Abstract
CPA is a class of isothermal amplification reactions that is carried out by a strand displacement DNA polymerase and does not require an initial denaturation step or the addition of a nicking enzyme. At the assay temperature of 63°C, the formation of a primer-template hybrid at transient, spontaneous denaturation bubbles in the DNA template is favored over re-annealing of the template strands by the high concentration of primer relative to template DNA. Strand displacement is encouraged by the annealing of cross primers with 5′ ends that are not complementary to the template strand and the binding of a displacement primer upstream of the crossing primer. The resulting exponential amplification of target DNA is highly specific and highly sensitive, producing amplicons from as few as four bacterial cells. Here we report on the basic CPA mechanism – single crossing CPA – and provide details on alternative mechanisms.
Sequence-based amplification of specific genes has many applications in molecular biology research and medical diagnostics. At the present time, there are two main strategies for amplifying a defined sequence of nucleic acid: polymerase chain reaction1 and isothermal amplification. The polymerase chain reaction relies upon instrument-based thermal cycling to denature template DNA, followed by the annealing of primers at specific sites in the denatured template and extension of the primers by a thermostable DNA polymerase in order to exponentially increase the amount of DNA. Isothermal amplification of DNA requires the same three steps to take place, but reagents and conditions are chosen to allow for the amplification of the DNA to take place at one defined temperature after an initial high temperature incubation to denature the template DNA.
A variety of isothermal amplification methods have been developed, for example strand displacement amplification2,3, nucleic acid sequence based amplification4, self-sustained sequence replication5, rolling circle amplification6, loop mediated amplification7, and helicase dependent amplification8. While overcoming some of the technical and cost barriers associated with PCR and providing amplification of target sequences on par with PCR, these methods are still relatively complex protocols requiring the use of multiple enzymes and/or special reagents. In particular, either a high temperature denaturation step or an enzyme based method for promoting nicking and strand displacement is required by these methods.
Cross priming amplification (CPA) is a class of isothermal nucleic acid amplification reactions that utilize multiple primers and probes, one or more of which is a cross primer. We have previously utilized a CPA mechanism involving two cross primers (double crossing CPA) to detect Mycobacterium Tuberculosis in clinical samples9. Here we unveil the mechanism and optimization of the basic CPA reaction, which uses one cross primer (single crossing CPA). This method is able to amplify as little as 4 copies of genomic DNA in less than an hour with a high degree of specificity. In addition the underlying principle of CPA offers tremendous flexibility in the design and use of the method in both basic and applied research.
Results
Single Crossing CPA Assay
The CPA method, like many other isothermal amplification methods, relies on the use of a DNA polymerase with strand displacement activity10. In other methods using strand displacement, additional proteins are required to nick the DNA to allow for amplification by strand displacement. In the CPA assay, the 5′ end of the cross primer is not complementary to the template, and will be displaced when DNA polymerase extends the upstream displacement primer (Figure 1a). This feature of the CPA assay simplifies the reagents necessary for the assay and provides for more flexibility in designing specific applications of the assay.
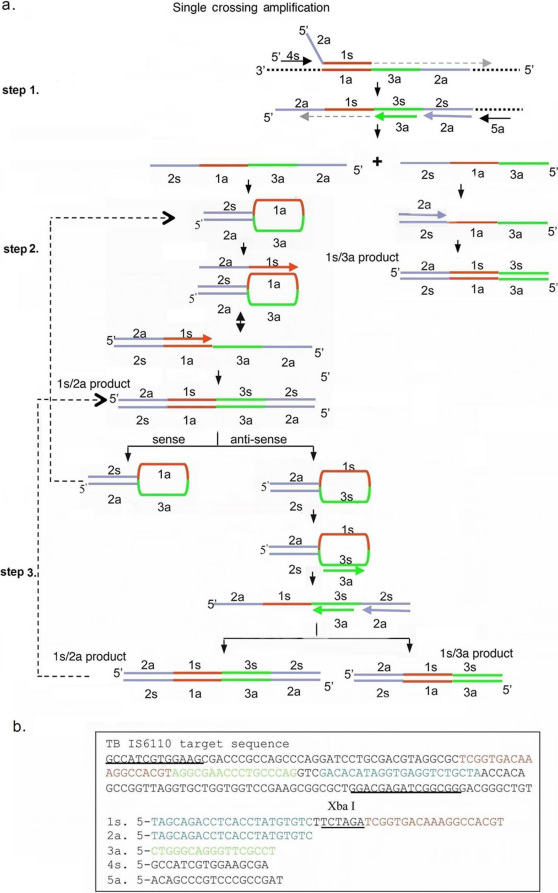
Figure 1. Single crossing CPA mechanism and primer design.
(a) Schematic showing the mechanism of single crossing CPA. (b) Target sequence, primer locations and primer design. Note that an Xba I site (TCTAGA) was inserted into the cross primer between 1s and 2a.
