Abstract
Among animals, urochordates (e.g., ascidians) are unique in their ability to biosynthesize cellulose. In ascidians cellulose is synthesized in the epidermis and incorporated into a protective coat know as the tunic. A putative cellulose synthase-like gene was first identified in the genome sequences of the ascidian Ciona intestinalis. We describe here a cellulose synthase gene from the ascidian Ciona savignyi that is expressed in the epidermis. The predicted C. savignyi cellulose synthase amino acid sequence showed conserved features found in all cellulose synthases, including plants, but was most similar to cellulose synthases from bacteria, fungi, and Dictyostelium discoidium. However, unlike other known cellulose synthases, the predicted C. savignyi polypeptide has a degenerate cellulase-like region near the carboxyl-terminal end. An expression construct carrying the C. savignyi cDNA was found to restore cellulose biosynthesis to a cellulose synthase (CelA) minus mutant of Agrobacterium tumefaciens, showing that the predicted protein has cellulose synthase activity. The lack of cellulose biosynthesis in all other groups of metazoans and the similarity of the C. savignyi cellulose synthase to enzymes from cellulose-producing organisms support the hypothesis that the urochordates acquired the cellulose biosynthetic pathway by horizontal transfer.
Although cellulose biosynthetic ability is a conspicuous trait of plants, a number of other organisms, including certain bacteria and cellular slime molds, are capable of synthesizing cellulose (1). Despite the diversity of species producing cellulose, only one group of animals has the ability to biosynthesize cellulose, the urochordates (2). The urochordates are ubiquitous marine invertebrate chordates that have a similar body plan, embryology, and physiology with their sister chordates, the cephalochordates and the vertebrates (3). Members of the three urochordate classes (ascidiacea, thaliacea, and appendicularia) have all been shown to produce type I cellulose microfibrils (4–6). In urochordates, cellulose is a product of the epidermis and is incorporated into a tough outer coat known as the tunic, in ascidians and thaliaceans, and as the house, in appendicularians. The presence of cellulose in all urochordates suggests an early origin for cellulose biosynthetic ability in the urochordates lineage, and that its advent may have been a key event favoring the widespread radiation of the urochordates.
The absence of cellulose in all other extant metazoans makes the presence of cellulose in urochordates particularly puzzling. It is unlikely that cellulose biosynthetic ability was present early in the metazoan lineage and subsequently lost in all animals except the urochordates. Minimally, loss of cellulose biosynthetic ability would have had to occur at the base of the protostomes and at the bases of the lineages leading to the echinoderms and hemichordates, as well as that leading to the cephalochordates and vertebrates. More likely scenarios are that either urochordates independently evolved cellulose biosynthetic ability, or that the urochordates acquired the ability by horizontal transfer from some other organism. If urochordates were to have independently evolved cellulose biosynthesis, for example, by duplication and modification of an existing glycosyltransferase, the resulting enzyme is unlikely to show extensive similarity to the previously characterized cellulose synthases. On the other hand, the presence of an enzyme in urochordates showing strong similarity to known cellulose synthases would be strong evidence for horizontal transfer. The presence of a predicted cellulose synthase-like gene in the genome of the ascidian Ciona intestinalis supports the horizontal transfer hypothesis (7). However, the family of processive glycosyltransferases, including those that do not produce cellulose, have similar primary sequences (8, 9). To investigate ascidian cellulose biosynthesis further, we have isolated a cellulose synthase cDNA from the related species Ciona savignyi. We report here that the cDNA encodes a protein with cellulose synthase activity. Furthermore, the cDNA differs considerably from that predicted in the C. intestinalis annotation and has unique features not found in other known cellulose synthases.
Materials and Methods
Collection of C. savignyi and Preparation of RNA. Adult C. savignyi were collected from the docks of the Santa Barbara yacht harbor. Eggs were dissected, fertilized, and cultured in sea water to the mid-tailbud stage for RNA isolation as described (10). cDNA was made from the total RNA by using the SMART RACE cDNA Amplification kit (Clontech).
