Abstract
In this study, both culture-dependent and culture-independent methods were used to determine whether the iron sulfide mineral- and nitrate-rich freshwater nature reserve Het Zwart Water accommodates anoxic microbial iron cycling. Molecular analyses (16S rRNA gene clone library and fluorescence in situ hybridization, FISH) showed that sulfur-oxidizing denitrifiers dominated the microbial population. In addition, bacteria resembling the iron-oxidizing, nitrate-reducing Acidovorax strain BrG1 accounted for a major part of the microbial community in the groundwater of this ecosystem. Despite the apparent abundance of strain BrG1-like bacteria, iron-oxidizing nitrate reducers could not be isolated, likely due to the strictly autotrophic cultivation conditions adopted in our study. In contrast an iron-reducing Geobacter sp. was isolated from this environment while FISH and 16S rRNA gene clone library analyses did not reveal any Geobacter sp.-related sequences in the groundwater. Our findings indicate that iron-oxidizing nitrate reducers may be of importance to the redox cycling of iron in the groundwater of our study site and illustrate the necessity of employing both culture-dependent and independent methods in studies on microbial processes.
Keywords: freshwater anoxic iron bacteria, cultivation, nitrate, FISH, clone library
Introduction
Redox cycling of iron has profound effects on the chemistry of soils and sediments due to the sheer abundance of iron, and the interdependence with the cycling of virtually all other biochemically relevant elements (Stumm and Sulzberger, 1992; Davison, 1993; Nealson et al., 2002). Iron species are subject to both chemical and microbial transformations. Iron can be abiotically reduced in various manners. Photoreduction, and interactions with sulfhydryl groups, organic acids, and reducing ligands, all result in reduction of ferric oxides (Nealson and Saffarini, 1994; Castillo-Gonzalez and Bruns, 2005). In sulfur-rich, mainly marine, environments, hydrogen sulfide reduces ferric iron oxides which results in the formation of iron sulfide (FeS), pyrite (FeS2), and elemental sulfur (Thamdrup et al., 1994; Carey and Taillefert, 2005). In non-sulfidogenic anoxic soils and sediments however, microbial iron reduction, coupled to organic carbon or hydrogen oxidation, is the dominant process (Sobolev and Roden, 2002). The importance of microbial iron reduction has been unequivocally demonstrated: in many environments it accounts for significant turnover of organic carbon, and has inhibitory effects on sulfate reduction and methanogenesis (Nealson and Saffarini, 1994; Thamdrup, 2000; Nealson et al., 2002). The first described iron-reducing bacterial species were fermenting bacteria that make use of the reduction as a means to dispose of electron equivalents (Lovley and Phillips, 1988). Nowadays, also a large variety of microorganisms, ranging from slightly psychrophilic to thermophilic, have been described that use energy generated through iron reduction for growth. Examples of such dissimilatory iron reducers are Geobacter sp., Rhodoferax ferrireducens, Geothrix fermentans, Ferribacterium limneticum, Geoglobus ahangari, and Shewanella sp. These microorganisms couple iron reduction to oxidation of a large variety of organic compounds, hydrogen, or elemental sulfur (Nealson and Saffarini, 1994; Cummings et al., 1999; Thamdrup, 2000; Tor et al., 2001; Kashefi et al., 2002; Nevin and Lovley, 2002; Finneran et al., 2003). Especially Geobacter species have received considerable attention because of their ability to convert pollutants such as uranium, aromatic hydrocarbons, and chlorinated solvents (Anderson et al., 1998; Lin et al., 2005; Nevin et al., 2005; Sung et al., 2006).
The microbial contribution to iron oxidation has long been subject of debate between microbiologists and geochemists (Ghiorse, 1984), especially because abiotic iron oxidation occurs rapidly in the presence of oxygen (Sobolev and Roden, 2002; Emerson and Weiss, 2004). Under anoxic conditions, nitrite is a chemical oxidant of ferrous iron (Moraghan and Buresh, 1977; Cooper et al., 2003; Coby and Picardal, 2005), although this may require millimolar amounts. In spite of these chemical reactions, it is presently accepted that microorganisms can contribute significantly to iron oxidation. Aerobic, acidophilic iron-oxidizing microorganisms play an important role in the generation of acid mine drainage (AMD; Baker and Banfield, 2003). Aerobic, neutrophilic, iron-oxidizing microorganisms, that have to compete with the rapid chemical oxidation, have been isolated from marine as well as freshwater environments, ranging from iron seeps, groundwater, and hydrothermal vents to root-plaque of wetland plants (Emerson and Revsbech, 1994; Emerson and Moyer, 1997; Emerson et al., 1999; Edwards et al., 2003, 2004). The ability to gain energy from anaerobic oxidation of iron is widespread among bacteria. Ferrous iron serves as an electron donor for phototrophic, purple, non-sulfur bacteria, and green sulfur bacteria under anoxic conditions (Ehrenreich and Widdel, 1994; Heising et al., 1999). Several bacterial species and an archeon, Ferroglobus placidus, have been described that couple ferrous iron oxidation to nitrate reduction (Hafenbradl et al., 1996; Benz et al., 1998; Straub et al., 2004; Senko et al., 2005; Kumaraswamy et al., 2006; Weber et al., 2006). It has been suggested that on a global scale nitrate-reducing, iron-oxidizing bacteria are more important than the phototrophs, since they are not limited by the availability of light (Straub et al., 2001).
Straub et al. (1998) used ferrihydrite produced by iron-oxidizing nitrate reducers to enrich iron-reducing bacteria. Indications for the coexistence of aerobic iron-oxidizing and anaerobic iron-reducing microorganisms in close proximity to each other were found by Weiss et al. (2003) and Roden et al. (2004). Furthermore, microbial anoxic iron cycling by a single organism (a Geobacter sp.) capable of both dissimilatory iron reduction and nitrate-dependent iron oxidation was proposed by Weber et al. (2005) and iron respiration with formate as electron donor as well as iron oxidation with nitrate as electron acceptor was demonstrated for the anaerobic ammonium-oxidizing bacterium Kuenenia stuttgartiensis (Strous et al., 2006).
Iron sulfide mineral- and nitrate-rich soils in freshwater ecosystems provide both a source of reduced iron and an electron acceptor that could promote growth of anaerobic, non-phototrophic iron-oxidizers. Oxidized iron species resultant from iron-oxidizing, nitrate-reducing activity could in turn serve as electron acceptors for microbial iron reducers. To determine whether the iron sulfide mineral- and nitrate-rich freshwater environment of the nature reserve Het Zwart Water (Venlo, The Netherlands) provides a suitable habitat for anoxic microbial iron cycling, groundwater from an iron sulfide/and nitrate-rich soil was examined using both culture-dependent and culture-independent methods.
