Abstract
Large heat shock proteins (HSPs), including hsp110 and grp170, are unique immunochaperones capable of carrying and introducing antigens into professional antigen presenting cells (APCs) for efficient cross-presentation. Therefore, reconstituted chaperone complexes of large HSPs and protein antigen may be exploited for augmentation of an antigen-specific immune response. The methods for the preparation of the recombinant protein antigen chaperone complex and characterization of its T-cell priming capability in both in vitro and in vivo settings are described.
Keywords: large heat shock protein, hsp110, grp170, chaperone vaccine, antigen presentation, T-cell priming
1. Introduction
Heat shock proteins (HSPs) are among the most abundant and ubiquitous intracellular proteins. As molecular chaperones, they are actively involved in almost every aspect of protein homeostasis, e.g., folding/refolding, assembly, translocation and degradation 1. The studies during the last decade have shown that certain tumor-derived HSPs can serve as effective cancer vaccines2–4, which has been attributed to a HSP-carried peptide antigenic ‘fingerprint’ of the tumor5. Indeed, it has been documented that exogenous HSPs are highly efficient in directing associated antigens into antigen-presenting cells (APCs) though the interactions with surface receptors 6–9, resulting in the cross-presentation of antigens on MHC class I molecules. In addition to promoting antigen processing and presentation, HSP interactions with certain signaling receptors, such as toll-like receptors, facilitate phenotypic and functional maturation of professional APCs, e.g., dendritic cells (DCs) or monocytes 10, 11. Thus, the properties of HSPs as antigen carriers and as activators of innate immune cells enable these chaperone molecules to be utilized as physiological adjuvants for development of various immunotherapeutic approaches against cancer or infectious diseases 12.
Large HSPs, called hsp110 and grp170, exhibit similar albeit distinct structural and functional features compared to other chaperone molecules 13. In light of their exceptional client protein-holding capacity and superior immunostimulatory activity 4, 14–17, we have created novel recombinant heat shock vaccines by complexing clinically relevant tumor protein antigens to these large HSPs in vitro. Since it has long been understood that HSPs chaperone full length protein substrates, these generated complexes are believed to resemble natural intracellular HSP-substrate chaperone complexes. We have demonstrated that these chaperone vaccines exhibit potent antitumor activities in various tumor models16, 18–21, and large HSPs are significantly more potent than complete Freund’s adjuvant (CFA) as an adjuvant19. The ‘chaperoning’ approach that we have developed clearly provides several advantages over autologous vaccines, including no requirement for a surgical tumor specimen, unlimited quantities of off-the-shelf vaccines with uniformity, broad applicability and easy immunomonitoring using well-defined antigens.
Here we describe the methods for preparing recombinant large HSPs and protein antigens using the baculovirus protein expression system, generating large HSP-protein antigen chaperone complexes by heat shock, and assessing the complex-stimulated T-cell activation using both in vitro and in vivo systems.