Isolation of C. savignyi Cellulose Synthase cDNA. Translations of the unassembled sequence reads from the genome project for the ascidian C. savignyi (www-genome.wi.mit.edu/annotation/ciona) were searched with the amino acid sequence of Arabidopsis thaliana cellulose synthase (AAC29067) by using a blast 2.2 search program (Blossum 62 matrix, gap penalty of 11, extension penalty of 1) (11) run against the raw genomic reads from C. savignyi. Several C. savignyi genomic sequence reads encoded predicted peptides with strong similarity to the core catalytic domain of cellulose synthase. Sequences within predicted exons were used to design two oligonucleotide primers (5′-GGCCACTCGGGAGGGTGCTGG-3′ and 5′-CGGTCTCTATGCCTTCCACCC-3′) for 5′-RACE by using the SMART RACE cDNA Amplification kit (Clontech). The two primers were offset in the predicted exon by ≈100 bp, and gave amplification products of ≈400 and 500 bp, respectively. The nucleotide sequence of the two 5′-RACE products was virtually identical within the 400-bp overlap, and the 5′ ends of both sequences contained exact matches to the 16-nucleotide Ciona transspliced 5′ leader (12). Sequence flanking of the 5′ leader was used to design an oligonucleotide primer (5′-GCAAGACGCCAAGAGTTGGCCTGG-3′) that, together with the SMART RACE cDNA Amplification kit 3′-RACE primer (Clontech), was used to amplify the full cDNA by using LA TAQ polymerase with cycling conditions as suggested by the manufacturer (Takara Shuzo, Kyoto). Two amplification products of ≈4.6 and 5.3 kbp were obtained and subcloned into the plasmid pCRII by using the TA Cloning Kit (Invitrogen). Nucleotide sequences were determined by using the BigDye chemistry (Applied Biosystems).
Expression Analysis. For in situ hybridization, sense and antisense digoxygenin-containing RNA probes were synthesized from a 557-bp fragment (nucleotides 2248–2804 of the full cDNA) corresponding to the core catalytic domain of the C. savignyi cellulose synthase. The in situ hybridization to tailbud stage embryos was as described (13). For Northern blot analysis, a 7-μg sample of total RNA from whole atrial siphons of adult C. savignyi was electrophoresed on a formaldehyde gel and transferred to a nylon filter. The filter was hybridized in Quickhyb (Stratagene) solution with a random-primed 32P-containing probe synthesized from the full-length cDNA.
Construction of Bacterial Strains and Growth of Bacteria. The C. savignyi cDNA was cloned behind the lac promoter into the broad host expression vector pTE3 (14), transformed into Escherichia coli, and conjugated into either wild-type Agrobacterium tumefaciens strain A1045 or its cellulose synthase-deficient derivative A1045celA by using pRK2013 as a mobilizing plasmid for the conjugation. Transconjugants were selected for resistance to tetracycline (10 mg/liter, carried by the plasmid) and rifampicin (50 mg/liter, carried by the A. tumefaciens strains). E. coli was grown in Luria broth or agar at 37°C. Expression of the cDNA in E. coli was induced by isopropylthio-β-galactoside (IPTG) added to a final concentration of 0.2 mg/ml. A. tumefaciens was grown to stationary phase at room temperature in Luria broth containing 10 mg/liter tetracycline. The A. tumefaciens endogenous cellulose synthesis genes were induced by the addition of soytone (final concentration 0.02%), and the cultures were grown an additional 24 h. To visualize the synthesis of cellulose, 20 mg/liter calcafluor (fluorescent brightener no. 2) was included in some plates.
Measurements of Cellulose Synthesis. Bacterial cell extracts were made as described (15) except that the sonication buffer contained 50 mM Tris·HCl (pH 7.5), 10 mM MgCl2, 1 mM EDTA (TME), and 20% polyethylene glycol 3,500. The extract was incubated in TME containing 1 mM CaCl2, 0.1 mM ZnSO4 300 pM UDP-glucose, and 1 μCi (1 Ci = 37 GBq) of UDP-14C-glucose per ml at room temperature for 20 min. The reaction was stopped by the addition of 0.5 M NaOH containing 0.05 M NaBH4 and 1 mg/ml Sigmacel carrier, and the mixture was heated at 65°C for 20 min. When cool, the pellet was collected on GF/C filter paper, washed three times with water and three times with ethanol, dried, and counted in a liquid scintillation counter.
GC-MS Linkage Analysis. After incubation, the cell extracts containing the washed radioactive product were dried and derivatized for glycosyl linkage analysis by methylation with n-butyllithium (16–19). Reaction products were dissolved in 0.5 ml of DMSO, and n-butyllithium was added slowly, followed by methyl iodide. Methylated samples were extracted with chloroform and washed with water, and the chloroform phase was evaporated to dryness. Samples were fully hydrolyzed with 2 M trifluoroacetic acid at 121°C for 1 h and reduced with NaBD4. Per-O-methylated alditols were acetylated with acetic anhydride and 1-methylimidazole as a catalyst (20). These partially methylated alditol acetates were introduced by a 3-min splitless injection on a 30 m × 0.75 mm SP-2330 fused silica capillary column (Supelco) at a temperature of 170°C. After a 3-min delay, column temperature was raised 4°C/min to 240°C, and derivative structures were deduced as described by Carpita and Shea (19).
