried 5–6 risk alleles had a 2.19-fold increased risk of developing prostate cancer compared with those who carrying 0–2 risk alleles, indicating the importance of the combined effects from independent risk loci in prostate carcinogenesis. However, the use of cumulative effects on these SNPs has to be confirmed in a prospective study before it could be used for the prostate cancer risk assessment.
Unlike the Japanese study, we did not observe any association between two genetic variants, rs13385191 and rs1983891 and the risk of prostate cancer. There are several possible reasons for our discrepant findings. First, the current sample size could have insufficient power to detect the modest effect sizes since many GWAS risk variants were found only after pooling several large GWAS. Second, different environmental exposures may affect the degree to which genes are activated and thereby modulating the extent of the association. Future studies with larger sample sizes and more detailed environmental exposure are needed to confirm this observation.
In conclusion, our study confirmed that three of the risk loci identified in Japanese men (rs12653946 at 5p15, rs339331 at 6q22 and rs9600079 at 13q22 but not rs13385191 at 2p24 and rs1983891 at 6p21) also affect susceptibility to prostate cancer in Chinese men.
These findings provide new insights into prostate cancer etiology, the causal variants/genes associated with these risk regions need to be characterized further fine-mapping and functional studies.
Supplementary material
Supplementary Table I can be found at http://carcin.oxfordjournals.org/
Funding
This study was partly supported by National Cancer Institute (1R01CA129684-01), National Natural Science Foundation of China (30872084, 30972444, 81102089), the Key Program of Natural Science Foundation of Jiangsu Province (BK2010080), Natural Science Foundation of Jiangsu Province (BK2011773) and the Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (Public Health and Preventive Medicine).
Supplementary Material
Acknowledgments
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- CI
confidence intervals
- GWAS
genome-wide association study
- OR
odds ratio
- SNP
single-nucleotide polymorphism
References
- 1.Jemal A, et al. Cancer statistics. CA Cancer J. Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Matsuda T, et al. Comparison of time trends in prostate cancer incidence (1973–2002) in Asia, from cancer incidence in five continents, Vols IV-IX. Jpn. J. Clin. Oncol. 2009;39:468–469. doi: 10.1093/jjco/hyp077. [DOI] [PubMed] [Google Scholar]
- 3.Jian L, et al. Protective effect of green tea against prostate cancer: a case-control study in southeast China. Int. J. Cancer. 2004;108:130–135. doi: 10.1002/ijc.11550. [DOI] [PubMed] [Google Scholar]
- 4.Gronberg H. Prostate cancer epidemiology. Lancet. 2003;361:859–864. doi: 10.1016/S0140-6736(03)12713-4. [DOI] [PubMed] [Google Scholar]
- 5.Manolio TA, et al. A HapMap harvest of insights into the genetics of common disease. J. Clin. Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rahim NG, et al. Genetic determinants of phenotypic diversity in humans. Genome Biol. 2008;9:215. doi: 10.1186/gb-2008-9-4-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Witte JS. Prostate cancer genomics: towards a new understanding. Nat. Rev. Genet. 2009;10:77–82. doi: 10.1038/nrg2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takata R, et al. Genome-wide association study identifies five new susceptibility loci for prostate cancer in the Japanese population. Nat. Genet. 2010;42:751–754. doi: 10.1038/ng.635. [DOI] [PubMed] [Google Scholar]
- 9.Gudmundsson J, et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat. Genet. 2009;41:1122–1126. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eeles RA, et al. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat. Genet. 2009;41:1116–1121. doi: 10.1038/ng.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu F, et al. Systematic confirmation study of reported prostate cancer risk-associated single nucleotide polymorphisms in Chinese men. Cancer Sci. 2011;102:1916–1920. doi: 10.1111/j.1349-7006.2011.02036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shao P, et al. Functional polymorphisms in cell death pathway genes FAS and FAS ligand and risk of prostate cancer in a Chinese population. Prostate. 2011;71:1122–1130. doi: 10.1002/pros.21328. [DOI] [PubMed] [Google Scholar]
- 13.Xu B, et al. A functional polymorphism in MSMB gene promoter is associated with prostate cancer risk and serum MSMB expression. Prostate. 2010;70:1146–1152. doi: 10.1002/pros.21149. [DOI] [PubMed] [Google Scholar]
- 14.Zheng SL, et al. Association of 17 prostate cancer susceptibility loci with prostate cancer risk in Chinese men. Prostate. 2010;70:425–432. doi: 10.1002/pros.21076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrett JC, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 16.Lee WC, et al. Data-dredging gene-dose analyses in association studies: biases and their corrections. Cancer Epidemiol. Biomarkers Prev. 2005;14:3004–3006. doi: 10.1158/1055-9965.EPI-05-0605. [DOI] [PubMed] [Google Scholar]
- 17.Ghoussaini M, et al. Multiple loci with different cancer specificities within the 8q24 gene desert. J. Natl Cancer Inst. 2008;100:962–966. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aftab S, et al. Identification and characterization of novel human tissue-specific RFX transcription factors. BMC Evol. Biol. 2008;8:226. doi: 10.1186/1471-2148-8-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith SB, et al. Rfx6 directs islet formation and insulin production in mice and humans. Nature. 2010;463:775–780. doi: 10.1038/nature08748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pi M, et al. Identification of a novel extracellular cation-sensing G-protein-coupled receptor. J. Biol. Chem. 2005;280:40201–40209. doi: 10.1074/jbc.M505186200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wellendorph P, et al. Molecular cloning, expression, and sequence analysis of GPRC6A, a novel family C G-protein-coupled receptor. Gene. 2004;335:37–46. doi: 10.1016/j.gene.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 22.Kuang D, et al. Cloning and characterization of a family C orphan G-protein coupled receptor. J. Neurochem. 2005;93:383–391. doi: 10.1111/j.1471-4159.2005.03025.x. [DOI] [PubMed] [Google Scholar]
- 23.Pi M, et al. GPRC6A null mice exhibit osteopenia, feminization and metabolic syndrome. PLoS One. 2008;3:e3858. doi: 10.1371/journal.pone.0003858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamada H, et al. Replication of prostate cancer risk loci in a Japanese case-control association study. J. Natl Cancer Inst. 2009;101:1330–1336. doi: 10.1093/jnci/djp287. [DOI] [PubMed] [Google Scholar]
- 25.Kader AK, et al. Individual and cumulative effect of prostate cancer risk-associated variants on clinicopathologic variables in 5,895 prostate cancer patients. Prostate. 2009;69:1195–1205. doi: 10.1002/pros.20970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kote-Jarai Z, et al. Multiple novel prostate cancer predisposition loci confirmed by an international study: the PRACTICAL Consortium. Cancer Epidemiol. Biomarkers Prev. 2008;17:2052–2061. doi: 10.1158/1055-9965.EPI-08-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gelmann EP. Complexities of prostate-cancer risk. N. Engl. J. Med. 2008;358:961–963. doi: 10.1056/NEJMe0708703. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.