Abstract
TOR is a conserved serine/threonine kinase that responds to nutrients, growth factors, the bioenergetic status of the cell and cellular stress to control growth, metabolism and ageing. A diverse group of small GTPases including Rheb, Rag, Rac1, RalA and Ryh1 play a variety of roles in the regulation of TOR. For example, while Rheb binds to and activates TOR directly, Rag and Rac1 regulate its localization and RalA activates it indirectly through the production of phosphatidic acid. Here, we review recent findings on the regulation of TOR by small GTPases.
Keywords: TOR, Rheb, Rag, RalA, Rac1, Ryh1
See Glossary for abbreviations used in this article.
Glossary.
AMPK AMP-activated protein kinase
Arf ADP-ribosylation factor
AVO adheres voraciously to TOR
CTD carboxy-terminal domain
Deptor DEP domain-containing mTOR-interacting protein
eIF4E eukaryotic initiation factor 4E
ERK extracellular signal-regulated kinase
GAP GTPase-activating protein
GEF guanine nucleotide exchange factor
HIF1 hypoxia-inducible factor 1
MAPK mitogen-activated protein kinase
MEK MAPK/ERK kinase
PA phosphatidic acid
PI3K phosphatidylinositol-3 kinase
PKC protein kinase C
PLD phospholipase D
PP2A protein phosphatase 2A
PRAK p38-regulated/activated kinase
PRAS40 proline-rich AKT substrate 40
Protor protein observed with Rictor
PRR5 proline-rich protein 5
Raptor regulatory associate protein of TOR
REDD1 regulated in development and DNA damage response 1
Rheb Ras homologous enriched in brain
Rictor rapamycin-insensitive companion of TOR
RSK ribosomal s6 kinase
SGK1 serum and gucocorticoid-induced kinase 1
TCTP translationally controlled tumour protein
TOR target of rapamycin
TORC1/2 TOR complex 1/2
tRNA transfer RNA
TSC tuberous sclerosis complex
Introduction
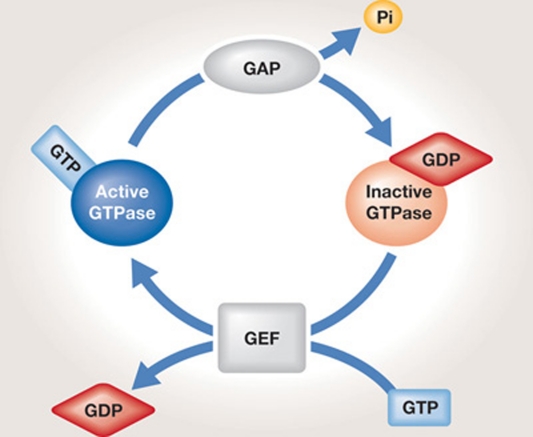
The growth-controlling TOR signalling pathway is structurally and functionally conserved from unicellular eukaryotes to humans. TOR, an atypical serine/threonine kinase, was originally discovered in Saccharomyces cerevisiae as the target of rapamycin (Heitman et al, 1991). It was later described in many other organisms including the protozoan Trypanosoma brucei, the yeast Schizosaccharomyces pombe, photosynthetic organisms such as Arabidopsis thaliana and Chlamydomonas reinhardtii, and in metazoans such as Caenorhabditis elegans, Drosophila melanogaster and mammals. TOR integrates various stimuli to control growth, metabolism and ageing (Avruch et al, 2009; Kim & Guan, 2011; Soulard et al, 2009; Wullschleger et al, 2006; Zoncu et al, 2011a). In mammals, mTOR is activated by nutrients, growth factors and cellular energy, and is inhibited by stress. Thus, the molecular regulation of TOR is complex and diverse. Among the increasing number of TOR regulators, small GTPases are currently garnering much attention. Small GTPases (20–25 kDa) are either in an inactive GDP-bound form or an active GTP-bound form (Bos et al, 2007). GDP–GTP exchange is regulated by GEFs, which mediate the replacement of GDP by GTP, and by GAPs, which stimulate the intrinsic GTPase activity of a cognate GTPase to convert GTP into GDP (Fig 1). Upon activation, small GTPases interact with effector proteins, thereby stimulating downstream signalling pathways. Small GTPases constitute a superfamily that comprises several subfamilies, such as the Rho, Ras, Rab, Ran and Arf families. Rheb, Rag, RalA, Rac1 and Ryh1, all members of the small GTPase superfamily, play a role in the concerted regulation of TOR by different stimuli. This review summarizes recent advances in the understanding of TOR regulation by these small GTPases.
Figure 1.
Regulation of small GTPases by GEFs and GAPs. A guanine nucleotide exchange factor (GEF) replaces GDP with GTP to activate the signalling function of the GTPase. Conversely, a GTPase-activating protein (GAP) stimulates hydrolysis of GTP into GDP to inactivate the small GTPase.
The TOR complexes
TOR is found in two functionally and structurally distinct multiprotein complexes, named TORC1 and TORC2 (Avruch et al, 2009; Kim & Guan, 2011; Soulard et al, 2009; Wullschleger et al, 2006; Zoncu et al, 2011a). TORC1 regulates several cellular processes including protein synthesis, ribosome biogenesis, nutrient uptake and autophagy. TORC2, in turn, regulates actin cytoskeleton organization, cell survival, lipid synthesis and probably other processes. TORC1 and TORC2 are rapamycin-sensitive and rapamycin-insensitive, respectively, although in some organisms, for example A. thaliana and T. brucei, this rule does not apply (Barquilla et al, 2008; Mahfouz et al, 2006). Nevertheless, long-term treatment with rapamycin can also indirectly inhibit TORC2 in mammalian cell lines (Sarbassov et al, 2006). Furthermore, there is accumulating evidence that not all TORC1 readouts are rapamycin-sensitive (Choo & Blenis, 2009; Dowling et al, 2010; Peterson et al, 2011).
Saccharomyces cerevisiae has two TORs encoded by two separate genes, TOR1 and TOR2. TOR1 is part of TORC1 only, whereas TOR2 is found in both TORC1 and TORC2. The core components of TORC1 in budding yeast are TOR (TOR1 or TOR2), KOG1 and LST8. The core components of TORC2 are TOR2, AVO1, AVO2, AVO3 and LST8. The fission yeast S. pombe also has two TORs (SpTOR1 and SpTOR2), but their numbering is reversed compared with those in budding yeast—SpTOR1 is the orthologue of TOR2 in S. cerevisiae, and SpTOR2 is orthologous to budding yeast TOR1. By contrast, mammals have a single mTOR. The core components of mTORC1 are mTOR, Raptor (KOG1 orthologue) and mLST8. Other proteins, such as the regulators PRAS40 and Deptor, interact directly with TORC1 to regulate its kinase activity (Peterson et al, 2009; Sancak et al, 2007; Thedieck et al, 2007; Vander Haar et al, 2007). However, most significant among the regulating interactors are the small GTPases Rheb and Rag (Avruch et al, 2009; Kim et al, 2008; Sancak et al, 2008). Mammalian mTORC2 is composed of mTOR, Rictor, mSIN1 (AVO1 orthologue), mLST8 and PRR5 (also known as Protor). Similarly to mTORC1, Deptor also binds to mTORC2 (Peterson et al, 2009).