2. Materials
2.1. Preparation of recombinant large HSPs and protein antigen
BacPAK baculovirus expression system (Clontech)
Baculovirus rapid titer kit (Clontech)
Ni2+-nitrilotriacetic acid (Ni-NTA)-agarose (Qiagen)
Lysis buffer: 20 mM Tris-HCl (pH7.9), 0.5M NaCl, 5 mM Imidazole (Sigma), 0.1% Nonidet P-40, and protease inhibitor cocktail tablets (Roche Molecular Biochemicals)
Binding buffer: 20 mM Tris-HCl (pH7.9), 0.5M NaCl, 5 mM Imidazole
Wash buffer: 20 mM Tris-HCl (pH7.9), 0.5M NaCl, 20–50 mM Imidazole
Elution buffer: 20 mM Tris-HCl (pH7.9), 0.5M NaCl, 300 mM Imidazole
2.2. Chaperone complex formation in vitro
Luciferase aggregation assay buffer: 25 mM HEPES (pH7.9), 5 mM magnesium acetate, 50 mM KCl, and 5 mM β-mercaptoethanol
Complexing buffer: Phosphate-buffered saline containing 20 mM HEPES, pH 7.2, 20 mM NaCl
GelCode Blue Stain Reagent (Pierce)
BCA (bicinchoninic acid) protein assay kit (Pierce)
Enhanced chemiluminescence detection system (Amersham Pharmacia)
2.3. Measuring antigen cross-presentation in vitro using dendritic cells
Complete BMDC medium: RPMI-1640 medium containing 10% heat-inactivated fetal bovine serum (FBS), 10 mM HEPES (PH7.3, Invitrogen), 20 ng/ml GM-CSF (R&D system), 2 mM L-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin and 50 µM β-ME (Sigma)
Red blood cell lysis buffer: 0.15 M NH4Cl, 10 mM KHCO3, 0.1 M Na2EDTA, pH 7.2
TruStain FcX™ (anti-mouse CD16/32) antibody (Clone 93, BioLegend)
FITC BrdU Flow kits (BD Pharmingen)
PE Rat anti-mouse CD8 antibody (clone 53–6.7, BioLegend)
PerCP/Cy5.5 anti mouse CD90.1 (clone OX-7, BioLegend)
FACS staining buffer: PBS containing 0.1% (m/v) bovine serum albumin, 0.1% Na3N
2.4. Measuring the immunogenicity of chaperone complex vaccines in vivo
Red blood cell lysis buffer: 0.15 M NH4Cl, 10 mM KHCO3, 0.1 M Na2EDTA, pH 7.2
RPMI-1640 complete medium containing 10% FBS, 2 mM L-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin and 50 µM β-ME
96-well nitrocellulose-backed microtiter plates (Millipore)
Rat anti-mouse IFN-γ (clone R4-6A2, BD Pharmingen)
Wash solution: PBS containing 0.05% (v/v) Tween 20
Biotinylated IFN-γ antibody (clone XMG1.2, BD Pharmingen)
Avidin-alkaline phosphatase D (Vector Laboratories)
5-bromo-4-chloro-3-indolyl phosphatase/Nitro Blue Tetrazolium (BCIP/NBT) (Boehringer Mannheim)
3. Methods
3.1. Preparation of recombinant large heat shock protein and protein antigen
The baculovirus-insect cell expression system, which not only facilitates proper protein folding and post-translational processing, but also greatly reduces endotoxin contamination, is used to generate recombinant large HSPs (i.e., hsp110 and grp170) and protein antigens (e.g., gp100). The full-length cDNAs for large HSPs or targeted antigens are first subcloned into a baculovirus transfer vector (pBacPAK) containing the promoter for transcription as well as the sequences for homologous recombination and selection. The 6×histidine (His) affinity tag is introduced into the N-terminus or C-terminus of the proteins to facilitate protein binding to Ni2+-NTA agarose for purification. The constructed plasmids are co-transfected along with replication-deficient baculovirus DNA into Sf21 insect cells for virus packaging.