Results
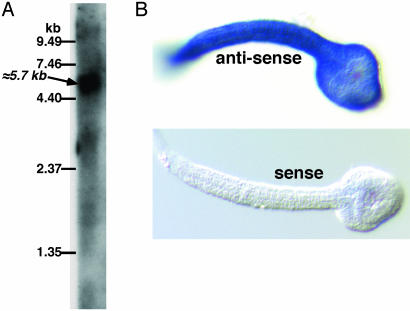
Identification and Cloning of a Putative Cellulose Synthase Gene. The amino acid sequence of the A. thaliana cellulose synthase was used to identify likely exons from the C. savignyi genomic sequences encoding the core catalytic domain of a putative cellulose synthase. To isolate a cDNA for the putative cellulose synthase, we first isolated the 5′ end of the transcript by 5′-RACE using oligonucleotides designed from a predicted exon upstream of the catalytic domain. The nucleotide sequence of the 5′-RACE product contained a perfect match for the Ciona transspliced 5′ leader sequence (12), indicating that we had identified the 5′ end of the transcript. cDNAs were then amplified from a 3′-RACE library by using a gene-specific oligonucleotide to the 5′ end and the 3′-RACE primer. Two PCR products of 4.6 and 5.3 kbp were obtained. Three isolates of the PCR products were sequenced, and the nucleotide sequences of the differently sized products were virtually identical except that the 4.6-kbp product was ≈700 bp shorter at the 3′ end. The 4.6-kbp product had the same long ORF as the 5.3-kbp product but had no stop codon and was lacking the C-terminal 30 codons. The 4.6-kbp product thus seemed to be the product of an up-stream priming during cDNA synthesis. The larger PCR product (5,342 bp), which contained a 5′ leader sequence, a consensus polyadenylation site, and a polyA+ tail, was designated the full-length clone. By Northern blotting, a single large transcript with a calculated molecular size of ≈5.7 kb was detected in RNA from whole adults by using the 5.3-kbp cDNA as a probe (Fig. 1A). The putative C. savignyi cellulose synthase transcript was also detected in tailbud stage embryos by RT-PCR (data not shown), and whole mount in situ hybridization to tailbud stage embryos showed that transcript was expressed uniformly in the epidermis (Fig. 1B). A search of the partially assembled C. savignyi genome identified a 23,000-bp contig that encompassed the entire cDNA in 32 putative exons (Fig. 2A).
Fig. 1.
(A) Northern blot analysis of RNA extracted from adult C. savignyi atrial siphons. A blot of total RNA resolved on formaldehyde-agarose gels was hybridized with a radioactive probe synthesized from the full-length putative cellulose-synthase cDNA. The relative positions of standard RNAs and the calculated size of the major hybridizing band are indicated. (B) In situ hybridization of digoxygenin-labeled antisense and sense (control) probes to tailbud stage C. savignyi embryos. Strong hybridization (blue) was observed with the antisense probe throughout the epidermis, which is the source of both the larval tunic (test) and the adult tunic (5).
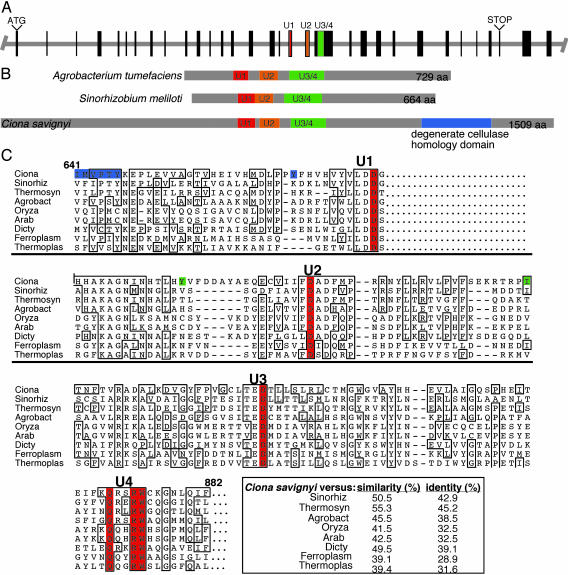
Fig. 2.