Upstream of TOR
Four main inputs regulate mTORC1: nutrients, growth factors, the bioenergetic status of the cell and oxygen availability. It is well established that growth factors activate mTORC1 through the PI3K–AKT pathway. Once activated, AKT phosphorylates and inhibits the heterodimeric complex TSC1–TSC2, a GAP for Rheb and thus an inhibitor of mTORC1 (Avruch et al, 2009). The TSC1–TSC2 heterodimer is a ‘reception centre’ for various stimuli that are then transduced to mTORC1, including growth factor signals transduced through the AKT and ERK pathways, hypoxia through HIF1 and REDD1, and energy status through AMPK (Wullschleger et al, 2006). In addition to the small GTPases Rheb and Rag (see below), PA also binds to and activates mTORC1 (Fang et al, 2001). Pharmacological or genetic inhibition of PA production, through the inhibition of PLD, impairs activation of mTORC1 by nutrients and growth factors (Fang et al, 2001). Moreover, elevated PLD activity leads to rapamycin resistance in human breast cancer cells (Chen et al, 2003), further supporting a role for PA as an mTORC1 regulator. As discussed below, the small GTPase RalA participates in the mechanism by which PA activates mTORC1 (Maehama et al, 2008; Xu et al, 2011).
In the case of nutrients, amino acids in particular, several elements mediate the activation of TORC1. As discussed below, the Rag GTPases are necessary to activate TORC1 in response to amino acids (Binda et al, 2009; Kim et al, 2008; Sancak et al, 2008). In mammals, it has also been proposed that amino acids stimulate an increase in intracellular calcium concentration, which in turn activates mTORC1 through the class III PI3K Vps34 (Gulati et al, 2008). However, this model has not been confirmed in other organisms (Juhasz et al, 2008). Furthermore, the Ste20-like kinase MAP4K3 plays a role in the activation mTORC1 in response to amino acids (Findlay et al, 2007; Yan et al, 2010). It is not yet clear how the above nutrient signalling components are functionally related—that is, whether they are in a single pathway or in parallel pathways. It has been proposed that amino acid signalling might involve both activation of kinases and inhibition of counteracting phosphatases, namely PP2A (Meijer & Dubbelhuis, 2004; Yan et al, 2010). Finally, tRNAs have been invoked as part of an amino acid sensing and signalling mechanism, but short-term amino acid deprivation does not affect aminoacyl-tRNA or free tRNA levels (Dennis et al, 2001).
By contrast, the regulation of TORC2 is poorly understood. In mammals, insulin stimulates mTORC2 through the PI3K pathway. mTORC2 interacts with the ribosome (Oh et al, 2010), and this direct interaction is required for the efficient activation of mTORC2 by insulin and PI3K (Zinzalla et al, 2011). As TORC2 appears to interact with the ribosome in both mammalian and yeast systems, this is probably a conserved mechanism of TORC2 activation. PA has also been proposed to positively regulate mTORC2, as inhibition of PLD blocks mTORC2-mediated phosphorylation of AKT-Ser 473 and PRAS40 (Toschi et al, 2009).
Downstream of TOR
TORC1 regulates growth-related processes such as transcription, ribosome biogenesis, protein synthesis, nutrient transport and autophagy (Wullschleger et al, 2006). In mammals, the best-characterized substrates of mTORC1 are S6K and 4E-BP1, through which mTORC1 stimulates protein synthesis. mTORC1 activates S6K, which is a positive regulator of protein synthesis, and inhibits 4E-BP1, which is a negative regulator of protein synthesis. Upon phosphorylation by mTORC1, 4E-BP1 releases eIF4E. Once released from 4E-BP1, eIF4E interacts with the eIF4G subunit of the eIF4F complex, allowing initiation of translation. In mammals, 4E-BP1 participates mainly in the regulation of cell proliferation and metabolism (Dowling et al, 2010). In S. cerevisiae, the main substrate of TORC1 is the S6K orthologue Sch9 (Urban et al, 2007). Sch9 is required for the activation of ribosome biogenesis and translation initiation stimulated by TORC1. Furthermore, it participates in TORC1-dependent inhibition of G0 phase entry.
In part due to the fact that TORC2 is rapamycin-insensitive, its downstream effects have been analysed to a lesser extent than those of TORC1. TORC2 controls organization of the actin cytoskeleton, and phosphorylation and activation of AGC kinase family members such as AKT, SGK1 and PKC in mammals (Cybulski & Hall, 2009) and the SGK1 orthologues YPK2 and GAD8 in S. cerevisiae and S. pombe, respectively. Phosphorylation of YPK2 by TORC2 in S. cerevisiae mediates sphingolipid biosynthesis, whereas phosphorylation of GAD8 by TORC2 in S. pombe controls entry into mitosis and G1 arrest in response to stress. In C. elegans, TORC2 phosphorylates SGK1 to control fat storage, body size and development (Soulard et al, 2009).
Regulation of TOR by Rheb
The small GTPase Rheb was first identified in 1994 in a screen for genes induced in neurons in response to synaptic activity (Yamagata et al, 1994), and was first described to interact with the Raf1 kinase (Yee & Worley, 1997). A later report showed that loss of Rhb1, the Rheb orthologue in S. pombe, causes a starvation-like growth arrest (Mach et al, 2000). In 2003, several independent groups working with mammalian cells in vitro and Drosophila in vivo demonstrated that Rheb is the target of the TSC1–TSC2 GAP and a TORC1 activator (Avruch et al, 2009).
Drosophila and fission yeast have a single Rheb gene, whereas mammals have two, termed RHEB1 and RHEB2 (Patel et al, 2003). RHEB1 mRNA is relatively widespread, whereas RHEB2 is expressed mainly in the brain (Yamagata et al, 1994). A recent study performed in mice demonstrated that RHEB1 is dominant over RHEB2 in the regulation of mTORC1 in vivo (Zou et al, 2011). The mechanism underlying TORC1 activation by Rheb is not completely understood. Rheb binds to TOR directly (Long et al, 2005a), although this interaction appears to be weak and has not been detected between endogenous proteins.
Interestingly, the Rheb–mTOR interaction both in vivo and in vitro does not depend on GTP loading of Rheb. This is unusual for GTPases as GTP loading usually regulates effector binding. However, GTP loading of Rheb is crucial for the activation of mTOR kinase activity (Sancak et al, 2007). Conversely, mTOR becomes inactive after association with a nucleotide-deficient Rheb (Long et al, 2005a; Fig 2). Similar results were obtained in S. pombe, making use of mutations that hyperactivate Rheb by increasing its overall GTP : GDP binding ratio (Urano et al, 2005). In contrast to the situation in mammals, interaction of Rheb with SpTOR2 in fission yeast is detected only with a hyperactive Rheb mutant. This suggests that, in S. pombe, Rheb binds to SpTOR2 in a GTP-dependent manner.
