Abstract
Helicobacter pylori has been strongly associated with gastritis, gastric and duodenal ulcers, and it is a risk factor for gastric cancer. Two major virulence factors of H. pylori have been described: the cytotoxin-associated gene product (cagA) and the vacuolating toxin (vacA). Since considerable geographic diversity in the prevalence of H. pylori virulence factors has been reported, the aim of this work was to determine if there is a significant correlation between different H. pylori virulence genes (cagA and vacA) in 68 patients, from Saudi Arabia, and gastric clinical outcomes. H. pylor was recognized in cultures of gastric biopsies. vacA and cagA genes were detected by polymerase chain reaction (PCR). The cagA gene was obtained with 42 isolates (61.8%). The vacA s- and m- region genotypes were determined in all strains studied. Three genotypes were found: s1/m1 (28%), s1/m2 (40%) and s2/m2 (26%). The s2/m1 genotype was not found in this study. The relation of the presence of cagA and the development of cases to gastritis and ulcer was statistically significant (P < 0.05). The study showed a significant correlation between the vacA s1/m2 genotype and gastritis cases, and a significant correlation between vacA s1/m1 genotype and peptic ulcer cases. The results of this study might be used for the identification of high-risk patients who are infected by vacA s1/m1 genotype of H. pylori strains. In conclusion, H. pylori strains of vacA type s1 and the combination of s1/m1 were associated with peptic ulceration and the presence of cagA gene.
Keywords: Helicobacter pylori, cagA, vacA, Gastritis, Peptic Ulcer
INTRODUCTION
Helicobacter pylori (H. pylori) is a Gram-negative spiral bacterium which colonizes the human stomach (1). Infections with H. pylori may induce chronic gastritis, peptic ulcer, gastric adenocarcinoma and gastric mucosa-associated lymphoid tissue lymphoma (MALT) (2). There is increasing evidence that the genetic variability of H. pylori may have a clinical importance (3). There are two bacterial virulence factors of H. pylori and their genes also serve as epidemiological markers. The vacA (vacuolating toxin) and the cagA (cytotoxin-associated gene) play a major role in determining the clinical outcome of Helicobacter infections (4). The vacA gene, encoding the vacuolating toxin, is considered an important virulence factor and it is present in all strains (5). Significant sequence polymorphisms within vacA can be found in the coding sequence for the signal peptide (referred to as the s-region) and in the middle of the gene, called the middle (m) region. There are two allelic types (m1 and m2) in the middle region while the signal region has either an s1 (s1a, s1b, and s1c) or an s2 allele. The strains of the s1/m1 subtype typically produced higher levels the vacuolating cytotoxin than other genotypes, while s2/m2 strains do not secrete vacA (6). Toxins with different m- genotypes also display a differential specificity for intoxicating the target mammalian cells, with vacA m1 variants affecting wider range of target cells than those with m1 (7).
The product of the cagA gene is introduced into gastric epithelial cells by the type IV secretion system, where it becomes phosphorylated and modulates various cellular processes and signal transduction pathways. The intracellular cagA activities associated with the development of gastric carcinoma include disruption of tight junctions and a the responses (8). The presence of the cagA gene is an important marker for the most virulent strains associated with peptic ulcer, atrophic gastritis and adenocarcinoma (9). The cagA gene is a marker for the presence of the cag pathogenicity island and this gene with others on the island is associated with more severe clinical outcomes (10, 11). Atherton et al. (6) first reported a strong association between cagA and vacA signal sequence type s1/m1.
The patterns of H. pylori genotypes have been recently analyzed in several patient populations worldwide (6, 12-18). However, no such study has been carried out in Saudi Arabia. Here I report the results of such study and for the first time, investigate the prevalence of the cagA and vacA genotypes of H. pylori isolates from gastric cultures and demonstrate their relationships with clinical outcomes.
MATERIALS AND METHODS
Biopsy samples were obtained over a 6 months-period (January through June 2010) from selected patients referred for endoscopy at different hospitals in Riyadh, Saudi Arabia. Sixty eight patients, who had H. pylori, were enrolled in this study. The mean age of the patients was 43 yr (range, 14-84) and 45% were female. Histologically, patients were classified into gastritis in 37 cases (54.4%) and peptic ulceration in 31 cases (45.6%). One biopsy specimen taken from the antrum was used for the culture. All data of the subjects were collected from patient's file.