The Bacfectin-DNA mixture is prepared by diluting transfer vector (0.5µg) with linear viral DNA (0.1µg) and the Bacfectin (4 µl) in 100µl steriled nuclease-free water and then added dropwise to a 35 mm culture dish containing Sf21 cells while gently swirling the dish to mix. 1.5 ml of BacPak complete medium is then added after culture at 27 °C for 5 h. When the signs of infection, such as irregular shapes and increased volumes, appear (approximately 5 days post-transfection), culture supernatants are harvested for plaque assay. Freshly prepared sf21 cells in 35mm dish are incubated with the serially diluted supernatant (10−2~ 10−4) at room temperature for 1 h. After removing the virus inoculums, pre-warmed 1% SeaPlaque agarose solution (FMC Bioproducts) is gently overlaid on the infected cell monolayer and BacPAK complete medium will be subsequently added on the solidified agarose. After culture in a humidified incubator for 5–7 days, the plates are stained with 0.03% neutral red solution (Sigma) in PBS and the well-isolated viral plaques are picked using sterile pasteur pipettes. Vortex of agarose plug in the medium will allow the viruses to diffuse out of the agarose plug. 0.1 ml of inoculum from each virus plaque will be amplified for 3–4 days using fresh sf21 cells until the cells appear grainy with irregularly shaped membranes. The supernatants are collected after centrifugation at 1000×g for 5 min at room temperature (passage I virus stock). Expression of large HSPs and targeted antigen should be determined at this step using the infected remaining cells by SDS-PAGE and immunoblotting analysis. The positive recombinant virus plaques are further amplified (0.1 ml passage I stock into 1.5×107 cells/ 30 ml medium in a 150 mm plate) for 4–6 days to produce passage II virus stock. Several aliquots should be kept at –70 °C for long-term storage and the remainder kept at 4 °C as the working stock. The titration of virus should be performed using passage II virus so that subsequent infections can be optimized to produce the maximal yield of recombinant proteins. Virus titers can be determined using a baculovirus rapid titer Kit (Clontech) and calculated as follows: The titer of the virus stock (pfu/ml) = (average plaques per dish) ×10× (dilution factor)–1. Passage II virus stocks for large HSPs and protein antigens (e.g., gp100) usually have a high-titer of 5×107 pfu/ml.
Recombinant proteins are produced in large quantities by infecting insect cells growing in suspension using higher multiplicity of infection (MOI, ~5 to 10). Adding glucose (4.5 mg/ml) to culture media was seen in our hands to increase the glycosylation of certain glycoproteins such as grp170 and gp100. 3–4 days after virus infection, a small aliquot of cells is subjected to SDS-PAGE to ensure protein expression before cells are collected for the scale-up of protein production. Cells are sonicated in lysis buffer (109 cells per 100 ml lysis buffer) and incubated for 30 min on ice. The supernatant of post-centrifugation at 10,000 × g for 1h will be incubated with Ni2+-NTA agarose beads under native conditions at 4°C (volume ratio ~20:1) and the resins are packed onto columns the next day. The columns are washed first with 10-fold bed volume of binding buffer and subsequently with wash buffer to remove non-specific protein binding. His-tagged recombinant proteins are eluted from column with 5–10 ml of elution buffer. The eluted proteins are dialyzed against phosphate-buffered saline (PBS) using Slide-A-Lyzer (Pierce) and concentrated with Centriplus (Milipore) or Vivaspin (Vivascience) ultrafiltration columns. The recovered proteins are quantified using BCA protein assay with bovine serum albumin as a standard. The estimated total protein yield is 1–2 mg per 109 cells.
3.2. Complex formation of large heat shock protein and protein antigen
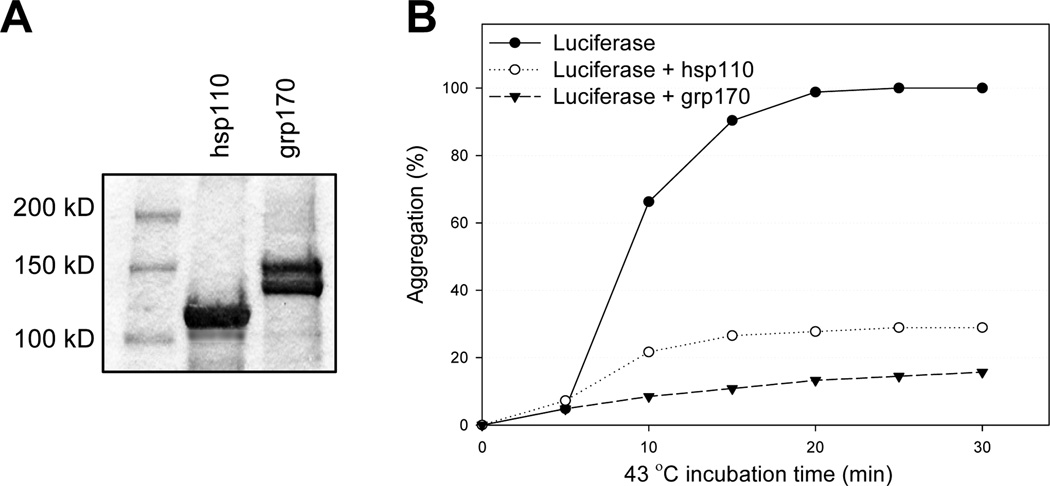