The schematic in Figure 1a outlines the series of primer extensions and products leading to amplification of the DNA. The primary function of cross primer 1s is to allow for strand displacement and to incorporate a second defined priming site in the 5′ end of the resulting product. Primers for the opposite strand each consist of a single sequence complementary to the template, and are chosen so that they bind in tandem on the template, in essence providing a region of nicked double stranded DNA that can be extended by a strand displacing DNA polymerase. After cross primer 1s anneals and extends displacement primer 4s anneals and extends, displacing the down-stream strand. The displaced strand is 5′ defined, with a new primer binding site added at the 5′ end. This strand is used as a template by primers 3a, 2a and displacement primer 5a which anneal and extend in tandem creating two different ssDNA products (step 1). The 3a product is quickly extended by either the 2a or 1s primer to form a 1s/3a product. The 2a product is complementary at its 5′ and 3′ ends by virtue of the introduction of the 2a sequence at the 5′ end of the top strand and the 2s sequence at the 3′ end. This product forms an intramolecular hairpin-like structure which is bound and extended by the cross primer 1s, forming a 1s/2a product (step 2). Both strands from the 1s/2a product also form hairpin-like structures which are favored over re-annealing and are stabilized by the 19 base pair double helix formed between 2a and 2c. These DNAs can act as templates for further primer extension by primers 1s, 3a and 2a leading to the production of 1s/3a and 1s/2a products (step 3). Figure 1b shows a set of five primers to the IS6110 region of Mycobacterium tuberculosis genomic DNA for a single crossing CPA reaction.
In order to demonstrate the mechanism of CPA, amplification reactions were carried out in the presence or absence of template DNA using various combinations of the 5 primers (Figure 2a). The two arrows point to the 1s/2a amplification product (top) and the 1s/3a amplification product (bottom), which are basic units as shown in figure 1a. Both products are obtained using solely the 1s, 2a and 3a primers (lane 1) but the yield is enhanced when primers 4s and 5a are also included in the reaction (lane 2). When primer 2a is omitted from the mix of primers, the 1s/2a product is missing (lane 3), as is the 1s/3a product if primer 3a is omitted from the mix (lane 4). If the cross primer 1s is not present in the reaction, there are no products produced from the mixture of the other four primers (lane 5). These results demonstrate that the system works with a minimum of three primers provided one of these is a cross primer (e.g. primer 1s).
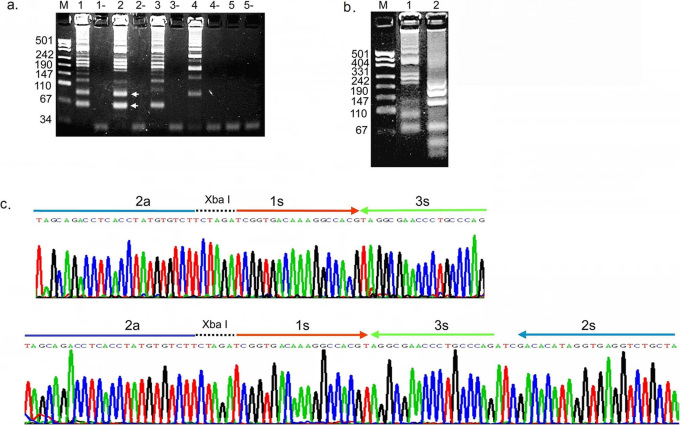
Figure 2. Agarose gel and sequencing analysis of single crossing CPA amplified TB IS6110 DNA.
(a) An agarose gel showing the amplification products obtained by single crossing CPA carried out with different primer combinations. Lane 1: 1s, 2a and 3a; Lane 2: 1s, 2a, 3a, 4s and 5a; Lane 3: 1s, 3a, 4s and 5a; Lane 4: 1s, 2a, 4s and 5a; Lane 5: 2a, 3a, 4s and 5a; (-) indicates no target control for the corresponding reaction. (b) An agarose gel demonstrating that the single crossing CPA products can be digested by the restriction enzyme Xba I. Lane1, single crossing CPA product prior to digestion; Lane 2, single crossing CPA product that has been digested by Xba I. (c) Sequencing of single crossing CPA products. The two bands indicated by the arrows in (a) were excised, cloned and sequenced.
The reaction scheme does not predict the formation of the higher molecular weight amplicons observed in the reaction products shown in Figure 2a. To determine whether these products represent tandem copies of the basic amplicons, the reaction products were digested with Xba I, which is introduced in the design of the 1s primer. The higher molecular weight products of the amplification reaction are reduced in size upon digestion with Xba I (Figure 2b), as are the 1s/3a and 1s/2a basic products. The incomplete digestion of the higher molecular weight fragments may be the result of their ability to form a variety of secondary structures which might interfere with restriction enzyme cleavage at some Xba I sites.
The two bands corresponding to the smallest amplification products (Figure 2a) were excised from the gel, ligated into a TA vector and sequenced. The sequences obtained correspond to the predicted 1s/3a and 1s/2a final products (Figure 2c). The amplification products with higher molecular weight were cloned and sequenced and are tandem repeats of the 1s/3a and 1s/2a basic units (data not shown).