Materials and Methods
Sampling procedure
In October 2004, a groundwater sample was collected in the freshwater nature reserve Het Zwart Water (51°24′N; 6°11′E, Venlo, The Netherlands) at 1.5 m depth, at a site situated 1.5 m from the edge of lake De Venkoelen. Previous measurements had shown increased concentrations of acid volatile sulfur (an indication for the presence of iron sulfide minerals) nitrate, soluble iron, and sulfate around the groundwater table of this system (Haaijer et al., 2006). The collected groundwater was enriched in iron(hydr)oxides (visible orange precipitates). The sample was transported to the lab in a glass serum bottle (10 ml) closed with an aluminum crimp seal. Immediately upon arrival at the lab, high molecular weight DNA was extracted by standard procedures from 0.2 ml of sample and 0.3 ml of the sample was fixed for fluorescence in situ hybridization (FISH) by addition of 4% w/v paraformaldehyde (0.9 ml), incubating on ice (2 h), centrifuging (15 min, 13.000 rpm), and washing the resulting pellet with phosphate buffered saline (PBS) and finally adding 100% EtOH and PBS (1:1) after which the sample was stored at −20°C until analysis. Furthermore, mineral media enrichment cultures (20 ml, for composition, see Haaijer et al., 2006) were established through 10-fold dilution and supplemented with either ferrous iron chloride and nitrate (iron oxidizer culture medium) or ferric iron citrate and acetate (iron reducer culture medium), from anaerobic sterile stock solutions (5 mM final concentrations). These enrichment cultures were incubated under a gas atmosphere of N2/CO2 (80/20%), and incubated at 30°C and 150 rpm for 1.5 months to determine whether the groundwater possessed anoxic iron-oxidizing respectively iron-reducing capacity. All further liquid culturing was performed under the same conditions for at least 3 weeks with final concentrations of 8 mM ferrous iron and 2 mM nitrate in the iron oxidizer culture medium and 8 mM ferric iron citrate and 1.5 mM acetate in the iron reducer culture medium unless specified otherwise.
Enrichment and isolation of anaerobic iron-oxidizing and iron-reducing bacteria
The initial iron-oxidizing and reducing enrichment cultures were transferred by 10-fold dilution and after 1.5 months of incubation used to inoculate iron oxidizer and iron reducer dilution series (10 ml tubes, 10-fold dilutions down to 1010 dilution). These dilution series were incubated for 1 month without agitation. The three highest dilutions (106, 107, and 108 for the iron reducer series, 103, 104, and 105 for the iron oxidizer series) exhibiting activity (changes in color: green to whitish-orange for the iron oxidizer, orange to white/green for the iron reducer series) were then used to inoculate new iron oxidizer and reducer cultures (20 ml volume, 20-fold dilution). Ferric and ferrous iron, nitrate, and nitrite concentrations were determined (see below) at least once a week. When a substrate was depleted, new additions were done from sterile stock solutions. The same procedure was applied to subsequent subcultures.
The 20-ml iron-oxidizing, nitrate-reducing culture obtained from the 105 dilution was used to inoculate a larger volume enrichment culture (50-fold dilution, 50 ml) that was transferred once (50-fold dilution, 50 ml). To obtain template DNA for PCR (see below), 25 ml of this last culture was centrifuged (40 × g, 1 min) to pellet ferrous iron precipitates, the supernatant taken and centrifuged (2600 × g, 5 min) to pellet the bacteria, and the resulting pellet was used to isolate high molecular weight DNA.
The 20-ml iron-reducing culture obtained from the 108 dilution was used to inoculate a larger volume enrichment culture (50-fold dilution, 50 ml). This culture was then transferred once (50-fold dilution, 50 ml) and from the resulting culture a 500-ml enrichment (100-fold dilution, incubation at room temperature, magnetic stirring: 150 rpm) was derived. Medium of this culture was replenished when ferric iron was depleted by allowing it to settle for at least 6 h and decanting after which fresh medium was added and the anaerobic gas atmosphere re-applied. The medium was replenished three times before DNA for PCR was extracted from 25 ml culture material as described above. The 500-ml culture was transferred six times (10-fold dilution). To exclude non-iron-reducing contaminants, the iron-reducing culture was checked by plating 1 ml portions on rich Luria Bertani (LB) medium supplemented with 20 mM nitrate and solidified with agar (15 g l−1). These plates were incubated at 30°C under a N2/CO2 80/20% gas atmosphere. As long as growth was observed on these plates, iron-reducing dilution series were repeated (3×). When no more contaminant, non-iron-reducing, growth was detected, concentrations of acetate, ferric iron, and ferrous iron were monitored in a 50-ml iron-reducing culture over a 90-h period. After this, 0.3 ml of the culture was fixed for FISH analyses as described before. To obtain single colonies of iron-reducing bacteria, liquid iron reducer mineral medium was solidified with agarose (15 g l−1) and inoculated with 1 ml aliquots from iron-reducing cultures and incubated at 30°C under a N2/CO2 80/20% gas atmosphere for at least 1 month.
Isolation of Thiobacillus denitrificans strain K6.2: A sulfur-oxidizing denitrifier
An enrichment culture capable of iron sulfide oxidation at the expense of nitrate had been obtained previously from an iron sulfide-rich soil layer (Haaijer et al., 2006) and was used to isolate a sulfur-oxidizing denitrifier (Thiobacillus denitrificans strain K6.2). In short, a dilution series (10 consecutive 10 ml volume 10-fold dilutions) in mineral medium (for composition: see Haaijer et al., 2006) amended with 5 mM thiosulfate and 8 mM nitrate per dilution was derived from the enrichment. After incubation at 30°C, 150 rpm under a gas atmosphere of N2/CO2 (80/20%) for 2 weeks, the highest dilution (105) exhibiting growth was identified by visual inspection (turbidity) and used to establish a 50-ml culture which was subsequently transferred twice (20-fold dilution after 1 month of incubation). Aliquots of culture material were checked for the presence of heterotrophic contaminants as described above for the iron reducer cultures. Finally, half of the final pure subculture’s volume was used to extract DNA for PCR and 16S rRNA gene sequence analysis as described below.
Chemical analysis
Nitrate, nitrite, ferrous iron, and total iron analyses were performed as described previously (Haaijer et al., 2006, 2007). Acetate was measured through injections (10 μl) of acidified (5% v/v formic acid) samples on a Hewlett–Packard model 5890 gas chromatograph equipped with a flame ionization detector (195°C) and a HP INNOwax column (30 m × 0.32 m × 0.5 μm). Column temperature was increased from 35 to 110°C. Isobutyric acid (0.4 mM final concentration) was added as an internal standard.