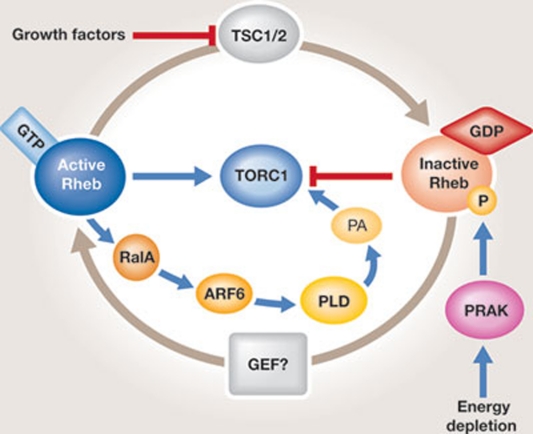
Figure 2.
Rheb activates TORC1 both directly and indirectly. GTP-bound Rheb interacts directly with TORC1 to activate TORC1 kinase. GTP-bound Rheb also activates RalA, which activates PLD to increase production of PA. PA in turn interacts with TORC1 to stimulate TORC1 kinase activity. Rheb is inactivated by TSC1–TSC2, which acts as a GAP for Rheb. GAP, GTPase-activating protein; PA, phosphatidic acid; PLD, phospholipase D; TORC1, TOR complex 1; TSC, tuberous sclerosis complex.
The TSC1–TSC2 complex plays a crucial role in the regulation of Rheb by growth factors. As the Rheb GAP, TSC1–TSC2 promotes conversion of GTP to GDP in Rheb. Growth factors stimulate several kinases, including AKT, ERK and RSK, that phosphorylate TSC2 and thereby inhibit the TSC1–TSC2 complex (Wullschleger et al, 2006). The phosphorylation status of TSC2 correlates with TORC1 activation. In the active unphosphorylated state, TSC1–TSC2 localizes at the membrane where it exerts its GAP activity to inhibit Rheb and ultimately TORC1. Phosphorylation of TSC2 promotes translocation of TSC2 to the cytosol, allowing for Rheb and TORC1 activation (Cai et al, 2006). The TSC1/2–Rheb pathway also participates in stress-mediated activation of mTORC1. Indeed, redox stress regulates mTORC1 activity independently of Rag GTPase. Instead, redox stress increases the GTP-bound state of Rheb (Yoshida et al, 2011). A role for TSC1–TSC2 upstream of Rheb has also been established in S. pombe, where disruption of TSC2 results in partial activation of TORC1 under nitrogen depletion, similarly to the activation of TORC1 observed on hyperactivation of Rheb (Nakashima et al, 2010). By contrast, much less is known about the GEF regulating the conversion of Rheb-GDP to Rheb-GTP. One possible reason for this is that Rheb does not require a GEF. This is supported by the finding that Rheb mutants (S20N, D60V and D60K) with a decreased affinity for nucleotides do not act as dominant negatives (Li et al, 2004), although these conclusions are still under debate (Long et al, 2005a). Another possible reason for the lack of a viable RhebGEF candidate is genetic redundancy among several possible GEFs.
One study in Drosophila suggested that the TCTP protein is the GEF for Rheb (Hsu et al, 2007; Fig 2). However, subsequent studies in mammalian cells showed that reducing TCTP levels did not affect mTORC1 signalling in amino-acid-replete/insulin-stimulated cells. Moreover, overexpression of TCTP does not rescue mTORC1 signalling in amino-acid-starved cells (Rehmann et al, 2008; Wang et al, 2008). These findings suggest that TCTP does not regulate mTORC1 signalling, at least in mammals.
In addition to the direct interaction between mTOR and Rheb, activation of PA production by Rheb is an additional mechanism by which Rheb might regulate mTORC1. Rheb binds to and activates PLD in a GTP-dependent manner (Sun et al, 2008). PLD produces PA, which binds directly to and upregulates mTORC1. This finding reveals cross-talk between the TSC–Rheb and the PA pathways in the regulation of mTORC1 signalling. A recent study by Yoon and colleagues further demonstrated the role of PLD in mTORC1 regulation (Yoon et al, 2011). They showed that amino acids activate PLD through translocation of PLD to the lysosomal compartment. This translocation is positively regulated by human Vps34 and is necessary for the activation of mTORC1 by amino acids. These authors propose the existence of a Vps34–PLD1 pathway that activates mTORC1 in parallel to the Rag pathway (Yoon et al, 2011).
Rheb is also regulated through direct phosphorylation by PRAK (Zheng et al, 2011). Using a mammalian cell culture approach, this report demonstrated that energy stress activates the p38β–PRAK cascade and that PRAK is essential for energy-depletion-induced inhibition of mTORC1. Furthermore, the action of PRAK on mTORC1 is independent of TSC2 and AMPK, but instead involves a direct interaction and subsequent phosphorylation of Rheb-Ser 130 by PRAK. This phosphorylation impairs the guanyl-nucleotide-binding ability of Rheb and therefore inhibits Rheb-mediated mTORC1 activation (Fig 2). These results are in contrast to previously reported findings demonstrating that p38 is an important positive regulator of TORC1. Indeed, phosphorylation of TSC2 at Ser 1210 by MK2, a kinase downstream from p38α, inhibits TSC2 and thereby activates Rheb and mTORC1 (Li et al, 2003), further inducing mTORC1 activation. Moreover, phosphorylation of Raptor by p38β participates in arsenite-induced mTORC1 activation in cultured mammalian cells (Wu et al, 2011). Thus, the cross-talk between MAPK and mTOR pathways is complex and operates at different levels, in both a Rheb-dependent and a Rheb-independent manner.
Although Rheb is required for the activation of mTORC1 by amino acids, Rheb itself does not participate in amino acid sensing, and GTP-loading of Rheb is not affected by amino acid depletion (Long et al, 2005b). Furthermore, amino acid depletion inhibits mTORC1 even in TSC2−/− fibroblasts (Roccio et al, 2006). Nevertheless, interaction of mTORC1 with Rheb depends on amino acid availability (Long et al, 2005b). As discussed below, the current model proposes that amino acids mediate translocation of mTORC1 to the lysosomal surface where mTORC1 interacts with and is activated by GTP-loaded Rheb (Sancak et al, 2008). The role of Rheb in development in mammals has also been investigated. Rheb is essential for murine development beyond embryonic day 12 (Goorden et al, 2011), and conditional knockout in the brain reduces brain size and impairs postnatal myelination (Zou et al, 2011). Rheb signalling also plays an important role in regulating mTORC1-dependent T-cell differentiation (Delgoffe et al, 2011).