(A) Structure of the putative C. savignyi cellulose synthase gene. Exons are indicated by boxes. Exons encoding the conserved cellulose synthase catalytic motifs are color-coded. (B) The characteristic spacing of the conserved cellulose synthase motifs (U1, U2, and U3/4) of the predicted C. savignyi cellulose synthase protein resembles that of prokaryotic cellulose synthases (e.g., A. tumefaciens and Sinorhizobium meliloti). Unlike previously characterized cellulose synthases, the predicted C. savignyi protein also contains a region showing similarity to cellulase. (C) Amino acid sequence alignment of the highly conserved U1, U2, and U3/4 regions of the predicted C. savignyi cellulose synthase with cellulose synthase sequences representing the two gene products from each major taxonomic group showing the highest blastp scores with the C. savignyi protein. In addition, the CelA protein from A. tumefaciens was included because the biochemical work reported here was carried out in this organism. The D, D, D35QXXRW motif is indicated in red. Differences in the predicted C. intestinalis sequence are indicated in the C. savignyi sequence. Residues highlighted in blue indicate nonconservative substitutions whereas green indicates conservative substitutions. The abbreviations, reference numbers, and taxonomic group of the organism for the amino acid sequences obtained from the National Center for Biotechnology Information web site are as follows: C. savignyi (Ciona), S. meliloti (Sinorhiz, A 95889, eubacterium), Thermosynechococcus elongatus (Thermosyn, NP 682585, eubacterium), A. tumefaciens (Agrobact, I 39714, eubacterium), Ferroplasma acidurmanus (Ferroplas, ZP 00000436, archaea), Thermoplasma acidophilum (Thermoplas, NP 393962, archaea), Dictyostelium discoideum (Dicty, AF 163835, eukaryotic slime mold), A. thaliana (Arab, NP 180039, plant), and Oryza sativa (Oryza, BAC 10759, plant).
Sequence Analysis of the Ciona Cellulose Synthase Gene. A consensus cDNA sequence was assembled from the three sequenced clones and the genomic sequence reads and found to encode a 1,509-aa polypeptide (Fig. 2B). When a blastp analysis was run using the predicted polypeptide sequence, two regions were identified with homology to known protein families. Amino acids 598–982 showed homology to COG 1215 (glycosyl transferases) with a value of e-34. Amino acids 1159–1349 showed homology to pfam 1341 (glycosyl hydrolyases family 6) with a value of 7e-11 (Fig. 2). Strong similarity was found between the predicted C. savignyi polypeptide and known or putative cellulose synthases from bacteria, fungi, plants, slime mold, and archaea. The highest scores on blast using blastp and the Blossum 62 matrix with a gap penalty of 11 and an extension penalty of 1 were with sequences from bacteria, slime mold, and fungi (Table 1). Lower degrees of homology were seen with plant and archaea sequences. No significant homology was seen with the only algal cellulose synthase sequences available [those from Mesotaenium caldariorum (21)]. The only metazoan sequence with significant homology was the sequence from the ascidian C. intestinalis. No other metazoan sequences with homology scores better than 0.01 were found by using blastp and tblastn and the nr, est, completed eukaryotic genomes, and unfinished eukaryotic genomes databases.
Table 1. Result of blast homology searches using the C. savignyi cellulose synthase gene.
| Organism | Gene | e value |
|---|---|---|
| Bacteria | ||
| S. meliloti | Putative cellulose synthase; AL603643 | 6e-39 |
| T. elongatus | Similar to cellulase synthase; AP005375 | 8e-38 |
| Bradyrhizobium japonicum | Possible cellulose synthase; AP005943 | 1e-34 |
| Slime mold | ||
| D. discoidium | Cellulose synthase; AF163835 | 6e-25 |
| Fungi | ||
| Neuospora crassa | Hypothetical protein; EAA30080 | 4e-18 |
| Aspergillis fumagitus* | Contig:4823 | 2e-22 |
| Archaea | ||
| Ferroplasma acidarmanus | Hypothetical protein; ZP 00000436 | 5e-15 |
| T. acidophilum* | Hypothetical protein; AL445064 | 7e-08 |
| Plants | ||
| O. sativa (rice) | Putative glucosyltransferase; BAC10759 | 2e-16 |
| A. thaliana | Unknown protein; NP 187389 | 8e-16 |
| Metazoans | ||
| C. intestinalis* | cDNA clone; AK115634 | 9e-107 |
All homologies were derived using the entire sequence of the C. savignyi gene and blastp (11) except those marked with an asterisk. The A. fumigatus sequence is from a tblastn of unfinished eukaryotic genomes, and the C. intestinalis and T. acidophilum sequences are a result of a tblastn of the nr data base.