16S rRNA gene sequence analysis
Because most samples contained a high amount of iron species, isolated high molecular weight DNA was purified prior to PCR (QIAEX II gel extraction kit, Qiagen). Hot-start PCR reactions were performed in a Tgradient PCR apparatus (Whatman Biometra, Göttingen, Germany). Primer specifications were described previously (Haaijer et al., 2006). The general bacterial 16S rRNA gene PCR primers 616F and 630R yielded products of about 1500 bp. Direct sequencing was performed on the PCR product obtained for the pure culture of T. denitrificans strain K6.2 using sequencing primers M13F, M13R, 1390R, and 610IIF. PCR products of the iron-reducing and oxidizing enrichments were purified once again prior to cloning. The TOPO TA cloning kit was used according to the instructions supplied by the manufacturer (Invitrogen, Groningen, The Netherlands). Isolation of plasmid DNA was performed with the FlexiPrep kit (Amersham Pharmacia P-L Biochemicals Inc.). Clones were checked by restriction analysis of plasmid DNA (EcoR1, Fermentas UAB, Vilnius, Lithuania). A clone library of the groundwater was constructed by partial sequencing (594–840 bp fragments) of 67 clones with primer 610IIF. Preliminary screening of the iron-oxidizing and iron-reducing cultures was performed by partial sequencing (10 clones and 5 clones respectively) with primer 610IIF. One of the clones from the iron-reducing culture was selected for full-sequencing with sequencing primers M13F, M13R, 1390R, and 610IIF. In addition, a persistent non-iron-reducing contaminant growing on rich medium plates inoculated with material from the iron-reducing culture was identified by using colony material suspended in MQ as a PCR template, followed by direct partial sequencing with primer 610IIF. 16S rRNA gene sequences were compared with their closest relatives by BLASTN searches1 or by pairwise alignment with selected relatives2. With the RDP classifier tool3 (Wang et al., 2007), clones were assigned to the taxonomical hierarchy proposed in Bergey’s Manual of Systematic Bacteriology, release 6.04. Further phylogenetic and molecular evolutionary analyses were conducted using the software program MEGA version 5.0 (Tamura et al., 2011). The clone library data was organized by defining operational taxonomic units (OTU’s) in which sequences exhibited ≥97% sequence identity. To evaluate the sampling effort in the clone library (Singleton et al., 2001), the coverage of the library (C) was calculated according to the equation C = [1 − (n1/N)] × 100%, where n1 is the number of OTUs representing a single clone, and N is the total number of clones analyzed.
Accession numbers
Sequences (16S rRNA gene) from the groundwater clone library are available in GenBank under accession numbers DQ997625–DQ997691. Sequences from cultures are available under accession number EF014493 (representative clone oxo8 from the iron-oxidizing, nitrate-reducing culture), EF014495 (nearly full-length sequence from the iron-reducing culture), EF014494 (contaminant in the iron-reducing culture), and EF079668 (nearly full-length sequence of T. denitrificans strain K6.2).
FISH and microscopic inspections
Fluorescence in situ hybridization analyses were performed as described before (Haaijer et al., 2006). Vectashield (Vector Laboratories, Inc., Burlingame, CA, USA) mounting medium with DAPI (4,6-diamidino-2-phenylindole) was used to enhance the fluorescent signal and stain all DNA. Probes were selected based on the sequencing results from the clone library and cultures. A bacterial probe mix of Cy-5-labeled probes EUB338, EUB338 II, and EUB338 III (Daims et al., 1999) was used to hybridize all Bacteria. Probes Geo 1423, EPS 681 (targeting Geobacter sp. respectively Epsilonproteobacteria, Haaijer et al., 2008), Betthio 1001 (betaproteobacterial Thiobacilli, Haaijer et al., 2006), BrG1-829, and BrG2-830 (iron-oxidizing denitrifiers, Straub and Buchholz-Cleven, 1998) were used to specifically detect organisms of interest. The total cell number (DAPI) in the groundwater was determined by analysis of 50 images. To quantify specific probe signals, 10 images were analyzed for each probe. As a control, background fluorescence was determined by analysis of 10 images of DAPI-stained groundwater without probes. Specific probe signals were only regarded as significant when they exceeded this number (5 ± 1.8% of the total DAPI count) in a statistically significant manner (Student’s t-test).
Results
Sample geochemistry
The groundwater sample was collected in the freshwater nature reserve Het Zwart Water at 1.5 m depth, at a site situated 1.5 m from the edge of lake De Venkoelen. High acid volatile sulfur (60 nmol g DW soil−1; an indication for the presence of iron sulfide minerals) nitrate (22 nmol g DW soil−1), soluble iron (25 nmol g DW soil−1), and sulfate (90 nmol g DW soil−1) concentrations were observed around the groundwater table (van der Welle, unpublished results). The collected groundwater was enriched in iron(hydr)oxides visible as orange precipitates.
Cell count and 16S rRNA clone library
The iron sulfide- and nitrate-rich freshwater wetland Het Zwart Water was assumed to provide a suitable environment for anoxic iron cycling bacteria. The groundwater collected from Het Zwart Water contained 8 ± 3 × 107 ml−1 bacteria based on the enumeration of DAPI-stained cells. The 16S rRNA gene sequence-based clone library derived from the bacteria present in the groundwater had a coverage of 66% and resulted in the identification of 30 separate OTU’s. Table 1 provides an overview of these OTU’s and provides information on the microbial processes that the first BLASTN hit with a cultured species is associated with for each OTU whenever such physiological information was available. If no published data was available for the microbial processes associated with the first BLASTN hit with a cultured species from GenBank, source information is listed in Table 1 instead. An exception was made for OTU 1 which encompasses 16S rRNA gene sequences within the genus Thiobacillus. This genus harbors both species converting nitrate only to nitrite (e.g., Thiobacillus thioparus) and T. denitrificans a species capable of complete denitrification to dinitrogen. These species share a high 16S rRNA gene sequence similarity of about 98%. Therefore, a comparison was made for OTU 1 sequences with the 16S rRNA gene sequence of the denitrifying, sulfur-oxidizing T. denitrificans strain K6.2 obtained within this study.
Table 1.