H. pylori culture and DNA extraction
Antral biopsies were cut into small pieces, homogenized and were smeared on the surface of H. pylori selective agar (Oxoid, Basingstoke, Hants, UK) then incubated at 37℃ in a BBL GasPak (Becton Dickinson Microbiology Systems, Cockeysville, MD, USA) containing a Campy-Pak Plus microaerophilic system generator (Becton Dickinson Microbiology Systems) for 7 days. The morphology of H. pylori clinical isolates colonies were smooth, translucent and small. Colonies that manifested the described characteristic morphologies were identified as H. pylori if they were Gram negative and shaped bacilli, and urease, catalase and oxidase positive. From the primary growth, seven or eight colonies were pooled together, and genomic DNA was extracted with the QIAamp DNA mini kit (Qiagen, Hilden, Germany) according to instructions of the manufacturer. The isolated DNA was eluted in 200 µL of 1 × TE buffer (10 mMTris-HCl, 1 mM EDTA [pH 8.0]) and stored at -20℃ until use.
H. pylori genotyping for cagA and vacA
After DNA samples extraction, polymerase chain reactions (PCR) were carried out in a volume of 50 µL containing 1 µM of each primers, 1 µL of genomic DNA (approximately 200 ng), 1 mM of dNTPs mix, 2 mM of MgCl2, and 0.05 U/µL Taq DNA polymerase.
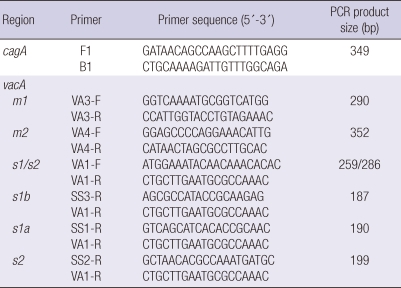
PCR amplifications were carried out in GeneAmp PCR system 9700 (Perkin Elmer, Norwalk, CT, USA). Table 1 summarized the primer sequences and the expected size of PCR products. The following cycle conditions were used: for cagA: 1 min at 94℃, 1 min at 56℃, and 1 min at 72℃ and for vacA: 35 cycles of 1 min at 94℃, 1 min at 53℃, and 1 min at 72℃. All runs included one negative and one positive DNA control. A 10 µL of amplified PCR products was then resolved by electrophoresis on 1.5% agarose gels run in acetate EDTA buffer and stained with ethidium bromide. The PCR product was visualized under a short wave length ultraviolet light source (Fig. 1).
Table 1.
PCR primers for amplification of cagA and vacA sequences (19)
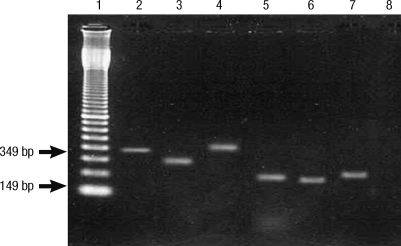
Fig. 1.
PCR genotyping of vacA and cagA status from different cases. Primers described in Table 1 were used for PCR reaction (Lanes- 1 = molecular weight marker; 2 = cagA+; 3 = m1; 4 = m2; 5 = s1a; 6 = s1b; 7 = s2 and 8 = Negative control [without DNA]).
Data analysis
Fisher's exact test was used for analysis of data. A P value of < 0.05 was accepted as statistically significant.
Ethics statement
Written informed consent was obtained from each participant before endoscopy.
RESULTS
Prevalence of cagA and vacA genotyping
The cagA gene was obtained with 42 isolates (61.8%) and 26 (38.2%) were negative. The vacA s- and m- region genotype were determined in all strains studied. For the s-region, in strains where a single vacA s allele was found, the minority 18 (26%) contained the s2 allele. In 46 isolates contained s1 allele (68%), 23 (34%) were subtype for each s1a and s1b. In the m-region, 2 strains contained both m1 and m2 alleles. In the strains containing one single vacA m allele, the m1 allele was found in 19 isolates (28%) and m2 in 45 ones (66%). Considering strains with only one single vacA genotype, and taking vacA s- and m-region together, three genotypes were found: s1/m1 (28%), s1/m2 (40%) and s2/m2 (26%). The s2/m1 genotype was not found in this study (Table 2).