Regulation of TOR by Rag
Rag GTPases have unique features among the Ras GTPase subfamily members: they form heterodimers and lack a membrane-targeting sequence (Nakashima et al, 1999; Sekiguchi et al, 2001). Gtr1 in S. cerevisiae was the first member of this GTPase subfamily to be identified (Bun-Ya et al, 1992). The mammalian RagA and RagB GTPases were later described as Gtr1 orthologues (Hirose et al, 1998). Gtr2 in yeast (Nakashima et al, 1999) and its mammalian orthologues RagC and RagD (Sekiguchi et al, 2001) were subsequently discovered due to their ability to form heterodimers with Gtr1 in yeast and RagA and RagB in mammals, respectively. The crystal structure of the Gtr1–Gtr2 complex has been determined recently (Gong et al, 2011). Gtr1 and Gtr2 have similar structures, organized in two domains: an amino-terminal GTPase domain (designated as the G domain) and a carboxy-terminal domain. The Gtr1–Gtr2 heterodimer presents a pseudo-twofold symmetry resembling a horseshoe. The crystal structure reveals that Gtr1–Gtr2 dimerization results from extensive contacts between the C-terminal domains of both proteins, while the G domains do not contact each other (Gong et al, 2011).
Gtr1 was initially implicated in phosphate uptake and the Ran/Gsp1–GTPase pathway (Bun-Ya et al, 1992; Hirose et al, 1998). The Rag family GTPases were only recently shown to have a role in TOR regulation. In yeast, Gtr1 and Gtr2 participate in the EGO complex, a vacuolar-membrane-associated protein complex that, in conjunction with TOR, positively regulates microautophagy (Dubouloz et al, 2005; Gao & Kaiser, 2006). Further studies demonstrated that the EGO complex functions upstream from TORC1, and that TORC1 activity is determined by the nucleotide-bound state of Gtr1 (Binda et al, 2009). In Drosophila and mammals, two independent reports published in 2008 demonstrated that the Rag proteins are necessary for the activation of TORC1 by amino acids (Kim et al, 2008; Sancak et al, 2008). In mammals, RagA or RagB interact with RagC or RagD to constitute a heterodimer. According to the current model of mTORC1 activation by amino acids through the Rag proteins (Fig 3), a Rag complex is constitutively anchored on the surface of the lysosome. The Rag heterodimer is anchored to the lysosome by the MP1–p14–p18 complex (also known as the ‘Ragulator’; Sancak et al, 2010). However, the Ragulator does not appear to act exclusively with Rags as it was previously shown to serve as an anchoring element also for MEK1 and ERK (Nada et al, 2009). In addition to the MP1–p14–p18 complex, the signalling adaptor p62 also interacts with Rags, thereby favouring the formation of the Rag complex and ultimately the activation of mTORC1 by Rags and amino acids (Duran et al, 2011). In the absence of amino acids, RagA or RagB, the dominant member of the Rag heterodimer (Binda et al, 2009), is GDP loaded and unable to recruit mTORC1 to the surface of the lysosome. As a result, the mTORC1 complex is dispersed throughout the cytosol in an inactive conformation (Kim et al, 2008; Sancak et al, 2008). The presence of amino acids inside the cell induces the exchange of GDP by GTP in RagA/B. The exchange mechanism is not well understood, but it has been proposed to be mediated by the Vam6 GEF in yeast (Binda et al, 2009). GTP-bound RagA/B binds to and recruits mTORC1 to the surface of the lysosome in a process that also requires the vacuolar H+-ATPase (Zoncu et al, 2011b). Once at the lysosome, mTORC1 is able to interact with active Rheb (activated by a separate growth factor input; Sancak et al, 2010). Structural data support the idea that Rag GTPase interacts with p18 in the MP1–p14–p18 complex. In a manner dependent on GTP-loading of RagA/B, the Rag heterodimer interacts with Raptor mainly through the G domain of RagA, although RagC/D is also required. Indeed, although the G domain of RagC/D plays a minor role in the interaction with Raptor, the nucleotide binding status of RagC/D still regulates the binding affinity of the complex (Gong et al, 2011). Unexpectedly, in this case it appears that RagC/D is bound to GDP and, consistent with this, expression of a GTP-bound mutant of RagB combined with a GDP-bound mutant of RagD activates mTORC1 (Sancak et al, 2008). Similar results were obtained in yeast (Binda et al, 2009).
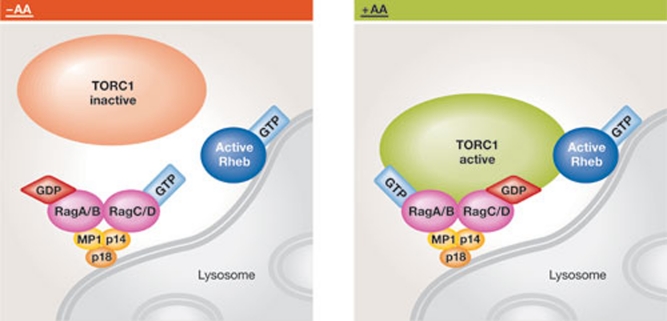
Figure 3.
Rag proteins mediate the activation of TORC1 in response to amino acids. The RagA/B–RagC/D heterodimer is anchored to the MP1–p14–p18 complex on the surface of the lysosome. In the absence of amino acids (left panel), RagA/B is GDP-bound whereas RagC/D is GTP-bound, and the Rag heterodimer is inactive and unable to recruit TORC1 to the lysosomal surface. In the presence of amino acids (right panel), RagA/B exchanges GDP for GTP and RagC/D converts its GTP to GDP. This Rag heterodimer acquires an active conformation that binds to and thereby recruits TORC1. Once recruited to the surface of the lysosome, TORC1 interacts with a GTP-loaded Rheb. TORC1, TOR complex 1.
The above model explains why a GDP-bound RagB mutant exerts a dominant negative effect on mTORC1. Such a mutant prevents mTORC1 from interacting with the lysosome. Furthermore, it explains how the interaction of Rheb with mTORC1, but not the GTP loading of Rheb, is stimulated by amino acids to activate mTORC1 (Long et al, 2005b). Finally, it also explains how overexpression of Rheb promotes mTORC1 activation even in the absence of amino acids. Overexpressed Rheb is mislocalized throughout the cell, and therefore interaction of mTORC1 with Rheb does not require amino-acid-induced translocation of mTORC1 to the lysosome. The model is further supported by observations in Drosophila showing that expression of a constitutively active mutant of RagA significantly increases the size of individual cells, whereas expression of a dominant negative mutant of RagA reduces cell size (Kim et al, 2008). Moreover, Rag plays a role in TORC1-mediated inhibition of autophagy both in Drosophila (Kim et al, 2008) and in human cells (Narita et al, 2011).