Overview clone library data from the groundwater of Het Zwart Water.
| OTU* | No. of clones | Taxonomical assignment# | Comparison species$ | Sequence identity to comparison species (%) | Association of comparison species@ |
|---|---|---|---|---|---|
| 1 | 28 | Thiobacillus | Thiobacillus denitrificans K6.2 (EF079668) | 98–100 | Reduced sulfur oxidation coupled to nitrate reduction |
| 2 | 2 | Thiobacillus | Thiobacillus thioparus THI111 (HM535226) | 95–97 | Aerobic reduced sulfur oxidation |
| 3 | 1 | Alcaligenaceae | Achromobacter xylosoxidans S18 (JF703286) | 100 | Nitrification |
| 4 | 1 | β-Proteobacteria | Nitrosospira Nsp62 (AY123790) | 94 | Aerobic ammonia oxidation |
| 5 | 3 | β-Proteobacteria | Sideroxydans lithotrophicus ES1 (CP001965) | 99 | Microaerophilic iron oxidation |
| 6 | 1 | β-Proteobacteria | C. Nitrotoga arctica (DQ839562) | 99 | Aerobic nitrite oxidation |
| 7 | 2 | β-Proteobacteria | C. Nitrotoga HAM-1 (FJ236061) | 97–98 | Aerobic nitrite oxidation |
| 8 | 1 | Aquabacterium | Aquabacterium parvum B6 (AF035052) | 97 | Organic acid oxidation |
| 9 | 1 | Comamonadaceae | Acidovorax P-9 (AM411932) | 98 | Oxic zone of unplanted paddy soil microcosms |
| 10 | 1 | Comamonadaceae | Rhodoferax antarcticus (GU233447) | 98 | Phototrophy |
| 11 | 6 | Acidovorax | Acidovorax BrG1 (U51101) | 99–100 | Iron oxidation coupled to nitrate reduction |
| 12 | 1 | Hydrogenophilus | Hydrogenophilus hirschii (FR749905) | 99 | Thermophilic, aerobic hydrogen oxidation |
| 13 | 1 | Klebsiella | Klebsiella pneumoniae L13 (HQ398862) | 99 | Biodegradation of cypermethrin |
| 14 | 2 | Serratia | Serratia marcescens AGPim1A (JF683415) | 99 | Capsicum annuum |
| 15 | 1 | γ-Proteobacteria | Ectothiorhodospira LO12 (EU908047) | 89 | Alkaline environment |
| 16 | 1 | Stenotrophomonas | Stenotrophomonas maltophilia YSP48 (JF894170) | 99 | Petroleum polluted soil |
| 17 | 1 | Rhizobium | Enrichment clone Y92 (HM597705) | 99 | Rumen |
| 18 | 1 | Bradyrhizobium | Bradyrhizobium IV-102 (AB531426) | 99 | Grassland soil |
| 19 | 1 | Proteobacteria | Enrichment clone 04SU4-P (HQ436504) | 86 | Freshwater wetland |
| 20 | 1 | Acetobacteraceae | Bacterium Ellin5134 (AY234551) | 95 | Pasture soil core |
| 21 | 1 | Proteobacteria | Desulfobacca acetoxidans DSM 11109 (CP002629) | 87 | Acetate degradation, sulfate reduction |
| 22 | 1 | Acidobacteria Gp1 | C. Koribacter versatilis Ellin345 (CP000360) | 95 | Aerobic heterotrophy |
| 23 | 1 | Acidobacteria Gp1 | Bacterium Ellin5058 (AY234475) | 97 | Pasture soil core |
| 24 | 1 | Bacteria | Enrichment clone AOM-SR-B36 (HQ405620) | 86 | Anaerobic methane-degrading bioreactor |
| 25 | 1 | Acidobacteria Gp3 | Bacterium Ellin6505 (HM748653) | 96 | Soil |
| 26 | 1 | Acidobacteria Gp3 | Bacterium Ellin6505 (HM748653) | 92 | Soil |
| 27 | 1 | Bacteria | C. Magnetobacterium bavaricum (FP929063) | 88 | Lake sediment |
| 28 | 1 | Actinobacteria | Actinobacterium TC4 (F510471) | 92 | Leachate chalcopyrite bio-heap |
| 29 | 1 | Acidobacteria Gp23 | Desulfacinum subterraneum (AF385080) | 86 | Thermophilic sulfate reduction |
| 30 | 1 | Clostridiales | Clostridium 6–31 (FJ808611) | 95 | Biocompost |
*An OTU consists of sequences exhibiting ≥97% sequence identity; #based on the RDP classifier tool (http://rdp.cme.msu.edu/classifier/; Wang et al., 2007); $in case of OTU 1 the 16S rRNA gene sequence of the denitrifying Thiobacillus denitrificans strain K6.2 was used, for the others the first BLASTN hit of a cultured species was used; @if an association with a specific process was found this process is mentioned, otherwise the environment from which the sequence was retrieved is mentioned.
As described below, the culture-independent and culture-dependent methods used in this study, yielded contrasting results for iron-oxidizing nitrate reducers, and iron reducers, respectively. In addition, sulfur-oxidizing denitrifiers were found the most abundant physiologically distinct group.
Iron-oxidizing, nitrate reducers
The clone library data from the groundwater (Table 1) contained multiple (OTU11, 9% of total clones) sequences strongly resembling (≥97% sequence identity) the iron-oxidizing, nitrate-reducing Acidovorax strain BrG1 (Buchholz-Cleven et al., 1997; Straub et al., 2004). FISH analysis of the groundwater sample, with a combination of probe BrG1-829, and competitor probe BrG2-830, showed the in situ abundance of strain BrG1-like microorganisms (25 ± 9% of the total DAPI count). The primary iron-oxidizing, nitrate-reducing enrichment culture derived from the groundwater exhibited activity (visible as a color change from green to whitish-orange due to the oxidation of ferrous iron). The dilution series derived from this primary enrichment demonstrated the presence of 105 ml−1 iron-oxidizing bacteria. Subsequent subcultures exhibited iron-oxidizing, nitrate-reducing activity. Stoichiometry of these cultures was in good agreement with the theoretical values of 10 mol ferrous oxidized at the expense of 2 mol of nitrate. In one of the subcultures for example 1.5 mM of ferrous iron was oxidized at the expense of 0.4 mM nitrate, accompanied by a minor formation of nitrite (0.05 mM).
Despite the abundance of Acidovorax strain BrG1 iron-oxidizing denitrifiers suggested by the clone library and FISH analyses data, these organisms were not detected in the final 50 ml volume enrichment culture by 16S rRNA analysis. Instead, a Klebsiella-like bacterium was detected. The consensus 16S rRNA gene sequence obtained from the enrichment (10 clones, 830 bp), was identical to the sequence of representative clone oxo8 and exhibited 99% sequence similarity to the Klebsiella pneumoniae strain described by Ludwig et al., 1995, accession number X87276). The clone library of the original sample contained a clone (Table 1; OTU13) with 99% sequence identity to the 16S rRNA gene sequence obtained from the culture, indicating the presence of this organism in the groundwater. Upon further transfer of the iron-oxidizing, nitrate-reducing culture, the activity was lost and addition of acetate as an organic co-substrate did not restore activity (data not shown).