Table 2.
Prevalence of H. pylori genotype detected in 68 isolates
Relationship between H. pylori genotyping and clinical outcomes
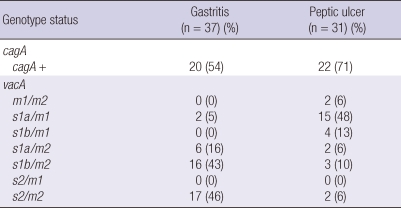
While estimating relationship between potentially virulent H. pylori strains and clinical outcomes, significant differences (P < 0.05) were found between isolates from gastritis and peptic ulcer cases (Table 3). The results showed a high percentage of cagA (70%) in peptic ulcer cases compared to gastritis cases (P < 0.05). The results showed a high percentage of m2/s1 with a distribution of 22 (59%) in gastritis cases (P < 0.05). In case of peptic ulcer, the highest rates were among m1/s1 with a frequency of 19 (61%) (P < 0.05).
Table 3.
Distribution of H. pylori genotype in gastritis and peptic ulcer patients
DISCUSSION
The geographic distribution of distinct H. pylori genotypes and the prevalence of virulent bacterial genotypes in several regions, particularly in Saudi Arabia, remain unknown. This study included 68 selected patients, who were infected with H. pylori. In the present study, the distribution of cagA and vacA genes and their relationship to clinical outcomes were examined. The cagA gene was obtained with 42 isolates (61.8%). These results were in agreement with other studies conducted in Europe, Central and South America, and East Asia where a higher prevalence (67% or more) of the cagA genotype was reported (20). For the vacA genotype, and when considering a single combined genotype, the results showed that the vacA s1 allele was predominant (68%) followed by the vacA s2 allele (26%). These are in contrary with a study in Kuwait reported that vacA s1 and s2 types were detected in approximately equal numbers in biopsies obtained from patients of Middle-Eastern origin, while North Africans were predominantly infected with the s2 type (21).
The distribution of cagA was 54% in gastritis and 71% in peptic ulcer cases. The relationship of the presence of cagA and the development of gastritis and peptic ulcer is statistically significant (P < 0.05), which further substantiate the role of cagA as a marker for increased virulence of H. pylori. These findings are in agreement with several previous studies (22, 23). A large number of studies have shown increased risk of gastric cancer in people with cagA positive H. pylori. However, other data have revealed that the occurrence of gastric malignancy is independent of cagA status. A few studies have also implicated pivotal roles of other virulence factors (cagE, cagT, vacA, babA, and hrgA) in the etiology of gastric cancer. Several genotypes of H. pylori have been reported to possess higher predictive value for the development of the severe form of the disease (24). Further study will be needed to determine the correlation between the H. pylori genotype and gastric cancer.
In this study, the prevalence of the vacA genotypes s1/m1 was detected in 28%, s1/m2 in 40% and s2/m2 in 26%. No single case for vacA s2/m1 genotype was detected in this study. This finding is in agreement with previous studies as this genotype was reported to be rare (12, 25). The most pathogenic vacA genotype (vacA s1/m1) was present in 61% of peptic ulceration cases, these are in agreement with previous studies in which association between this genotype and severe gastric outcomes were recognized. These findings support the role of vacA s1/m1 genotype in severe clinical outcomes (13, 14, 26).
In conclusion, the genotype may be used to identify patients who are at high risk for gastroduodenal disease. H. pylori strains with vacA type s1 and combination of s1/m1 are associated with peptic ulceration and the presence of cagA gene. This study suggests that further investigation is necessary to better understand the genetic diversity of this pathogen in our region.
Footnotes
This study was supported by the Deanship of Scientific Research and Research center, College of Applied Medical Sciences, King Saud University.