It is recommended that aggregation protection assays using luciferase as a reporter protein are performed to assess the chaperoning capability of recombinant large HSPs prior to the complex reconstitution 22. 150 nM luciferase and hsp110 or grp170 at a molar ratio of 1:1 are incubated in the aggregation assay buffer at 43°C for 30 min, and protein aggregation is monitored by measuring the optical density at 320 nm using a spectrophotometer that is connected to a water bath for temperature control. Hsp110 or grp170 should be able to protect 70–90% of luciferase from heat shock-induced denaturation (Fig. 1). For the generation of chaperone vaccines, recombinant large HSPs and targeted protein antigens (e.g., melanoma antigen gp100) are incubated under heat shock conditions for 30 min, and then incubated at 37°C for another 1 hr. Since the ‘melting’ temperatures differ for individual protein antigens, a pilot study should be carried out to determine the approximate temperature at which the protein starts to denature as indicated by aggregation and precipitation of the protein. The addition of hsp110 or grp170 to the protein antigen should then inhibit aggregation as a result of complex formation. While this model is generally applicable, there is an important caveat. There is an upper temperature limit of approximately 65°C that can be used to induce aggregation since hsp110 itself begins to aggregate at about 70°C. While most proteins examined do aggregate in the available temperature range and therefore adhere to this simple model, not all do. Some proteins are simply thermostable and not suitable for complexing, e.g. Ovalbumin. However, we have had experience with protein antigens, which were purified from bacterial inclusion bodies, that did not aggregate but readily complexed with hsp110 at higher temperature as indicated by co-immunoprecipitation (e.g. the intracellular domain of Her-2/neu18), yielding potent vaccines. Therefore, while a simple precipitation assay is an initial way to set parameters of complexing, failure of this assay should not be reason to abandon the antigen as not suitable. Most protein antigens that we have examined do aggregate in the available temperature range and complex with hsp110 or grp170. In addition, the molar ratios of large HSPs and the antigen can be adjusted to achieve the maximal complexing efficiency, although a one to one molar ratio has been found to be effective in most instances.
Figure 1.
In vitro chaperoning activity assay using luciferase as a reporter protein. A. Purified hsp110 and grp170 were analyzed by SDS–PAGE and subsequent gel staining. B. Luciferase was incubated in the presence or absence of hsp110 or grp170 at 43°C. The optical density of reaction samples at 320 nm was monitored using a spectrophotometer. Aggregation of heated luciferase alone was set as 100%.
The complex formation will be confirmed using immunoprecipitation assays as previously described 18. Anti-hsp110 (1:200) or grp170 (1:100) antibodies are incubated with the complexes to pull down chaperone proteins. Normal rabbit sera are used as negative controls. The immune complexes are then precipitated by Protein-A Sepharose CL-4B (Amersham Pharmacia) and subjected to SDS-PAGE followed by either Gel-blue staining (Pierce) or immunoblotting analysis with antibodies against the targeted antigen (e.g. gp100). Visualization of co-precipitated hsp110 (or grp170) and the protein antigen by Gel-blue staining also allows for an estimation of the molar ratio of complexing. This can be quantitated from a gel scan by adjusting for molecular weight differences and by assuming similar amino acid compositions. For example, gp100 has a molecular weight of about 75 kDa, so a 1 to 1 molar ratio with hsp110 would be seen as a ratio of about 0.70 based of Gel-blue band intensities. In practice, this ratio is closer to 0.60 indicating that most hsp110 molecules are occupied with gp100 protein antigens. However, simple visualization of both bands by Gel-blue staining appears to guarantee sufficient complexing to generate a significant immune response.