Iron reducers
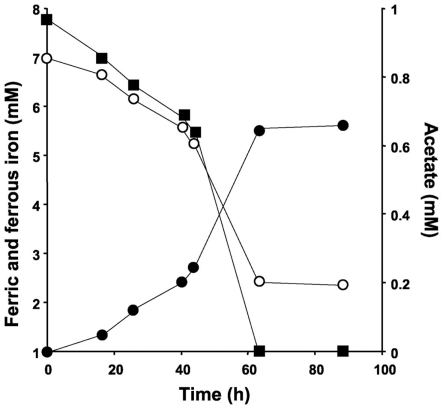
No 16S rRNA gene sequences of known iron-reducing species could be identified within the clone library data. FISH analysis of the groundwater using Geobacter-specific probe Geo1423 did not yield significant hybridization (<5 ± 1.8% of the total DAPI count cells hybridized). The iron-reducing dilution series of the initial enrichment culture however showed that this culture contained 108 ml−1 iron-reducing bacteria. Subcultures were readily obtained and very active: the medium of all subcultures needed to be replenished at least once a week. 16S rRNA gene sequence analysis demonstrated the presence of a Geobacter sp. in the final enrichment. The consensus sequence (five clones, 804 bp) and the nearly full-length 16S rRNA gene sequence (1526 bp) generated exhibited 100% sequence identity. Both sequences exhibited 99% sequence identity to the Geobacter sp. previously described by Cummings et al., 2000, accession number Y19191). Plating on rich medium showed a persistent contamination with a Citrobacter freundii-like organism. The partial 16S rRNA gene sequence of 684 bp obtained from this organism exhibited 99% sequence identity to C. freundii strain DSM 30039 (accession number AJ233408, Sproer et al., 1999). The C. freundii-like organism did not possess any iron-reducing capacity as demonstrated by a lack of iron reduction after 2 months of incubation of 250 ml iron reducer medium inoculated with this organism. Incubation of mineral media plates containing ferric citrate and acetate and inoculated with iron reducer enrichment material yielded growth of the contaminant bacterium only. The activity of the, through repetitive serial dilutions, obtained pure Geobacter sp. culture is shown in Figure 1. Over a period of 63 h, 1 mM of acetate was consumed and 5 mM ferric iron reduced. FISH analysis further confirmed the pure culture status because all DAPI-stained cells hybridized with the Geobacter-specific probe Geo1423.
Figure 1.
Activity of the pure iron-reducing Geobacter sp. culture. The closed squares indicate acetate, the closed circles ferrous iron, and the open circles ferric iron.
Sulfur-oxidizing nitrate reducers
The clone library data (Table 1; OTU 1) revealed 42% of the clones exhibited a sequence identity higher than 98% to the 16S rRNA gene sequence of T. denitrificans strain K6.2. FISH analysis of the groundwater material from Het Zwart Water with betaproteobacterial Thiobacilli-specific probe Betthio1001 corroborated the abundance of these microorganisms (23 ± 9% of the total DAPI count). Iron sulfide- and nitrate-rich environments like Het Zwart Water will be enriched in sulfide in addition to reduced iron as a possible electron donor for microbial growth. In addition to betaproteobacterial Thiobacilli, likely candidates for sulfide oxidation at the expense of nitrate are epsilonproteobacterial species like Sulfurimonas denitrificans (previously known as Thiomicrospira denitrificans; Schippers and Jørgensen, 2002; Takai et al., 2006) and members of the genus Sulfuricurvum (Kodama and Watanabe, 2004). Significant numbers of Epsilonproteobacteria were not detected (probe EPS 681) in the groundwater. In addition, the clone library did not contain epsilonproteobacterial sequences (Table 1).
Discussion
Iron cycle bacteria in the groundwater of Het Zwart Water
Although T. denitrificans possesses iron-oxidizing capacity in the presence of nitrate, specific reaction rates are much lower, weeks opposed to days, than those of the specific iron-oxidizing nitrate reducers previously described by Straub et al. (1996). Given the abundance of strain BrG1-like bacteria, it therefore seems most likely that the betaproteobacterial Thiobacilli detected in the groundwater are primarily involved in the oxidation of reduced sulfur compounds in Het Zwart Water as was demonstrated before (Haaijer et al., 2006, 2007).
Based on the abundance of Acidovorax strain BrG1-like bacteria, suggested by the clone library and FISH analyses data, these microorganisms are proposed as the mediators for anoxic iron oxidation in the groundwater from Het Zwart Water. In the iron sulfide- and nitrate-rich environment of Het Zwart Water these bacteria could be of particular importance. Edwards et al. (2004) demonstrated accelerated iron sulfide mineral dissolution in the presence of iron-oxidizing bacteria. This hypothesis is supported by earlier findings of the presence of this type of bacterium in an iron sulfide- and nitrate-fed enrichment culture (Haaijer et al., 2006). However, despite their abundance, strain BrG1-like bacteria were not detected in the final 50 ml volume enrichment culture. Instead, a Klebsiella-like bacterium was detected. Iron-oxidizing, nitrate-reducing activity by Klebsiella sp. cells has been described by Senko et al. (2005) under non-growth conditions. It therefore seems likely the K. pneumoniae-like organism detected in the iron-oxidizing nitrate-reducing culture did not possess the capacity of generating sufficient energy for growth by coupled iron oxidation and nitrate reduction. This fact together with the absence of any obvious iron-reducing bacteria in the groundwater, as demonstrated with the molecular methods, shows that complete microbial iron cycling could not be convincingly demonstrated in the groundwater of Het Zwart Water. The isolation of the iron-reducing Geobacter sp., however, indicates that bacterial iron reduction at the least could be important in other compartments (e.g., the surrounding soil) of Het Zwart Water.
The iron-reducing Geobacter isolate converted 1 mM of acetate at the expense of 5 mM ferric iron and FISH analysis confirmed the pure culture status. This ratio does not agree well with the theoretical stoichiometry of 8 mM ferric iron reduced at the expense of 1 mM acetate (Lovley et al., 1987). The contribution of assimilation to the total amount of acetate consumed, however, can be high: 46% of total acetate metabolized was assimilated by Geobacter sulfurreducens grown in batch culture on fumarate and acetate (Galushko and Schink, 2000). This phenomenon may also explain the observed low amount of ferric iron reduced in the Geobacter sp. culture described presently.