Although the amino acid sequences of cellulose synthases vary considerably, all, including the C. savignyi polypeptide, contain the invariant motif D, D, D35QXXRW (Fig. 2C) (1, 9, 22). The regions surrounding the D, D, D35QXXRW residues show varying degrees of sequence similarity between the cellulose synthases and are commonly referred to as the U1, U2, and U3/4 domains, respectively (Fig. 2). An alignment of the most highly conserved regions surrounding the invariant D, D, D35QXXRW residues from the C. savignyi polypeptide (with the two most similar sequences from eubacteria, plants, archaea, and the single sequence from cellular slime mold) is shown in Fig. 2C. Plants have many genes that show similarity to cellulose synthase, and many of the best matching genes to the C. savignyi cellulose synthase are members of the CSLC subfamily from A. thaliana and Oryza sativa (Table 1) (22). The spacing within the D, D, D35QXXRW motifs of these two plant sequences is similar to that of the C. savignyi and bacterial sequences, unlike the plant CesA sequences, which have large inserts within the motifs (1). The higher plant and algal cellulose synthases also have a conserved zinc binding region near the amino terminus (1, 21). This region is lacking in all other cellulose synthases, including the predicted C. savignyi protein.
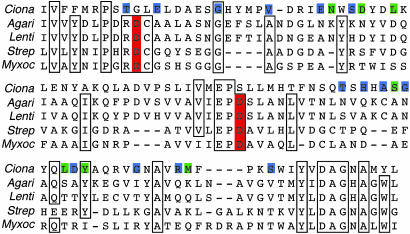
In addition to the predicted cellulose synthase domain, a second region near the C terminus of the C. savignyi gene shows similarity to cellulase (Fig. 2B), a feature not found in other known processive glycosyltransferases. The C. savignyi sequence is most similar to cellulases in the glycosyl hydrolase family 6, members of which are found in both eukaryotes and prokaryotes. An alignment of the similar domains from the putative C. savignyi protein and four cellulases that show strong similarity to it is shown in Fig. 3. Note that, whereas there is extensive identity of amino acid sequences, the predicted C. savignyi protein is lacking aspartic acid residues implicated in catalysis (23). Because of this result, the predicted C. savignyi protein may not have cellulase activity. Thus, it is unclear what role, if any, this cellulase-like domain has in cellulose synthesis.
Fig. 3.
Amino acid sequence alignment of the cellulase-like domains of the predicted C. savignyi cellulose synthase protein with closely matching examples of the glycosyl hydrolase family 6 cellulases from both prokaryotes [Streptomyces coelicolor, AL939114 (Strep) and Myxococcus xanthus, X76640.2 (Myxoc)] and eukaryotes [Agaricus bisporus, P49075 (Agari) and Lentinula edodes, AF411251 (Lenti)]. Aspartic acid residues in red indicate residues thought to be important for catalysis. Differences in the predicted C. intestinalis sequence are indicated in the C. savignyi sequence. Residues highlighted in blue indicate nonconservative substitutions whereas green indicates conservative substitutions.
Activity of the C. savignyi Cellulose Synthase Gene Expressed in Bacteria. Although the predicted C. savignyi protein has strong similarity to cellulose synthases, the conserved catalytic core residues highlighted in Fig. 2 are common to many glycosyltransferases with differing catalytic activities (for example, see the A. tumefaciens curdlan synthase) (9). To assess the enzymatic activity of the putative C. savignyi cellulose synthase, we chose a bacterial expression system (A. tumefaciens) because the predicted C. savignyi protein is most similar to bacterial cellulose synthases, and therefore might be expected to more successfully interact with essential host enzymes in the cellulose synthesis pathway in bacteria. In addition, the availability of a cellulose synthase-minus mutant of A. tumefaciens allowed us to undertake complementation studies in this bacterium (24). We also determined whether the C. savignyi gene product could catalyze cellulose synthesis without the help of other genes required for cellulose synthesis in bacteria by examining the ability of the gene product to catalyze cellulose synthesis in E. coli K12. Although some pathogenic strains of E. coli can make cellulose, the K12 strain LE392 does not (25).