3.3. Measuring the chaperone complex-facilitated antigen cross-presentation in vitro
In order to test the ability of large HSPs to enhance the cross-presentation of chaperoned protein antigen, mouse bone marrow-derived DCs (BMDC) are prepared and used as APCs. The mouse tibiae and femurs will be removed and cut with scissors to expose the marrow cavity, and the bone marrow cells are flushed out from the bones using a 3 cc syringe (VWR) attached to a 27-G needle (Becton Dickinson) with ice-cold serum-free RPMI1640. Clusters within the marrow suspension should be disintegrated by vigorous pipetting, followed by the removal of red blood cells from the cell suspension using lysis buffer. Cells are washed and passed through a cell strainer (Becton Dickinson) to remove small pieces of bone and debris. Yields are routinely 5~7×107 mononuclear cells per mouse (two tibiae and two femurs) with a viability of >99%. Cells in complete BMDCs culture medium are seeded at 2×106 cells/well into 12-well plates (Corning) on day 0. The loosely adherent granulocytes should be carefully depleted on day 3 and replaced with fresh complete medium. On days 5 and 7 half of the culture supernatant is harvested and centrifuged. The cell pellet is re-suspended in fresh medium and given back into the original wells. After 7 d cultures, the clustering adherent DCs will dislodge from the stromal cells and float in the culture medium. On day 9, the non- and semi-adherent cells are collected by gentle pipetting using a Pasteur pipet. The cells will at least 80% of DCs as identified by surface marker CD11c, MHC class II and B7.1/B7.2.
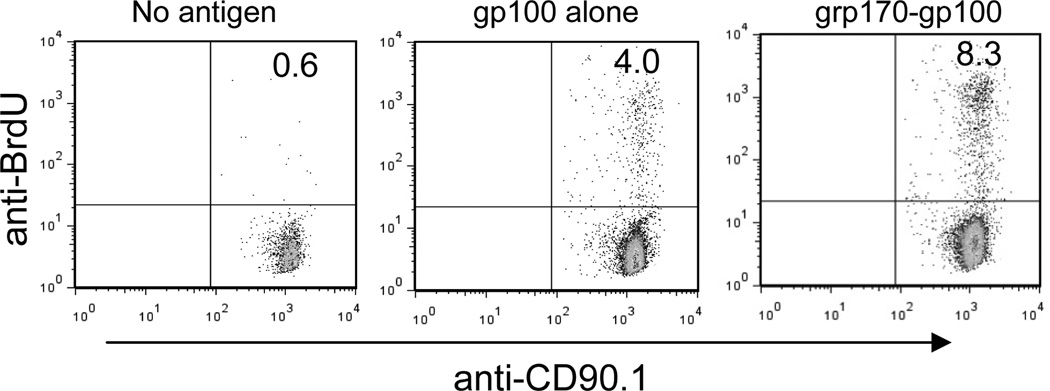
In vitro antigen-specific T-cell proliferation assay will be used to measure large HSP-enhanced antigen cross-presentation. Bromodeoxyuridine (BrdU), a synthetic nucleoside that is an analogue of thymidine, can be incorporated into the newly synthesized DNA of dividing cells. Therefore, BrdU incorporation assay involving immunofluorescent staining of incorporated BrdU and flow cytometric analysis provides an alternative non-radioisotope-based method for assessing the complex-promoted T-cell activation. The procedure described here is to determine the frequency of BrdU incorporating T-cells following co-culture with BMDCs that have been loaded with recombinant large HSP-protein antigen complexes (e.g., grp170-gp100).