Little is known about the mechanism of activation of Rag GTPases in response to an amino acid signal. In yeast, Vam6 is a GEF for Gtr1 (Binda et al, 2009). Vam6 was identified in a genome-wide synthetic lethal screen using a mutant of Gtr1 that preferentially binds to GDP. Loss of Vam6 and Gtr1 similarly affect TORC1. Furthermore, overexpression of Vam6 renders wild-type but not Gtr1-deficient cells resistant to rapamycin. Finally, Vam6 stimulates GDP release from Gtr1, and loss of Vam6 severely reduces the interaction between Gtr1 and Ego1 (Binda et al, 2009). These results suggest that Vam6 is a GEF for Gtr1 and explain the finding that class C vps mutants (such as vam6) fail to recover from rapamycin-induced growth arrest or to survive nitrogen starvation (Zurita-Martinez et al, 2007). Vam6 appears to be evolutionarily conserved (26% identity with human Vam6), suggesting that the regulation of Rag by the mammalian Vam6 orthologue could also be conserved.
Regulation of TOR by RalA
PA acts upstream of both mTORC1 and mTORC2, as suggested by the fact that inhibition of PLD impairs activation of both complexes (Fang et al, 2001; Toschi et al, 2009). PA is produced by PLD-mediated hydrolysis of phosphatidylcholine. The small GTPase RalA plays a role in this process (Fig 2; Xu et al, 2011) through interaction with PLD and its subsequent activation (Jiang et al, 1995). However, RalA does not activate PLD directly, but instead promotes the association of PLD with ARF6, a member of the ARF family of GTPases, which in turn activates PLD (Xu et al, 2003). As PLD is a positive regulator of both mTORC1 and mTORC2, a potential connection between RalA and the mTOR pathway has been proposed and indeed proven for mTORC1 (Maehama et al, 2008). The amount of GTP-bound RalA in cultured human cells increases when amino acids are added to the cells, suggesting that RalA responds to amino acid availability. Furthermore, RalA is indispensable for the Rheb-dependent activation of mTORC1 induced by extracellular nutrients. Inhibition of RalA abolishes amino-acid- and glucose-induced mTORC1 activation even after overexpression of a hyperactive Rheb mutant (Maehama et al, 2008). Therefore, RalA—similarly to PLD—seems to act downstream of Rheb to activate mTORC1 (Sun et al, 2008). In support of this model, activation of both PLD and mTORC1 in human cancer cells by nutrients is dependent on RalA and ARF6 (Xu et al, 2011). In conclusion, a RalA–ARF6–PLD complex appears to promote activation of mTORC1 in response to nutrients.
Regulation of TOR by Rac1
The Rho subfamily of GTPases is present in all eukaryotic cells, from yeast to mammals, regulating both actin organization and morphogenesis. In yeast, Rho proteins mediate polarized growth, cell integrity, cytokinesis and mating (Park & Bi, 2007). In mammals, the most prominent members of the Rho family are RhoA, Rac1 and CDC42, which regulate actin cytoskeleton reorganization to promote cell growth, exert anti-apoptotic functions and regulate gene expression through the activation of different signalling pathways (Wennerberg & Der, 2004). Activation of Rac1 promotes actin polymerization and the formation of lamellipodia (Hall, 1998). P-Rex1 is the GEF for Rac1 and, interestingly, P-Rex1 interacts directly with and is subsequently activated by mTORC2 (Hernandez-Negrete et al, 2007). Indeed, activated Rac1 suppresses actin cytoskeleton defects owing to a loss of mTORC2 function, suggesting that mTORC2 might signal to the actin cytoskeleton through Rac1 (Jacinto et al, 2004). Accordingly, mTORC2 deficiency causes a 20–30% decrease in GTP-bound Rac1, further suggesting that mTORC2 regulates Rac1 activity and thus signals through Rac1 (Jacinto et al, 2004). Besides its function as an mTORC2 target, Rac1 has been shown recently to act upstream of mTOR. Inhibition of Rac1 in mammalian cells blocks the activation of both mTORC1 and mTORC2 by growth factors (Saci et al, 2011). Similarly to P-Rex1, Rac1 also interacts directly with mTOR. This interaction has been proposed to be involved in mTORC1 and mTORC2 localization, as deletion of Rac1 affects the subcellular distribution of mTOR. Surprisingly, the regulation of mTORC1 and mTORC2 by Rac1 is independent of the GTP loading of Rac1 (Saci et al, 2011). Thus, Rac1 seems to have an intricate relationship with mTOR: it interacts directly with mTORC1 and mTORC2 and is both upstream and downstream of them (Fig 4).
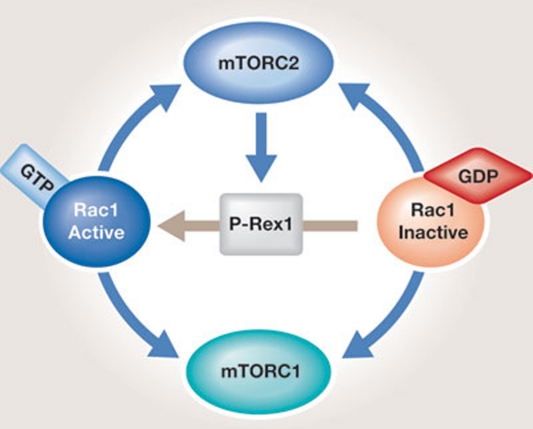
Figure 4.
In mammalian cells, Rac1 functions both upstream and downstream of mTOR. Once activated by TORC2, P-Rex1 (the GEF for Rac1) promotes the exchange of GDP for GTP in Rac1. Rac1 in turn interacts with and activates both mTORC1 and mTORC2 independently of its GTP loading. GEF, guanine nucleotide exchange factor; TOR, target of rapamycin; TORC1/2, target of rapamycin complex 1/2.
Regulation of TOR by Rab family members
A recent study in S. pombe revealed that the Rab GTPase subfamily is also involved in the regulation of the TOR pathway (Tatebe et al, 2010). TORC2 in fission yeast phosphorylates and activates Gad8, a member of the AGC kinase family orthologue of mammalian SGK1 (see above). Ablation of Gad8 sensitizes cells to osmotic stress and high temperature, causes a mating deficiency and prevents G1 arrest under nitrogen starvation, a global phenotype similar to that observed after SpTOR1 deletion. Furthermore, overexpression of Gad8 suppresses the stress sensitivity caused by SpTOR1 deletion (Matsuo et al, 2003). A genetic screen in fission yeast showed that deletion of Ryh1, a GTPase homologous to human RAB6, prevents phosphorylation of Gad8 by TORC2. Similar results were obtained after ablation of SAT1 and SAT4, the putative GEFs for Ryh1 (Tatebe et al, 2010). GTP-bound Ryh1 interacts with SpTOR1 and enhances the interaction of TORC2 with its substrate Gad8, stimulating the phosphorylation of Gad8 by mTORC2. Although Ryh1 is not implicated in the subcellular distribution of mTORC2, the SAT1/4–Ryh1–TORC2–GAD8 pathway seems to be implicated in vacuolar integrity in S. pombe.