References
- 1.Marshall BJ. Helicobacter pylori. Am J Gastroenterol. 1994;89:S116–S128. [PubMed] [Google Scholar]
- 2.Park SM, Park J, Kim JG, Yoo BC. Relevance of vacA genotypes of Helicobacter pylori to cagA status and its clinical outcome. Korean J Intern Med. 2001;16:8–13. doi: 10.3904/kjim.2001.16.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim SY, Woo CW, Lee YM, Son BR, Kim JW, Chae HB, Youn SJ, Park SM. Genotyping cagA, vacA subtype, iceA1, and babA of Helicobacter pylori isolates from Korean patients, and their association with gastroduodenal diseases. J Korean Med Sci. 2001;16:579–584. doi: 10.3346/jkms.2001.16.5.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaser MJ, Perez-Perez GI, Kleanthous H, Cover TL, Peek RM, Chyou PH, Stemmermann GN, Nomura A. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 1995;55:2111–2115. [PubMed] [Google Scholar]
- 5.Atherton JC. The clinical relevance of strain types of Helicobacter pylori. Gut. 1997;40:701–703. doi: 10.1136/gut.40.6.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atherton JC, Cao P, Peek RM, Jr, Tummuru MK, Blaser MJ, Cover TL. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 7.Ji X, Fernandez T, Burroni D, Pagliaccia C, Atherton JC, Reyrat JM, Rappuoli R, Telford JL. Cell specificity of Helicobacter pylori cytotoxin is determined by a short region in the polymorphic midregion. Infect Immun. 2000;68:3754–3757. doi: 10.1128/iai.68.6.3754-3757.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wen S, Moss SF. Helicobacter pylori virulence factors in gastric carcinogenesis. Cancer Lett. 2009;282:1–8. doi: 10.1016/j.canlet.2008.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Figura N, Guglielmetti P, Rossolini A, Barberi A, Cusi G, Musmanno RA, Russi M, Quaranta S. Cytotoxin production by Campylobacter pylori strains isolated from patients with peptic ulcers and from patients with chronic gastritis only. J Clin Microbiol. 1989;27:225–226. doi: 10.1128/jcm.27.1.225-226.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akopyants NS, Clifton SW, Kersulyte D, Crabtree JE, Youree BE, Reece CA, Bukanov NO, Drazek ES, Roe BA, Berg DE. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol Microbiol. 1998;28:37–53. doi: 10.1046/j.1365-2958.1998.00770.x. [DOI] [PubMed] [Google Scholar]
- 11.Rudi J, Rudy A, Maiwald M, Kuck D, Sieg A, Stremmel W. Direct determination of Helicobacter pylori vacA genotypes and cagA gene in gastric biopsies and relation to gastrointestinal disease. Am J Gastroenterol. 1999;94:1525–1531. doi: 10.1111/j.1572-0241.1999.1138_a.x. [DOI] [PubMed] [Google Scholar]
- 12.Martínez A, González C, Kawaguchi F, Montoya R, Corvalán A, Madariaga J, Roa J, García A, Salgado F, Solar H, Palma M. Helicobacter pylori: cagA analysis and vacA genotyping in Chile. Detection of a s2/m1 strain. Rev Med Chil. 2001;129:1147–1153. [PubMed] [Google Scholar]
- 13.Gatti LL, Fagundes e Souza EK, Leite KR, Bastos EL, Vicentini LR, Silva LC, Smith Mde A, Payão SL. cagA, vacA alleles and babA2 genotypes of Helicobacter pylori associated with gastric disease in Brazilian adult patients. Diagn Microbiol Infect Dis. 2005;51:231–235. doi: 10.1016/j.diagmicrobio.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 14.Wu CC, Chou PY, Hu CT, Liu ZC, Lin CY, Tseng YH, Lin NT. Clinical relevance of the vacA, iceA, cagA, and flaA genes of Helicobacter pylori strains isolated in Eastern Taiwan. J Clin Microbiol. 2005;43:2913–2915. doi: 10.1128/JCM.43.6.2913-2915.