Differences in the detection of iron-oxidizing nitrate reducers and iron reducers
Molecular analyses (16S rRNA gene clone library and FISH) showed that the Acidovorax strain BrG1-like, iron-oxidizing nitrate reducers accounted for a major portion of the microbial community of the groundwater sample from Het Zwart Water. In contrast to the abundance demonstrated by the molecular data, the cultivation procedure did not lead to the enrichment or isolation of this type of bacterium. A probable cause is the completely autotrophic culturing conditions adopted in the iron-oxidizing enrichments. Iron-oxidizing nitrate-reducing activity was first reported (Straub et al., 1996) for a solely lithotrophic enrichment culture. Isolated strains, however, seem dependent on the presence of organic co-substrates (Straub et al., 2004). In contrast to the results for iron-oxidizers, an iron-reducing Geobacter sp. culture was readily obtained. The molecular analyses, however, suggested the contribution of this bacterium was of little numerical relevance in the groundwater sample. Together, these results, conflicting as they are for the functional groups of interest in this study, illustrate the necessity to employ both culture-dependent and culture-independent methods in studies on microbial activity. To choose only one approach is to disregard the biases inherent to this approach, such as primer-induced biases in clone libraries (Forney et al., 2004), or selection of fast-growing microorganisms in culture media (Grosskopf et al., 1998). Inventory descriptive studies like ours, allow development of more direct approaches, necessary to understand ecosystem functioning.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Fons Smolders, Jack Van de Vossenberg, and Marcus Schmid for advice and discussion.
Footnotes
References
- Anderson R. T., Rooney-Varga J. N., Gaw C. V., Lovley D. R. (1998). Anaerobic benzene oxidation in the Fe(III) reduction zone of petroleum-contaminated aquifers. Environ. Sci. Technol. 32, 1222–1229 10.1021/es9704949 [DOI] [Google Scholar]
- Baker B. J., Banfield J. F. (2003). Microbial communities in acid mine drainage. FEMS Microbiol. Ecol. 44, 139–152 10.1016/S0168-6496(03)00028-X [DOI] [PubMed] [Google Scholar]
- Benz M., Brune A., Schink B. (1998). Anaerobic and aerobic oxidation of ferrous iron at neutral pH by chemoheterotrophic nitrate-reducing bacteria. Arch. Microbiol. 169, 159–165 10.1007/s002030050555 [DOI] [PubMed] [Google Scholar]
- Buchholz-Cleven B. E. E., Rattunde B., Straub K. L. (1997). Screening for genetic diversity of isolates of anaerobic Fe(II)-oxidizing bacteria using DGGE and whole-cell hybridization. Syst. Appl. Microbiol. 20, 301–309 10.1016/S0723-2020(97)80077-X [DOI] [Google Scholar]
- Carey E., Taillefert M. (2005). The role of soluble Fe(III) in the cycling of iron and sulfur in coastal marine sediments. Limnol. Oceanogr. 50, 1129–1141 10.4319/lo.2005.50.4.1129 [DOI] [Google Scholar]
- Castillo-Gonzalez H. A., Bruns M. A. (2005). Dissimilatory iron reduction and odor indicator abatement by biofilm communities in swine manure microcosms. Appl. Environ. Microbiol. 71, 4972–4978 10.1128/AEM.71.9.4972-4978.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coby A. J., Picardal F. W. (2005). Inhibition of NO3- and NO2- reduction by microbial Fe(III) reduction: evidence of a reaction between NO2- and cell surface-bound Fe2+. Appl. Environ. Microbiol. 71, 5267–5274 10.1128/AEM.71.9.5267-5274.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper D. C., Picardal F. W., Schimmelmann A., Coby A. J. (2003). Chemical and biological interactions during nitrate and goethite reduction by Shewanella putrefaciens 200. Appl. Environ. Microbiol. 69, 3517–3525 10.1128/AEM.69.6.3517-3525.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings D. E., Caccavo F., Spring S., Rosenzweig R. F. (1999). Ferribacterium limneticum, gen. nov., sp. nov., an Fe(III)-reducing microorganism isolated from mining-impacted freshwater lake sediments. Arch. Microbiol. 171, 183–188 10.1007/s002030050697 [DOI] [Google Scholar]
- Cummings D. E., March A. W., Bostick B., Spring S., Caccavo F., Fendorf S., Rosenzweig R. F. (2000). Evidence for microbial Fe(III) reduction in anoxic, mining-impacted lake sediments (lake Coeur d’Alene, Idaho). Appl. Environ. Microbiol. 66, 154–162 10.1128/AEM.66.1.154-162.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daims H., Bruhl A., Amann R. I., Schleifer K. H., Wagner M. (1999). The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22, 434–444 10.1016/S0723-2020(99)80053-8 [DOI] [PubMed] [Google Scholar]
- Davison W. (1993). Iron and manganese in lakes. Earth Sci. Rev. 34, 119–163 10.1016/0012-8252(93)90029-7 [DOI] [Google Scholar]
- Edwards K. J., Bach W., McCollom T. M., Rogers D. R. (2004). Neutrophilic iron-oxidizing bacteria in the ocean: their habitats, diversity, and role in mineral deposition, rock alteration, and biomass production in the deep-sea. Geomicrobiol. J. 21, 393–404 10.1080/01490450490485863 [DOI] [Google Scholar]
- Edwards K. J., Rogers D. R., Wirsen C. O., McCollom T. M. (2003). Isolation and characterization of novel psychrophilic, neutrophilic, Fe-oxidizing, chemolithoautotrophic α- and γ-Proteobacteria from the deep sea. Appl. Environ. Microbiol. 69, 2906–2913 10.1128/AEM.69.5.2906-2913.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich A., Widdel F. (1994). Anaerobic oxidation of ferrous iron by purple bacteria, a new type of phototrophic metabolism. Appl. Environ. Microbiol. 60, 4517–4526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson D., Moyer C. (1997). Isolation and characterization of novel iron-oxidizing bacteria that grow at circumneutral pH. Appl. Environ. Microbiol. 63, 4784–4792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson D., Revsbech N. P. (1994). Investigation of an iron-oxidizing microbial mat community located near Aarhus, Denmark: field studies. Appl. Environ. Microbiol. 60, 4023–4031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson D., Weiss J. V. (2004). Bacterial iron oxidation in circumneutral freshwater habitats: findings from the field and the laboratory. Geomicrobiol. J. 21, 405–414 10.1080/01490450490485881 [DOI] [Google Scholar]