As mentioned above, Ryh1 is homologous to human RAB6, which regulates transport pathways in and out of the Golgi. Several RAB6 effectors have been identified, including subunits of the dynein–dynactin complex, Rab kinesin-6/MKLP2, GAPCenA, TMF/ARA160, mint3, Rab6IP1 and Rab6IP2/ELKS, all involved in cellular compartmentalization (Fernandes et al, 2009). At present, no experimental evidence links RAB6 with the mTOR pathway in mammals, but interestingly, human RAB6 can stimulate TORC2 when expressed in S. pombe, suggesting that the role of RAB6 as a positive regulator of TOR might be conserved (Tatebe et al, 2010). As Rab proteins play a role in the establishment of compartmental specificity within the eukaryotic endomembrane system (Zerial & McBride, 2001), it has been proposed that Ryh1 GTPase, as well as its GEF and GAP, confer spatial regulation on TORC2 activity (Tatebe & Shiozaki, 2010).
In addition to TORC2, Rab family members also regulate TORC1. Inhibition of Rab1, Rab5 or Rab11 reduces the phosphorylation of S6K in Drosophila S2 cells, suggesting an important role of Rab proteins in TORC1 activation. In addition, constitutively active mutants of Rab5, Rab7, Rab10 or Rab31 potently inhibit S6K phosphorylation in human cells (Li et al, 2010). Rab5 appears to play a particularly important role in mTORC1 activation, as a constitutively active mutant of Rab5 inhibits Rag- and amino-acid-dependent phosphorylation of S6K without interfering with activation of mTORC1 by Rheb (Li et al, 2010). However, Rab5 is not sufficient to activate mTORC1 and it does not interact with mTORC1. These results suggest that disruption of the GTP–GDP cycling of Rab proteins can disrupt normal cellular transport and inhibit TORC1 activation. Furthermore, these results suggest concerted regulation of intracellular transport and mTOR activation.
Concluding remarks
mTOR and small GTPases are therapeutic targets in the treatment of cancer (Berndt et al, 2011; Dazert & Hall, 2011). Aberrant activation of GTPases, including Ras, Rho, Rab or Ran GTPases, promotes cell transformation and cancer (Agola et al, 2011; Ly et al, 2010; Pylayeva-Gupta et al, 2011), in some cases by acting in the mTOR pathway. Targeting GTPases by using farnesyltransferase inhibitors or geranylgeranyltransferase inhibitors affects signal transduction pathways, cell cycle progression, proliferation and cell survival. Both types of inhibitor are currently under investigation for cancer therapy, although only a small subset of patients responds to these inhibitors (Berndt et al, 2011). A better understanding of the relationship between GTPases and mTOR is essential for the design of combined therapies.
From a mechanistic point of view, research on TOR in different systems is continually adding new insight on the role of TOR in cell biology. However, what is lacking is an integration of the various proposed regulators of TOR, in particular small GTPases (see Sidebar A). Future work should clarify how the various proposed mechanisms that exert an effect on TOR are coordinately regulated and integrated. In the case of small GTPases, further understanding of how the GEF/GAP systems are regulated for each GTPase incoming signal and how TOR discerns the various inputs it receives from different GTPases is essential.
Sidebar A | In need of answers.
How are amino acids sensed by the cell?
What is the mechanism by which amino acids regulate the GTP-loading of Rag proteins? What are the GEF and GAP for the Rag proteins?
Is there a GEF that regulates the GTP-loading of Rheb?
What is the molecular mechanism by which Rheb activates TORC1?
How is the dual effect of Rac1 being both upstream and downstream from TOR regulated?
How are the diverse GTPases that impinge on TOR integrated?
Acknowledgments
We acknowledge support from the Swiss National Science Foundation. We apologize for the many primary publications we could not cite due to space limitations.
Footnotes
The authors declare that they have no conflict of interest.
References
- Agola J, Jim P, Ward H, Basuray S, Wandinger-Ness A (2011) Rab GTPases as regulators of endocytosis, targets of disease and therapeutic opportunities. Clin Genet June 08 2011. doi:; DOI: 10.1111/j.1399-0004.2011.01724.x [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avruch J, Long X, Ortiz-Vega S, Rapley J, Papageorgiou A, Dai N (2009) Amino acid regulation of TOR complex 1. Am J Physiol Endocrinol Metab 296: E592–E602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barquilla A, Crespo JL, Navarro M (2008) Rapamycin inhibits trypanosome cell growth by preventing TOR complex 2 formation. Proc Natl Acad Sci USA 105: 14579–14584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt N, Hamilton AD, Sebti SM (2011) Targeting protein prenylation for cancer therapy. Nat Rev Cancer 11: 775–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binda M, Peli-Gulli MP, Bonfils G, Panchaud N, Urban J, Sturgill TW, Loewith R, De Virgilio C (2009) The Vam6 GEF controls TORC1 by activating the EGO complex. Mol Cell 35: 563–573 [DOI] [PubMed] [Google Scholar]
- Bos JL, Rehmann H, Wittinghofer A (2007) GEFs and GAPs: critical elements in the control of small G proteins. Cell 129: 865–877 [DOI] [PubMed] [Google Scholar]
- Bun-Ya M, Harashima S, Oshima Y (1992) Putative GTP-binding protein, Gtr1, associated with the function of the Pho84 inorganic phosphate transporter in Saccharomyces cerevisiae. Mol Cell Biol 12: 2958–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai SL, Tee AR, Short JD, Bergeron JM, Kim J, Shen J, Guo R, Johnson CL, Kiguchi K, Walker CL (2006) Activity of TSC2 is inhibited by AKT-mediated phosphorylation and membrane partitioning. J Cell Biol 173: 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zheng Y, Foster DA (2003) Phospholipase D confers rapamycin resistance in human breast cancer cells. Oncogene 22: 3937–3942 [DOI] [PubMed] [Google Scholar]
- Choo AY, Blenis J (2009) Not all substrates are treated equally: implications for mTOR, rapamycin-resistance and cancer therapy. Cell Cycle 8: 567–572 [DOI] [PubMed] [Google Scholar]
- Cybulski N, Hall MN (2009) TOR complex 2: a signaling pathway of its own. Trends Biochem Sci 34: 620–627 [DOI] [PubMed] [Google Scholar]
- Dazert E, Hall MN (2011) mTOR signaling in disease. Curr Opin Cell Biol 23: 744–755 [DOI] [PubMed] [Google Scholar]
- Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, Xiao B, Worley PF, Powell JD (2011) The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol 12: 295–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis PB, Jaeschke A, Saitoh M, Fowler B, Kozma SC, Thomas G (2001) Mammalian TOR: a homeostatic ATP sensor. Science 294: 1102–1105 [DOI] [PubMed] [Google Scholar]
- Dowling RJ et al. (2010) mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science 328: 1172–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouloz F, Deloche O, Wanke V, Cameroni E, De Virgilio C (2005) The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol Cell 19: 15–26 [DOI] [PubMed] [Google Scholar]