BMDCs are incubated with recombinant gp100 protein, grp170-gp100 protein complexes or left untreated in 200µl RPMI complete medium overnight at 37 °C. BMDCs are washed and seeded into 96-well U-bottom cell culture cluster as stimulators (1−2×104/well). Gp100-specific CD8 T-cells are purified from Pmel17 TCR transgenic mice (Jackson Laboratory) as responders using mouse CD8 cell recovery column kit (Cedarlane laboratories Limited) and co-cultured with BMDCs at different ratios in the 96-well plates. BrdU solution is added at a final concentration of 10µM. After 72 hours culture in a humidified 37 °C, 5% CO2 incubator, cells are washed and blocked with CD16/CD32 antibodies in FACS staining buffer, followed by staining using anti-CD8-PE and CD90.1-PerCP/Cy5.5 antibodies at 4 °C for 30 minutes. Cells are fixed, treated with DNase I (300µg/ml), and stained with FITC-conjugated anti-BrdU antibodies (BD Pharmingen). Cells are analyzed on a flow cytometer (e.g. BD FACS Calibur) for the frequency of BrdU+ CD90.1+ T-cells (Fig. 2).
Figure 2.
Recombinant grp170 promotes gp100 cross-presentation and activation of gp100-specific T-cells in vitro. Purified CD8+ Pmel T-cells were co-cultured with BMDCs pulsed with gp100 or grp170-gp100 chaperone complexes at a molar ratio of 10:1 (T-cells: DCs) for 72h. BrdU (10µM) was added at the beginning of the co-culture. Cells were stained with anti-CD8, CD90.1 and BrdU antibodies conjugated with various fluorophores. BrdU+ cells were analyzed using FACS gating in CD8+CD90.1+ cells.
3.4. Measuring the chaperone complex induced T-cell activation in vivo
The most reliable readout for the immunogenicity of protein antigen-targeted chaperone complex vaccines is to determine their ability to generate functional T-cell responses in vivo. Several T-cell assays have been established, which include enzyme-linked immunosorbent spot (ELISPOT) assay, in vitro cytolytic assay, in vivo CTL killing assay, intracellular cytokine staining, and MHC class I-peptide tetramer assay. Here we will briefly discuss the ELISPOT assay, which measures the frequency of cytokine producing T-cells in response to antigen stimulation on a single cell level. The ELISPOT takes advantage of the relatively high concentration of the cytokines in the environment surrounding the cytokine-secreting cell, which can be captured and detected using high-affinity cytokine antibodies. A total of 25–30 μg of large HSP-protein antigen (e.g., gp100) complexes is injected in a volume of 100 μl intradermally to naïve mice. Mice immunized with large HSP alone, protein antigen with or without heat shock treatment will serve as controls. One week later, a second vaccination is given to boost the immune responses. Spleen or draining lymph nodes are collected after an additional week and a single cell suspension is prepared. Red blood cells are removed by lysis buffer. A 96-well, nitrocellulose-backed microtiter plates (Millipore) are pre-coated with 10 µg/ml rat anti-mouse IFN-γ in PBS overnight at 4 °C or 2 h at room temperature. Splenocytes, lymph node cells or purified CD8+ T-cells purified using magnetic beads (Myltenyi Biotec) are plated at a concentration of 2–10 × 105 cells/well in RPMI1640 complete medium containing antigens (e.g. 20 µg/ml gp100 protein or 1 µg/ml gp10025–32 peptide) on a level surface. Following culture in a humidified 37 °C, 5% CO2 incubator for 24 h and extensive rinse with wash buffer, the plates are incubated sequentially with 5 µg/ml biotinylated IFN-γ antibody and 0.2 U/ml avidin-alkaline phosphatase D. Spots will be developed by adding 5-bromo-4-chloro-3-indolyl phosphatase /Nitro Blue Tetrazolium (BCIP/NBT) to each well and incubating at room temperature until color develops. The number of the spots are counted with a Zeiss ELISPOT reader and presented as the number of IFN-γ spots per 1 × 106 cells.
Acknowledgments
This work was supported by NIH research grants CA12911, CA154708, CA099326 and ACS RSG-08-187-01-LIB
Footnotes
Given the inhibitory effect of FBS on the transfection, it is necessary to wash the insect cell once with BacPAK Grace basic medium (GIBCO) and replace the normal medium with serum-free medium before adding the Bacfectin-DNA mixture to the cells.