- Emerson D., Weiss J. V., Megonigal J. P. (1999). Iron-oxidizing bacteria are associated with ferric hydroxide precipitates (Fe-plaque) on the roots of wetland plants. Appl. Environ. Microbiol. 65, 2758–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finneran K. T., Johnsen C. V., Lovley D. R. (2003). Rhodoferax ferrireducens sp. nov., a psychrotolerant, faculatively anaerobic bacterium that oxidizes acetate with the reduction of Fe(III). Int. J. Syst. Evol. Microbiol. 53, 669–673 10.1099/ijs.0.02298-0 [DOI] [PubMed] [Google Scholar]
- Forney L. J., Zhou X., Brown C. J. (2004). Molecular microbial ecology: land of the one-eyed king. Curr. Opin. Microbiol. 7, 1–11 10.1016/j.mib.2004.04.015 [DOI] [PubMed] [Google Scholar]
- Galushko A. S., Schink B. (2000). Oxidation of acetate through reactions of the citric acid cycle by Geobacter sulfurreducens in pure culture and in syntrophic coculture. Arch. Microbiol. 174, 314–321 10.1007/s002030000208 [DOI] [PubMed] [Google Scholar]
- Ghiorse W. C. (1984). Biology of iron-depositing and manganese-depositing bacteria. Annu. Rev. Microbiol. 38, 515–550 10.1146/annurev.micro.38.1.515 [DOI] [PubMed] [Google Scholar]
- Grosskopf R., Janssen P. H., Liesack W. (1998). Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl. Environ. Microbiol. 64, 960–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaijer S. C. M., Harhangi H. R., Meijerink B. B., Strous M., Pol A., Smolders A. J. P., Verwegen K., Jetten M. S. M., Op den Camp H. J. M. (2008). Bacteria associated with iron seeps in a sulfur-rich, neutral pH freshwater ecosystem. ISME J. 2, 1231–1242 10.1038/ismej.2008.75 [DOI] [PubMed] [Google Scholar]
- Haaijer S. C. M., Lamers L. P. M., Smolders A. J. P., Jetten M. S. M., Op den Camp H. J. M. (2007). Iron sulfide and pyrite as potential electron donors for microbial nitrate reduction in freshwater wetlands. Geomicrobiol. J. 24, 391–401 10.1080/01490450701436489 [DOI] [Google Scholar]
- Haaijer S. C. M., van der Welle M. E. W., Schmid M. C., Lamers L. P. M., Jetten M. S. M., Op den Camp H. J. M. (2006). Evidence for the involvement of betaproteobacterial Thiobacilli in the nitrate-dependent oxidation of iron sulfide minerals. FEMS Microbiol. Ecol. 58, 439–448 10.1111/j.1574-6941.2006.00178.x [DOI] [PubMed] [Google Scholar]
- Hafenbradl D., Keller M., Dirmeier R., Rachel R., Rossnagel P., Burggraf S., Huber H., Stetter K. O. (1996). Ferroglobus placidus gen nov, sp nov, a novel hyperthermophilic archaeum that oxidizes Fe2+ at neutral pH under anoxic conditions. Arch. Microbiol. 166, 308–314 10.1007/s002030050388 [DOI] [PubMed] [Google Scholar]
- Heising S., Richter L., Ludwig W., Schink B. (1999). Chlorobium ferrooxidans sp. nov., a phototrophic green sulfur bacterium that oxidizes ferrous iron in coculture with a “Geospirillum” sp. strain. Arch. Microbiol. 172, 116–124 10.1007/s002030050748 [DOI] [PubMed] [Google Scholar]
- Kashefi K., Tor J. M., Holmes D. E., Van Praag C. V., Reysenbach A. L., Lovley D. R. (2002). Geoglobus ahangari gen. nov., sp. nov., a novel hyperthermophilic archeon capable of oxidizing organic acids and growing autotrophically on hydrogen with Fe(III) serving as the sole electron donor. Int. J. Syst. Evol. Microbiol. 52, 719–728 10.1099/ijs.0.01953-0 [DOI] [PubMed] [Google Scholar]
- Kodama Y., Watanabe K. (2004). Sulfuricurvum kujiense gen. nov., sp nov., a facultatively anaerobic, chemolithoautotrophic, sulfur-oxidizing bacterium isolated from an underground crude-oil storage cavity. Int. J. Syst. Evol. Microbiol. 54, 2297–2300 10.1099/ijs.0.63243-0 [DOI] [PubMed] [Google Scholar]
- Kumaraswamy R., Sjollema K., Kuenen G., van Loosdrecht M., Muyzer G. (2006). Nitrate-dependent [Fe(II)EDTA](2-) oxidation by Paracoccus ferrooxidans sp nov., isolated from a denitrifying bioreactor. Syst. Appl. Microbiol. 29, 276–286 10.1016/j.syapm.2005.08.001 [DOI] [PubMed] [Google Scholar]
- Lin B., Braster M., van Breukelen B. M., van Verseveld H. W., Westerhoff H. V., Röling W. F. M. (2005). Geobacteraceae community composition is related to hydrochemistry and biodegradation in an iron-reducing aquifer polluted by a neighbouring landfill. Appl. Environ. Microbiol. 71, 5983–5991 10.1128/AEM.71.12.8558-8564.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley D. R., Phillips E. J. P. (1988). Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl. Environ. Microbiol. 54, 1472–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley D. R., Stolz J. F., Nord G. L., Phillips E. J. P. (1987). Anaerobic production of magnetite by a dissimilatory iron-reducing microorganism. Nature 330, 252–254 10.1038/330252a0 [DOI] [Google Scholar]
- Ludwig W., Rosselo-Mora R., Aznar R., Klugbauer S., Spring S., Reetz K., Beimfohr C., Brockmann E., Kirchhof G., Dorn S., Bachleitner M., Klugbauer N., Springer N., Lane D., Nietupsky R., Weizenegger M., Schleifer K.-H. (1995). Comparative sequence analysis of 23S rRNA from Proteobacteria. Syst. Appl. Microbiol. 18, 164–188 10.1016/S0723-2020(11)80407-8 [DOI] [Google Scholar]
- Moraghan J. T., Buresh R. J. (1977). Chemical reduction of nitrite and nitrous-oxide by ferrous iron. Soil Sci. Soc. Am. J. 41, 47–50 10.2136/sssaj1977.03615995004100010017x [DOI] [Google Scholar]
- Nealson K. H., Belz A., McKee B. (2002). Breathing metals as a way of life: geobiology in action. Antonie Van Leeuwenhoek 81, 215–222 10.1023/A:1020518818647 [DOI] [PubMed] [Google Scholar]
- Nealson K. H., Saffarini D. (1994). Iron and manganese in anaerobic respiration: environmental significance, physiology, and regulation. Annu. Rev. Microbiol. 48, 311–343 10.1146/annurev.mi.48.100194.001523 [DOI] [PubMed] [Google Scholar]
- Nevin K. P., Holmes D. E., Woodard T. L., Hinlein E. S., Ostendorf D. W., Lovley D. R. (2005). Geobacter bemidjiensis sp. nov. and Geobacter psychrophilus sp. nov., two novel Fe(III)-reducing subsurface isolates. Int. J. Syst. Evol. Microbiol. 55, 1667–1674 10.1099/ijs.0.63417-0 [DOI] [PubMed] [Google Scholar]