When the plasmid carrying the C. savignyi cellulose synthase cDNA was introduced into wild-type A. tumefaciens A1045, we observed that the bacteria grew as larger aggregates or clumps in Luria broth containing soytone when compared with control bacteria, which is a characteristic of bacteria producing high levels of cellulose (not shown). On plates, the colonies had a matte surface as opposed to the shiny surface of the wild-type cells and showed increased staining with calcafluor, which stains β-linked carbohydrates including cellulose. The CelA mutants grew as uniformly suspended cells in liquid culture. When the C. savignyi cellulose synthase cDNA clone was introduced into a CelA mutant, small aggregates were observed in liquid culture, and on plates the colonies showed increased staining with calcafluor. These results suggest, but do not prove, that the bacteria carrying the cloned gene from C. savignyi synthesize cellulose. To determine whether the bacteria carrying this clone were synthesizing cellulose, in vitro synthesis of β-1,4-glucan was examined.
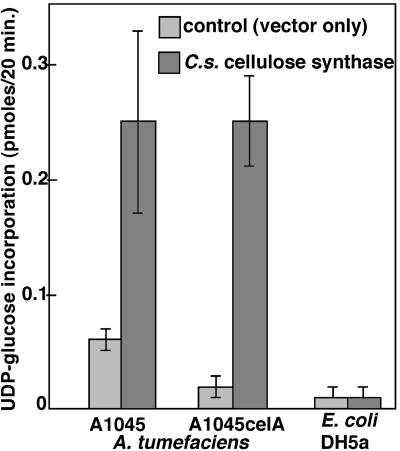
Using an assay that measures incorporation of UDP-14C-glucose into cellulose by cell-free bacterial extracts (24), we detected an ≈4-fold increase in cellulose biosynthesis by extracts from wild-type A. tumefaciens expressing the C. savignyi cDNA in comparison with cells carrying the vector alone (Fig. 4). The CelA mutant carrying the empty vector was unable to synthesize cellulose. When the vector carrying the cDNA clone was introduced into the CelA mutant, the incorporation of UDP-glucose into cellulose was the same as that of the wild-type strain carrying the cDNA clone. The radioactive material synthesized by the A. tumefaciens CelA mutant carrying the plasmid with and without the Ciona celluloase synthase gene was pelleted and washed with with water and ethanol. It was then derivatized for glycosyl linkage analysis and separated by GC-MS into the resulting partially methylated alditol acetates. The CelA mutant carrying the Ciona gene produced a total of 54 mol% (± 7%) glucose, of which 7.6 mol% (± 4%) was terminally linked glucosyl residues, and the remainder 46.4 mol% (± 5%) was 4-linked glucosyl residues. When the plasmid lacked the C. savignyi gene, no radioactive glucosyl residues were detected. When A. tumefaciens containing the C. savignyi cellulose synthase gene was grown in media without the soytone inducer for the A. tumefaciens cel genes, only 14 mol% (± 3%) of total glucose was detected. However, the ratio of terminally linked glucosyl residues to 4-linked glucosyl residues was the same as the data obtained from bacteria grown in the presence of soytone. No 3-linked glucosyl residues were detected. In addition, the radioactive product from the CelA mutant carrying the Ciona gene could be digested to water soluble oligosaccharides with β-(1, 4)-endoglucanase-II (EC 3.2.1.4) (0.5 mg of enzyme in 50 mM glycine buffer at pH 3.0 for 4 h at 50°C) or with cellulase (Worthington PB, 0.05 mg/ml enzyme in acetate buffer, pH 5.0, for 2 h at 37°C). Thus, it seems that the radioactive product of the reaction was indeed β1,4-linked glucose, i.e., cellulose. Hence, the C. savignyi cDNA was at least partially able to complement the celA mutation in vivo and completely able to complement the mutation in vitro, indicating that the C. savignyi gene product does indeed possess cellulose synthase activity.
Fig. 4.
The putative C. savignyi cellulose synthase gene functions as a cellulose synthase in A. tumefaciens. The C. savignyi cDNA was cloned into the broad host expression vector pTE3 (14), transformed into E. coli, and conjugated into either wild-type A. tumefaciens strain A1045 or its cellulose synthase-deficient derivative A1044celA. The graph shows incorporation of UDP-14C-glucose into cellulose by an extract of ≈107 bacteria. The bars indicate the SEM.
The plasmid carrying the C. savignyi cellulose synthase cDNA behind a lac promoter was introduced into wild-type E. coli K12 strain LE392. The bacteria were grown in Luria broth at room temperature, and the expression of the cDNA was induced by the addition of isopropylthio-β-galactoside. No phenotypic change was observed in the E. coli cultures after gene induction nor was any difference seen between the bacteria carrying the empty vector and those carrying the cDNA clone. Extracts of the induced E. coli were assayed for the ability to incorporate UDP-14C-glucose into cellulose. No significant cellulose synthesis was observed (Fig. 4), suggesting that the C. savignyi protein is not able to catalyze cellulose synthesis in cells that do not normally make cellulose. The substances and activities found in the extracts of A. tumefaciens (and absent in the extracts of E. coli), which result in enzymatic activity only in the case of A. tumefaciens, are unknown and may include other proteins such as the products of the celB, celC, celD, and celE genes and small molecules such as cyclic-di-guanylic acid.