To achieve maximal protein expression, log phase Sf21 cells that are at least 98% viable should be used for infections. Both the quality of virus plaques and the level of protein production are highly dependent on the viability of the cells. In addition, Sometimes a protein antigen can be engineered to improve solubility and expression, e.g. removal of a transmembrane domain as in the case of melanoma antigen gp100.
Including a low concentration of imidazole in the lysis and wash buffers can help minimize non-specific binding. Drying of the columns should be always avoided during washes, since this will result in the early elution of His-tagged proteins.
Male mice are preferred for the generation of DCs because of their larger bone size compared to female mice. We flush out the bone marrow cells using ice-cold serum-free RPMI 1640 instead of the complete medium, because FBS in the complete medium can generate a lot of foam during the repeated flushes that may be harmful to the cells. To increase the purity of DCs, the loosely adherent granulocytes should be removed thoroughly without disturbing the large clusters of DCs.
A diffuse darkening of the nitrocellulose membrane is commonly observed when too many cells per well are seeded. Inadequate removal of cells from the ELISPOT plates after antibody incubation may cause a high background. Washing the plate at least once with distilled water will help reduce the ‘noise’.
References
- 1.Welch WJ. Heat shock proteins functioning as molecular chaperones: their roles in normal and stressed cells. Philos Trans R Soc Lond B Biol Sci. 1993;339:327–333. doi: 10.1098/rstb.1993.0031. [DOI] [PubMed] [Google Scholar]
- 2.Tamura Y, Peng P, Liu K, Daou M, Srivastava PK. Immunotherapy of tumors with autologous tumor-derived heat shock protein preparations. Science. 1997;278:117–120. doi: 10.1126/science.278.5335.117. [DOI] [PubMed] [Google Scholar]
- 3.Graner M, Raymond A, Romney D, He L, Whitesell L, Katsanis E. Immunoprotective activities of multiple chaperone proteins isolated from murine B-cell leukemia/lymphoma. Clin Cancer Res. 2000;6:909–915. [PubMed] [Google Scholar]
- 4.Wang XY, Kazim L, Repasky EA, Subjeck JR. Characterization of heat shock protein 110 and glucose-regulated protein 170 as cancer vaccines and the effect of fever-range hyperthermia on vaccine activity. J Immunol. 2001;166:490–497. doi: 10.4049/jimmunol.166.1.490. [DOI] [PubMed] [Google Scholar]
- 5.Srivastava P. Interaction of heat shock proteins with peptides and antigen presenting cells: chaperoning of the innate and adaptive immune responses. Annu Rev Immunol. 2002;20:395–425. doi: 10.1146/annurev.immunol.20.100301.064801. [DOI] [PubMed] [Google Scholar]
- 6.Binder RJ, Han DK, Srivastava PK. CD91: a receptor for heat shock protein gp96. Nat Immunol. 2000;1:151–155. doi: 10.1038/77835. [DOI] [PubMed] [Google Scholar]
- 7.Delneste Y, Magistrelli G, Gauchat J, et al. Involvement of LOX-1 in dendritic cell-mediated antigen cross-presentation. Immunity. 2002;17:353–362. doi: 10.1016/s1074-7613(02)00388-6. [DOI] [PubMed] [Google Scholar]
- 8.Berwin B, Hart JP, Rice S, et al. Scavenger receptor-A mediates gp96/GRP94 and calreticulin internalization by antigen-presenting cells. Embo J. 2003;22:6127–6136. doi: 10.1093/emboj/cdg572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Facciponte JG, Wang XY, Subjeck JR. Hsp110 and Grp170, members of the Hsp70 superfamily, bind to scavenger receptor-A and scavenger receptor expressed by endothelial cells-I. Eur J Immunol. 2007;37:2268–2279. doi: 10.1002/eji.200737127. [DOI] [PubMed] [Google Scholar]