- Nevin K. P., Lovley D. R. (2002). Mechanisms for accessing insoluble Fe(III) oxide during dissimilatory Fe(III) reduction by Geothrix fermentans. Appl. Environ. Microbiol. 68, 2294–2299 10.1128/AEM.68.5.2294-2299.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden E. E., Sobolev D., Glazer B., Luther G. W., (III) (2004). Potential for microscale bacterial Fe redox cycling at the aerobic-anaerobic interface. Geomicrobiol. J. 21, 379–391 10.1080/01490450490485872 [DOI] [Google Scholar]
- Schippers A., Jørgensen B. B. (2002). Biogeochemistry of pyrite and iron sulfide oxidation in marine sediments. Geochim. Cosmochim. Acta 66, 85–92 10.1016/S0016-7037(01)00745-1 [DOI] [Google Scholar]
- Senko J. M., Dewers T. A., Krumholz L. R. (2005). Effect of oxidation rate and Fe(II) state on microbial nitrate-dependent Fe(III) mineral formation. Appl. Environ. Microbiol. 71, 7172–7177 10.1128/AEM.71.11.7172-7177.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton D. R., Furlong M. A., Rathbun S. L., Whitman W. B. (2001). Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67 4374–4376 10.1128/AEM.67.9.4374-4376.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolev D., Roden E. E. (2002). Evidence for the rapid microscale bacterial redox cycling of iron in circumneutral environments. Antonie Van Leeuwenhoek 81, 587–597 10.1023/A:1020569908536 [DOI] [PubMed] [Google Scholar]
- Sproer C., Mendrock U., Swiderski J., Lang E., Stackebrandt E. (1999). The phylogenetic position of Serratia, Buttiauxella and some other genera of the family Enterobacteriaceae. Int. J. Syst. Bacteriol. 49, 1433–1438 10.1099/00207713-49-4-1433 [DOI] [PubMed] [Google Scholar]
- Straub K. L., Benz B., Schink B., Widdel F. (1996). Anaerobic, nitrate dependent microbial oxidation of ferrous iron. Appl. Environ. Microbiol. 62, 1458–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub K. L., Benz M., Schink B. (2001). Iron metabolism in anoxic environments at near neutral pH. FEMS Microbiol. Ecol. 34, 181–186 10.1111/j.1574-6941.2001.tb00768.x [DOI] [PubMed] [Google Scholar]
- Straub K. L., Buchholz-Cleven B. E. E. (1998). Enumeration and detection of anaerobic ferrous iron oxidizing, nitrate reducing bacteria from diverse European sediments. Appl. Environ. Microbiol. 64, 4846–4856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub K. L., Hanzlik M., Buchholz-Cleven B. E. E. (1998). The use of biologically produced ferrihydrite for the isolation of novel iron-reducing bacteria. Syst. Appl. Microbiol. 21, 442–449 10.1016/S0723-2020(98)80054-4 [DOI] [PubMed] [Google Scholar]
- Straub K. L., Schönhuber W. A., Buchholz-Cleven B. E. E., Schink B. (2004). Diversity of ferrous iron-oxidizing, nitrate-reducing bacteria and their involvement in oxygen-independent iron cycling. Geomicrobiol. J. 21, 371–378 10.1080/01490450490485854 [DOI] [Google Scholar]
- Strous M., Pelletier E., Mangenot S., Rattei T., Lehner E., Taylor M. W., Horn M., Daims H., Bartol-Mavel D., Wincker P., Barbe V., Fonknechten N., Vallenet D., Segurens B., Schenowitz-Truong C., Médigue C., Collingro A., Snel B., Dutilh B. E., Op den Camp H. J. M., van der Drift C., Cirpus I., van de Pas-Schoonen K. T., Harhangi H. R., van Niftrik L., Schmid M., Keltjens J., van de Vossenberg J., Kartal B., Meier H., Frishman D., Huynen M. A., Mewes H.-W., Weissenbach J., Jetten M. S. M., Wagner M., Le Paslier D. (2006). Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440, 790–794 10.1038/nature04647 [DOI] [PubMed] [Google Scholar]
- Stumm W., Sulzberger B. (1992). The cycling of iron in natural environments – considerations based on laboratory studies of heterogeneous redox processes. Geochim. Cosmochim. Acta 56, 3233–3257 10.1016/0016-7037(92)90301-X [DOI] [Google Scholar]
- Sung Y., Fletcher K. E., Ritalahti K. M., Apkarian R. P., Ramos-Hernández N., Sanford R. A., Mesbah N. M., Löffler F. E. (2006). Geobacter lovleyi sp. nov. strain SZ, a novel metal-reducing and tetrachloroethene-dechlorinating bacterium. Appl. Environ. Microbiol. 72, 2775–2782 10.1128/AEM.72.3.1980-1987.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai K., Suzuki M., Nakagawa S., Miyazaki M., Suzuki Y., Inagaki F., Horikoshi K. (2006). Sulfurimonas paralvinellae sp nov., a novel mesophilic, hydrogen- and sulfur-oxidizing chemolithoautotroph within the Epsilonproteobacteria isolated from a deep-sea hydrothermal vent polychaete nest, reclassification of Thiomicrospira denitrificans as Sulfurimonas denitrificans comb. nov and emended description of the genus Sulfurimonas. Int. J. Syst. Evol. Microbiol. 56, 1725–1733 10.1099/ijs.0.64255-0 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thamdrup B. (2000). Bacterial manganese and iron reduction in aquatic sediments. Adv. Microb. Ecol. 16, 41–84 10.1007/978-1-4615-4187-5_2 [DOI] [Google Scholar]
- Thamdrup B., Fossing H., Jørgensen B. B. (1994). Manganese, iron, and sulfur cycling in a coastal marine sediment, Aarhus Bay, Denmark. Geochim. Cosmochim. Acta 58, 5115–5129 10.1016/0016-7037(94)90446-4 [DOI] [Google Scholar]
- Tor J. M., Kashefi K., Lovley D. R. (2001). Acetate oxidation coupled to Fe(III) reduction in hyperthermophilic microorganisms. Appl. Environ. Microbiol. 67, 1363–1365 10.1128/AEM.67.3.1363-1365.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K. A., Pollock J., Cole K. A., O’Connor S. M., Achenbach L. A., Coates J. D. (2006). Anaerobic nitrate-dependent iron(II) bio-oxidation by a novel lithoautotrophic betaproteobacterium, strain 2002. Appl. Environ. Microbiol. 72, 686–694 10.1128/AEM.72.1.686-694.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K. A., Urrutia M. M., Churchill P. F., Kukkadapu R. K., Roden E. E. (2005). Anaerobic redox cycling of iron by freshwater sediment microorganisms. Environ. Microbiol. 8, 100–113 10.1111/j.1462-2920.2005.00873.x [DOI] [PubMed] [Google Scholar]
- Weiss J. V., Emerson D., Backer S. M., Megonigal J. P. (2003). Enumeration of Fe(II)-oxidizing and Fe(III)-reducing bacteria in the root zone of wetland plants: implications for a rhizosphere iron cycle. Biogeochemistry 64, 77–96 10.1023/A:1024953027726 [DOI] [Google Scholar]