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamazaki S, Yamakawa A, Okuda T, Ohtani M, Suto H, Ito Y, Yamazaki Y, Keida Y, Higashi H, Hatakeyama M, Azuma T. Distinct diversity of vacA, cagA, and cagE genes of Helicobacter pylori associated with peptic ulcer in Japan. J Clin Microbiol. 2005;43:3906–3916. doi: 10.1128/JCM.43.8.3906-3916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagiyev T, Yula E, Abayli B, Koksal F. Prevalence and genotypes of Helicobacter pylori in gastric biopsy specimens from patients with gastroduodenal pathologies in the Cukurova Region of Turkey. J Clin Microbiol. 2009;47:4150–4153. doi: 10.1128/JCM.00605-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yakoob J, Abid S, Abbas Z, Jafri W, Ahmad Z, Ahmed R, Islam M. Distribution of Helicobacter pylori virulence markers in patients with gastroduodenal diseases in Pakistan. BMC Gastroenterol. 2009;9:87. doi: 10.1186/1471-230X-9-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jang S, Jones KR, Olsen CH, Joo YM, Yoo YJ, Chung IS, Cha JH, Merrell DS. Epidemiological link between gastric disease and polymorphisms in vacA and cagA. J Clin Microbiol. 2010;48:559–567. doi: 10.1128/JCM.01501-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ben Mansour K, Fendri C, Zribi M, Masmoudi A, Labbene M, Fillali A, Ben Mami N, Najjar T, Meherzi A, Sfar T, Burucoa C. Prevalence of Helicobacter pylori vacA, cagA, iceA and oipA genotypes in Tunisian patients. Ann Clin Microbiol Antimicrob. 2010;9:10. doi: 10.1186/1476-0711-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Doorn LJ, Figueiredo C, Mégraud F, Pena S, Midolo P, Queiroz DM, Carneiro F, Vanderborght B, Pegado MD, Sanna R, De Boer W, Schneeberger PM, Correa P, Ng EK, Atherton J, Blaser MJ, Quint WG. Geographic distribution of vacA allelic types of Helicobacter pylori. Gastroenterology. 1999;116:823–830. doi: 10.1016/s0016-5085(99)70065-x. [DOI] [PubMed] [Google Scholar]
- 21.Al Qabandi A, Mustafa AS, Siddique I, Khajah AK, Madda JP, Junaid TA. Distribution of vacA and cagA genotypes of Helicobacter pylori in Kuwait. Acta Trop. 2005;93:283–288. doi: 10.1016/j.actatropica.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 22.Blaser MJ, Berg DE. Helicobacter pylori genetic diversity and risk of human disease. J Clin Investig. 2001;107:767–773. doi: 10.1172/JCI12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stephens JC, Stewart JA, Folwell AM, Rathbone BJ. Helicobacter pylori cagA status, vacA genotypes and ulcer disease. Eur J Gastroenterol Hepatol. 1998;10:381–384. doi: 10.1097/00042737-199805000-00005. [DOI] [PubMed] [Google Scholar]
- 24.Tiwari SK, Manoj G, Kumar GV, Sivaram G, Hassan SI, Prabhakar B, Devi U, Jalaluddin S, Kumar K, Ahmed S, Abid Z, Habeeb MA, Khan AA, Habibullah CM. Prognostic significance of genotyping Helicobacter pylori infection in patients in younger age groups with gastric cancer. Postgrad Med J. 2008;84:193–197. doi: 10.1136/pgmj.2007.065060. [DOI] [PubMed] [Google Scholar]
- 25.Letley DP, Lastovica A, Louw JA, Hawkey CJ, Atherton JC. Allelic diversity of the Helicobacter pylori vacuolating cytotoxin gene in South Africa: rarity of the vacA s1a genotype and natural occurrence of an s2/m1 allele. J Clin Microbiol. 1999;37:1203–1205. doi: 10.1128/jcm.37.4.1203-1205.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aydin F, Kaklikkaya N, Ozgur O, Cubukcu K, Kilic AO, Tosun I, Erturk M. Distribution of vacA alleles and cagA status of Helicobacter pylori in peptic ulcer disease and non-ulcer dyspepsia. Clin Microbiol Infect. 2004;10:1102–1104. doi: 10.1111/j.1469-0691.2004.00989.x. [DOI] [PubMed] [Google Scholar]