- 10.Asea A, Kraeft SK, Kurt-Jones EA, et al. HSP70 stimulates cytokine production through a CD14-dependant pathway, demonstrating its dual role as a chaperone and cytokine. Nat Med. 2000;6:435–442. doi: 10.1038/74697. [DOI] [PubMed] [Google Scholar]
- 11.Vabulas RM, Ahmad-Nejad P, Ghose S, Kirschning CJ, Issels RD, Wagner H. HSP70 as endogenous stimulus of the Toll/interleukin-1 receptor signal pathway. J Biol Chem. 2002;277:15107–15112. doi: 10.1074/jbc.M111204200. [DOI] [PubMed] [Google Scholar]
- 12.Wang XY, Facciponte JG, Subjeck JR. Molecular chaperones and cancer immunotherapy. Handb Exp Pharmacol. 2006;172:305–329. doi: 10.1007/3-540-29717-0_13. [DOI] [PubMed] [Google Scholar]
- 13.Easton DP, Kaneko Y, Subjeck JR. The hsp110 and Grp1 70 stress proteins: newly recognized relatives of the Hsp70s. Cell Stress Chaperones. 2000;5:276–290. doi: 10.1379/1466-1268(2000)005<0276:thagsp>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oh HJ, Chen X, Subjeck JR. Hsp110 protects heat-denatured proteins and confers cellular thermoresistance. J Biol Chem. 1997;272:31636–31640. doi: 10.1074/jbc.272.50.31636. [DOI] [PubMed] [Google Scholar]
- 15.Park J, Easton DP, Chen X, MacDonald IJ, Wang XY, Subjeck JR. The chaperoning properties of mouse grp170, a member of the third family of hsp70 related proteins. Biochemistry. 2003;42:14893–14902. doi: 10.1021/bi030122e. [DOI] [PubMed] [Google Scholar]
- 16.Park J, Facciponte JG, Chen X, et al. Chaperoning Function of Stress Protein grp170, a Member of the hsp70 Superfamily, Is Responsible for its Immunoadjuvant Activity. Cancer Res. 2006;66:1161–1168. doi: 10.1158/0008-5472.CAN-05-2609. [DOI] [PubMed] [Google Scholar]
- 17.Wang XY, Arnouk H, Chen X, Kazim L, Repasky EA, Subjeck JR. Extracellular targeting of endoplasmic reticulum chaperone glucose-regulated protein 170 enhances tumor immunity to a poorly immunogenic melanoma. J Immunol. 2006;177:1543–1551. doi: 10.4049/jimmunol.177.3.1543. [DOI] [PubMed] [Google Scholar]
- 18.Manjili MH, Henderson R, Wang XY, et al. Development of a recombinant HSP110-HER-2/neu vaccine using the chaperoning properties of HSP110. Cancer Res. 2002;62:1737–1742. [PubMed] [Google Scholar]
- 19.Wang XY, Chen X, Manjili MH, Repasky E, Henderson R, Subjeck JR. Targeted immunotherapy using reconstituted chaperone complexes of heat shock protein 110 and melanoma-associated antigen gp100. Cancer Res. 2003;63:2553–2560. [PubMed] [Google Scholar]
- 20.Manjili MH, Wang XY, Chen X, et al. HSP110-HER2/neu chaperone complex vaccine induces protective immunity against spontaneous mammary tumors in HER-2/neu transgenic mice. J Immunol. 2003;171:4054–4061. doi: 10.4049/jimmunol.171.8.4054. [DOI] [PubMed] [Google Scholar]
- 21.Kim H, Sun X, Subjeck J, Wang X-Y. Evaluation of renal cell carcinoma vaccines targeting carbonic anhydrase IX using heat shock protein 110. Cancer Immunology, Immunotherapy. 2007;56:1097–1105. doi: 10.1007/s00262-006-0258-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oh HJ, Easton D, Murawski M, Kaneko Y, Subjeck JR. The chaperoning activity of hsp110. Identification of functional domains by use of targeted deletions. J Biol Chem. 1999;274:15712–15718. doi: 10.1074/jbc.274.22.15712. [DOI] [PubMed] [Google Scholar]