Discussion
We have identified and sequenced a putative cellulose synthase gene from C. savignyi. The cDNA of this gene seems to encode a product with cellulose synthase enzymatic activity because a clone of this cDNA is able complement a cellulose synthase mutant of A. tumefaciens. This cDNA does not seem by itself to be sufficient for cellulose synthesis activity because its expression did not result in cellulose synthesis in an E. coli strain that does not ordinarily make cellulose. This result suggests that A. tumefaciens supplied additional proteins and/or factors required for cellulose synthesis and that these factors are lacking in the E. coli strain. Hybridization and RT-PCR analysis suggest that this gene is expressed in C. savignyi at times and locations in which cellulose is synthesized in tunicates. Expression was detected in the epidermis of tailbud embryos and in whole atrial siphons of adults.
In addition to the region of homology to cellulose synthase, the C. savignyi gene contains a region of homology to family 6 glycosylhydrolases. The role, if any, of this region of the gene in cellulose synthesis activity is unclear. The protein product of this gene may not have glycosylhydrolyase activity because the predicted amino acid sequence lacks aspartate residues, which are thought to be required for enzymatic activity. Cellulase activity is known to be essential for both eukaryotic and prokaryotic cellulose synthesis although the precise role is controversial, and requirements for cellulase in cleaving the initiation product, elongation, proof reading, and chain termination have been proposed (1, 24, 26). It is interesting to note that, in many prokaryotes, the cellulose synthase and endoglucanase genes required for cellulose synthesis are contained within a single operon (25). Further study is required to determine whether urochordate cellulose synthesis requires cellulase activity. However, these bacterial endoglucanases belong to family 8 of the glycosylhydrolases unlike the C. savignyi cellulase homologue, which belongs to family 6. The C. savignyi and C. intestinalis genomes encode several other predicted cellulases. However, these predicted cellulases are not necessarily involved in cellulose biosynthesis and may play roles in intestinal digestion of cellulose or tunic remodeling.
Urochordates are the only metazoans known to make cellulose. This finding raises the interesting question of the origin of the cellulose synthase gene in this group. If the gene had evolved independently from the cellulose synthases found in microorganisms and plants, it is unlikely that it would show extensive homology with cellulose synthases from these sources. However, the identified gene has high homology to the cellulose synthases from a wide variety of bacteria, a slime mold, and a few fungi. It shows lesser, but still quite significant, homology to the cellulose synthases from archaea and higher plants. C. intestinalis, the only other urochordate for which sequence data are available, seems to have a similar gene. Thus, it seems likely that urochordates acquired this gene by lateral transfer from one of the groups of microorganisms containing similar genes. It is probable that the sequence of the cellulose synthase gene of the organism from which the urochordates acquired the gene is not in the database, either because the particular group of organisms has not been sequenced or because the relevant organisms are now extinct. The distances between the sequenced groups of organisms carrying a cellulose synthase gene is so large that is not possible to make a robust tree of the phylogeny of the gene (data not shown).
The core catalytic domain of the C. savignyi cDNA is nearly identical to that predicted from the C. intestinalis genome (Fig. 2C). Therefore, the horizontal transfer event that we hypothesize resulted in the acquisition of a cellulose synthase-like gene in C. savignyi preceded the split of C. savignyi and C. intestinalis. We cannot conclude with certainty whether a single event is responsible for the acquisition of cellulose biosynthetic ability by the entire urochordate lineage. However, there is no trace of noneukaryotic codon bias or unusual G/C content in the putative C. savignyi cellulose synthase gene, arguing, as does the multiexon structure of the gene, that the horizontal transfer was an ancient event. Sequence information on the appendicularian cellulose synthase gene could help resolve this question because 18S rRNA sequence analysis suggests that they diverged much earlier than did the ascidians and thaliacea (27, 28).
Acknowledgments
We thank Drs. Arend Sidow and Jim Cooper for assistance and advice and Dr. Steve Poole for critical reading of the manuscript. We thank Dr. David Fenstermacher for assistance with computer analysis of the gene sequence and for general advice. This work was funded by National Institutes of Health Grant HD38701 (to W.C.S.) and National Science Foundation Grants IBN-0080060 (to W.C.S.), DE-FG02-00ER-15074 (to A.G.M.), and DE-FG02-00ER15073 (to A.R.W.).