- Duran A, Amanchy R, Linares JF, Joshi J, Abu-Baker S, Porollo A, Hansen M, Moscat J, Diaz-Meco MT (2011) p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell 44: 134–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J (2001) Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science 294: 1942–1945 [DOI] [PubMed] [Google Scholar]
- Fernandes H, Franklin E, Recacha R, Houdusse A, Goud B, Khan AR (2009) Structural aspects of Rab6–effector complexes. Biochem Soc Trans 37: 1037–1041 [DOI] [PubMed] [Google Scholar]
- Findlay GM, Yan L, Procter J, Mieulet V, Lamb RF (2007) A MAP4 kinase related to Ste20 is a nutrient-sensitive regulator of mTOR signalling. Biochem J 403: 13–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Kaiser CA (2006) A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat Cell Biol 8: 657–667 [DOI] [PubMed] [Google Scholar]
- Gong R, Li L, Liu Y, Wang P, Yang H, Wang L, Cheng J, Guan KL, Xu Y (2011) Crystal structure of the Gtr1p–Gtr2p complex reveals new insights into the amino acid-induced TORC1 activation. Genes Dev 25: 1668–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goorden SM, Hoogeveen-Westerveld M, Cheng C, van Woerden GM, Mozaffari M, Post L, Duckers HJ, Nellist M, Elgersma Y (2011) Rheb is essential for murine development. Mol Cell Biol 31: 1672–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati P, Gaspers LD, Dann SG, Joaquin M, Nobukuni T, Natt F, Kozma SC, Thomas AP, Thomas G (2008) Amino acids activate mTOR complex 1 via Ca2+/CaM signaling to hVps34. Cell Metab 7: 456–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A (1998) Rho GTPases and the actin cytoskeleton. Science 279: 509–514 [DOI] [PubMed] [Google Scholar]
- Heitman J, Movva NR, Hall MN (1991) Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253: 905–909 [DOI] [PubMed] [Google Scholar]
- Hernandez-Negrete I, Carretero-Ortega J, Rosenfeldt H, Hernandez-Garcia R, Calderon-Salinas JV, Reyes-Cruz G, Gutkind JS, Vazquez-Prado J (2007) P-Rex1 links mammalian target of rapamycin signaling to Rac activation and cell migration. J Biol Chem 282: 23708–23715 [DOI] [PubMed] [Google Scholar]
- Hirose E, Nakashima N, Sekiguchi T, Nishimoto T (1998) RagA is a functional homologue of S. cerevisiae Gtr1p involved in the Ran/Gsp1–GTPase pathway. J Cell Sci 111: 11–21 [DOI] [PubMed] [Google Scholar]
- Hsu YC, Chern JJ, Cai Y, Liu M, Choi KW (2007) Drosophila TCTP is essential for growth and proliferation through regulation of dRheb GTPase. Nature 445: 785–788 [DOI] [PubMed] [Google Scholar]
- Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN (2004) Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol 6: 1122–1128 [DOI] [PubMed] [Google Scholar]
- Jiang H, Luo JQ, Urano T, Frankel P, Lu Z, Foster DA, Feig LA (1995) Involvement of Ral GTPase in v-Src-induced phospholipase D activation. Nature 378: 409–412 [DOI] [PubMed] [Google Scholar]
- Juhasz G, Hill JH, Yan Y, Sass M, Baehrecke EH, Backer JM, Neufeld TP (2008) The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J Cell Biol 181: 655–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL (2008) Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol 10: 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guan KL (2011) Amino acid signaling in TOR activation. Annu Rev Biochem 80: 1001–1032 [DOI] [PubMed] [Google Scholar]
- Li L, Kim E, Yuan H, Inoki K, Goraksha-Hicks P, Schiesher RL, Neufeld TP, Guan KL (2010) Regulation of mTORC1 by the Rab and Arf GTPases. J Biol Chem 285: 19705–19709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Inoki K, Vacratsis P, Guan KL (2003) The p38 and MK2 kinase cascade phosphorylates tuberin, the tuberous sclerosis 2 gene product, and enhances its interaction with 14-3-3. J Biol Chem 278: 13663–13671 [DOI] [PubMed] [Google Scholar]
- Li Y, Inoki K, Guan KL (2004) Biochemical and functional characterizations of small GTPase Rheb and TSC2 GAP activity. Mol Cell Biol 24: 7965–7975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J (2005a) Rheb binds and regulates the mTOR kinase. Curr Biol 15: 702–713 [DOI] [PubMed] [Google Scholar]
- Long X, Ortiz-Vega S, Lin Y, Avruch J (2005b) Rheb binding to mammalian target of rapamycin (mTOR) is regulated by amino acid sufficiency. J Biol Chem 280: 23433–23436 [DOI] [PubMed] [Google Scholar]
- Ly TK, Wang J, Pereira R, Rojas KS, Peng X, Feng Q, Cerione RA, Wilson KF (2010) Activation of the Ran GTPase is subject to growth factor regulation and can give rise to cellular transformation. J Biol Chem 285: 5815–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach KE, Furge KA, Albright CF (2000) Loss of Rhb1, a Rheb-related GTPase in fission yeast, causes growth arrest with a terminal phenotype similar to that caused by nitrogen starvation. Genetics 155: 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehama T, Tanaka M, Nishina H, Murakami M, Kanaho Y, Hanada K (2008) RalA functions as an indispensable signal mediator for the nutrient-sensing system. J Biol Chem 283: 35053–35059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfouz MM, Kim S, Delauney AJ, Verma DP (2006) Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell 18: 477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T, Kubo Y, Watanabe Y, Yamamoto M (2003) Schizosaccharomyces pombe AGC family kinase Gad8p forms a conserved signaling module with TOR and PDK1-like kinases. EMBO J 22: 3073–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer AJ, Dubbelhuis PF (2004) Amino acid signalling and the integration of metabolism. Biochem Biophys Res Commun 313: 397–403 [DOI] [PubMed] [Google Scholar]
- Nada S, Hondo A, Kasai A, Koike M, Saito K, Uchiyama Y, Okada M (2009) The novel lipid raft adaptor p18 controls endosome dynamics by anchoring the MEK–ERK pathway to late endosomes. EMBO J 28: 477–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima A, Sato T, Tamanoi F (2010) Fission yeast TORC1 regulates phosphorylation of ribosomal S6 proteins in response to nutrients and its activity is inhibited by rapamycin. J Cell Sci 123: 777–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima N, Noguchi E, Nishimoto T (1999) Saccharomyces cerevisiae putative G protein, Gtr1p, which forms complexes with itself and a novel protein designated as Gtr2p, negatively regulates the Ran/Gsp1p G protein cycle through Gtr2p. Genetics 152: 853–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M et al. (2011) Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science 332: 966–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh WJ, Wu CC, Kim SJ, Facchinetti V, Julien LA, Finlan M, Roux PP, Su B, Jacinto E (2010) mTORC2 can associate with ribosomes to promote cotranslational phosphorylation and stability of nascent Akt polypeptide. EMBO J 29: 3939–3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HO, Bi E (2007) Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol Mol Biol Rev 71: 48–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel PH, Thapar N, Guo L, Martinez M, Maris J, Gau CL, Lengyel JA, Tamanoi F (2003) Drosophila Rheb GTPase is required for cell cycle progression and cell growth. J Cell Sci 116: 3601–3610 [DOI] [PubMed] [Google Scholar]
- Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM (2009) DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 137: 873–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TR et al. (2011) mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 146: 408–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D (2011) RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer 11: 761–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmann H, Bruning M, Berghaus C, Schwarten M, Kohler K, Stocker H, Stoll R, Zwartkruis FJ, Wittinghofer A (2008) Biochemical characterisation of TCTP questions its function as a guanine nucleotide exchange factor for Rheb. FEBS Lett 582: 3005–3010 [DOI] [PubMed] [Google Scholar]
- Roccio M, Bos JL, Zwartkruis FJ (2006) Regulation of the small GTPase Rheb by amino acids. Oncogene 25: 657–664 [DOI] [PubMed] [Google Scholar]
- Saci A, Cantley LC, Carpenter CL (2011) Rac1 regulates the activity of mTORC1 and mTORC2 and controls cellular size. Mol Cell 42: 50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM (2007) PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell 25: 903–915 [DOI] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM (2008) The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320: 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM (2010) Ragulator–Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141: 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM (2006) Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell 22: 159–168 [DOI] [PubMed] [Google Scholar]
- Sekiguchi T, Hirose E, Nakashima N, Li M, Nishimoto T (2001) Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J Biol Chem 276: 7246–7257 [DOI] [PubMed] [Google Scholar]
- Soulard A, Cohen A, Hall MN (2009) TOR signaling in invertebrates. Curr Opin Cell Biol 21: 825–836 [DOI] [PubMed] [Google Scholar]
- Sun Y, Fang Y, Yoon MS, Zhang C, Roccio M, Zwartkruis FJ, Armstrong M, Brown HA, Chen J (2008) Phospholipase D1 is an effector of Rheb in the mTOR pathway. Proc Natl Acad Sci USA 105: 8286–8291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebe H, Shiozaki K (2010) Rab small GTPase emerges as a regulator of TOR complex 2. Small GTPases 1: 180–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebe H, Morigasaki S, Murayama S, Zeng CT, Shiozaki K (2010) Rab-family GTPase regulates TOR complex 2 signaling in fission yeast. Curr Biol 20: 1975–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thedieck K, Polak P, Kim ML, Molle KD, Cohen A, Jeno P, Arrieumerlou C, Hall MN (2007) PRAS40 and PRR5-like protein are new mTOR interactors that regulate apoptosis. PLoS ONE 2: e1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toschi A, Lee E, Xu L, Garcia A, Gadir N, Foster DA (2009) Regulation of mTORC1 and mTORC2 complex assembly by phosphatidic acid: competition with rapamycin. Mol Cell Biol 29: 1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano J, Comiso MJ, Guo L, Aspuria PJ, Deniskin R, Tabancay AP Jr, Kato-Stankiewicz J, Tamanoi F (2005) Identification of novel single amino acid changes that result in hyperactivation of the unique GTPase, Rheb, in fission yeast. Mol Microbiol 58: 1074–1086 [DOI] [PubMed] [Google Scholar]
- Urban J et al. (2007) Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell 26: 663–674 [DOI] [PubMed] [Google Scholar]
- Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH (2007) Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol 9: 316–323 [DOI] [PubMed] [Google Scholar]
- Wang X, Fonseca BD, Tang H, Liu R, Elia A, Clemens MJ, Bommer UA, Proud CG (2008) Re-evaluating the roles of proposed modulators of mammalian target of rapamycin complex 1 (mTORC1) signaling. J Biol Chem 283: 30482–30492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wennerberg K, Der CJ (2004) Rho-family GTPases: it's not only Rac and Rho (and I like it). J Cell Sci 117: 1301–1312 [DOI] [PubMed] [Google Scholar]
- Wu XN, Wang XK, Wu SQ, Lu J, Zheng M, Wang YH, Zhou H, Zhang H, Han J (2011) Phosphorylation of Raptor by p38β participates in arsenite-induced mammalian target of rapamycin complex 1 (mTORC1) activation. J Biol Chem 286: 31501–31511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN (2006) TOR signaling in growth and metabolism. Cell 124: 471–484 [DOI] [PubMed] [Google Scholar]
- Xu L, Frankel P, Jackson D, Rotunda T, Boshans RL, D'Souza-Schorey C, Foster DA (2003) Elevated phospholipase D activity in H-Ras- but not K-Ras-transformed cells by the synergistic action of RalA and ARF6. Mol Cell Biol 23: 645–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Salloum D, Medlin PS, Saqcena M, Yellen P, Perrella B, Foster DA (2011) Phospholipase D mediates nutrient input to mammalian target of rapamycin complex 1 (mTORC1). J Biol Chem 286: 25477–25486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata K, Sanders LK, Kaufmann WE, Yee W, Barnes CA, Nathans D, Worley PF (1994) Rheb, a growth factor- and synaptic activity-regulated gene, encodes a novel Ras-related protein. J Biol Chem 269: 16333–16339 [PubMed] [Google Scholar]
- Yan L, Mieulet V, Burgess D, Findlay GM, Sully K, Procter J, Goris J, Janssens V, Morrice NA, Lamb RF (2010) PP2A T61 epsilon is an inhibitor of MAP4K3 in nutrient signaling to mTOR. Mol Cell 37: 633–642 [DOI] [PubMed] [Google Scholar]
- Yee WM, Worley PF (1997) Rheb interacts with Raf-1 kinase and may function to integrate growth factor- and protein kinase A-dependent signals. Mol Cell Biol 17: 921–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon MS, Du G, Backer JM, Frohman MA, Chen J (2011) Class III PI-3-kinase activates phospholipase D in an amino acid-sensing mTORC1 pathway. J Cell Biol 195: 435–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Hong S, Suzuki T, Nada S, Mannan AM, Wang J, Okada M, Guan KL, Inoki K (2011) Redox regulates mammalian target of rapamycin complex 1 (mTORC1) activity by modulating the TSC1/TSC2–Rheb GTPase pathway. J Biol Chem 286: 32651–32660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial M, McBride H (2001) Rab proteins as membrane organizers. Nat Rev Mol Cell Biol 2: 107–117 [DOI] [PubMed] [Google Scholar]
- Zheng M et al. (2011) Inactivation of Rheb by PRAK-mediated phosphorylation is essential for energy-depletion-induced suppression of mTORC1. Nat Cell Biol 13: 263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzalla V, Stracka D, Oppliger W, Hall MN (2011) Activation of mTORC2 by association with the ribosome. Cell 144: 757–768 [DOI] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM (2011a) mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 12: 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM (2011b) mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H-ATPase. Science 334: 678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J et al. (2011) Rheb1 is required for mTORC1 and myelination in postnatal brain development. Dev Cell 20: 97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurita-Martinez SA, Puria R, Pan X, Boeke JD, Cardenas ME (2007) Efficient Tor signaling requires a functional class C Vps protein complex in Saccharomyces cerevisiae. Genetics 176: 2139–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]