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY504665).
References
- 1.Delmer, D. P. (1999) Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 245–276. [DOI] [PubMed] [Google Scholar]
- 2.Brown, R. M. (2003) Pure Appl. Chem. 71, 767–776. [Google Scholar]
- 3.Corbo, J. C., Di Gregorio, A. & Levine, M. (2001) Cell 106, 535–538. [DOI] [PubMed] [Google Scholar]
- 4.Hirose, E., Kimura, S., Itoh, T. & Nishikawa, J. (1999) Biol. Bull. 196, 113–120. [DOI] [PubMed] [Google Scholar]
- 5.Satoh, N. (1994) Developmental Biology of Ascidians (Cambridge Univ. Press, Cambridge, U.K.).
- 6.Kimura, S., Ohshima, C., Hirose, E., Nishikawa, J. & Itoh, T. (2001) Protoplasma 216, 71–74. [DOI] [PubMed] [Google Scholar]
- 7.Dehal, P., Satou, Y., Campbell, R. K., Chapman, J., Degnan, B., De Tomaso, A., Davidson, B., Di Gregorio, A., Gelpke, M., Goodstein, D. M., et al. (2002) Science 298, 2157–2167. [DOI] [PubMed] [Google Scholar]
- 8.Stasinopoulos, S. J., Fisher, P. R., Stone, B. A. & Stanisich, V. A. (1999) Glycobiology 9, 31–41. [DOI] [PubMed] [Google Scholar]
- 9.Saxena, I. M., Brown, R. M. & Dandekar, T. (2001) Phytochemistry 57, 1135–1148. [DOI] [PubMed] [Google Scholar]
- 10.Davis, S. W. & Smith, W. C. (2002) Dev. Genes Evol. 212, 81–86. [DOI] [PubMed] [Google Scholar]
- 11.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 24, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vandenberghe, A. E., Meedel, T. H. & Hastings, K. E. (2001) Genes Dev. 15, 294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wada, S., Katsuyama, Y., Yasugi, S. & Saiga, H. (1995) Mech. Dev. 51, 115–126. [DOI] [PubMed] [Google Scholar]
- 14.Egelhoff, T. T. & Long, S. R. (1985) J. Bacteriol. 164, 591–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matthysse, A. G., White, S. & Lightfoot, R. (1995) J. Bacteriol. 177, 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.York, W. S., Darvill, A. G., McNeil, M., Stevenson, T. T. & Albersheim, P. (1985) Methods Enzymol. 118, 3–40. [Google Scholar]
- 17.White, A. R., Xin, Y. & Pezeshk, V. (1993) Physiol. Plant. 87, 31–38. [Google Scholar]
- 18.White, A. R., Xin, Y. & Pezeshk, V. (1993) Biochem. J. 294, 231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carpita, N. C. & Shea, E. M. (1989) in Analysis of Carbohydrates by GLC and MS, eds. Biermann, C. J. & McGinnis, G. D. (CRC, Boca Raton, FL), pp. 157–216.
- 20.Blakeney, A. B., Harris, P. J., Henry, R. J. & Stone, B. A. (1983) Carbohydr. Res. 113, 291–299. [DOI] [PubMed] [Google Scholar]
- 21.Roberts, A. W., Roberts, E. M. & Delmer, D. P. (2002) Eukaryot. Cell 1, 847–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richmond, T. A. & Somerville, C. R. (2000) Plant Physiol. 14, 495–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koivula, A., Reinikainen, T., Ruohonen, L., Valkeajarvi, A., Claeyssens, M., Teleman, O., Kleywegt, G. J., Szardenings, M., Rouvinen, J., Jones, T. A. & Teeri, T. T. (1996) Protein Eng. 9, 691–699. [DOI] [PubMed] [Google Scholar]
- 24.Matthysse, A. G., Thomas, D. L. & White, A. R. (1995) J. Bacteriol. 177, 1076–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romling, U. (2002) Res. Microbiol. 153, 205–212. [DOI] [PubMed] [Google Scholar]
- 26.Peng, L., Kawagoe, Y., Hogan, P. & Delmer, D. (2002) Science 295, 147–150. [DOI] [PubMed] [Google Scholar]
- 27.Swalla, B. J., Cameron, C. B., Corley, L. S. & Garey, J. R. (2000) Syst. Biol. 49, 52–64. [DOI] [PubMed] [Google Scholar]
- 28.Wada, H. & Satoh, N. (1994) Proc. Natl. Acad. Sci. USA 91, 1801–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]