Optimization and Characterization of the single crossing CPA Assay
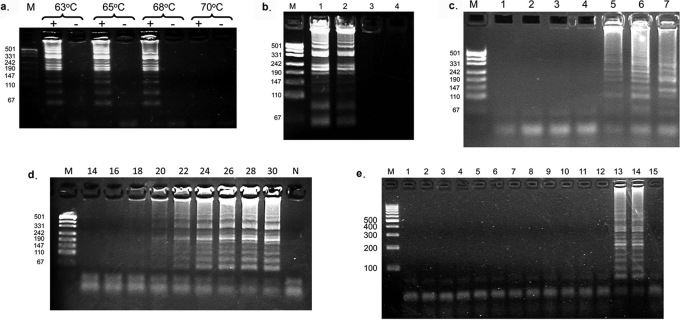
The optimal temperature for an isothermal amplification assay must balance binding of the primer to the template strand with elongation activity of the DNA polymerase. Reactions were carried out at different temperatures using M. tuberculosis DNA as a template with the appropriate primers. Roughly equivalent levels of amplicons were observed to form in a template-dependent manner at temperatures ranging from 63 to 68°C, but failed to form at 70°C (Figure 3a). Temperatures below 58°C gave little to no product (data not shown). A reaction temperature of 63°C was selected as the standard temperature for the rest of the CPA assays conducted in this study.
Figure 3. Optimization of the single crossing CPA assay.
(a) Effect of assay temperature on single crossing CPA. Agarose gel of single crossing CPA amplification products as a function of the reaction temperatures indicated above the lanes. A (+) denotes a reaction containing template, and (-) denotes a reaction without template. (b) Effect of template concentration on CPA. Agarose gel of single crossing CPA amplification products as a function of template concentration. M, size markers; lane 1, 40 M. tuberculosis cells; lane 2, 4 M. tuberculosis cells; lane 3, 0.4 M. tuberculosis cells; lane 4, buffer (60 minute reaction). (c) Effect of assay time on production of non-specific amplification products. Agarose gel showing products of single crossing CPA assays carried out in the absence of template. Lane 1, 60 mins; lane 2, 90 mins; lane 3,120 mins; lane 4, 150 mins; lane 5, 180 mins; lane 6, 210 mins; lane 7, positive control reaction with template incubated for 60 mins. Non specific amplification products appeared after 180 minutes (lane 5 and 6). Note that the gel patterns in these lanes are different from the pattern of CPA products obtained with the intended TB template (lane 7). (d) Time course of the single crossing CPA assay. Agarose gel showing the products of single crossing amplification reactions using 104 copies of a cloned TB plasmid as template. The assay time was varied from 14–30 minutes as indicated above each lane. M: marker; N: Negative control reaction lacking a template incubated for 60 minutes. (e) Test of the specificity of the single crosing CPA assay. M, size marker; lane 1, Mycobacterium marinum; lane 2, M. gordonae; lane 3, M. Simiae; lane 4, M. scrofulaceum; lane 5, M. ranae; lane 6, M. intracellulare; lane 7, M. Phlei ; lane 8, M. smegmatis; lane 9, M. vaccae; lane 10, M. fortuitmn; lane 11, M. chelonae subs.abscessus ; lane 12, M. chelonae subs.chelonae; lane 13, M. tuberculosis; lane 14, plasmid template containing the target TB sequence; lane 15, no template. Non-tuberculosis mycobacterium strains were purchased from China National Institute for the Control of Pharmaceutical and Biological Products.
The minimal level of template required for detection by the single crossing CPA assay was determined by conducting the assay with serial dilutions of M. tuberculosis bacteria cells. As the results in Figure 3b show, amplicons were observed when as few as 4 bacterial cells were present in the amplification reaction, indicating that the CPA assay is able to produce specific amplicons from just a few cells.
In the polymerase chain reaction, the reduction in the background noise of nonspecific products derived from mis-priming events, or primer-primer interactions is controlled by restricting the number of thermal cycles. In an isothermal amplification reaction, total incubation time will influence the production of specific vs. nonspecific amplicons. Single crossing CPA reactions were incubated from 60 minutes to 210 minutes in the absence of template (Figure 3c). Non-specific amplicons appeared after 180 minutes with a banding pattern (lane 5) distinct from the pattern obtained after a 60 minute amplification in the presence of the M. tuberculosis DNA template (lane 7). Figure 3d shows a time course for the production of specific amplicons in a single crossing CPA assay conducted with 104 copies of template DNA. From the intensity of the banding pattern it is apparent that the reaction reaches completion after 28 minutes, while 60-minute incubation in the absence of template DNA (lane N) does not yield amplicons.
The specificity of the single crossing CPA reaction was tested using bacterial cells from a variety of mycobacterial species (Figure 3e). Amplification products were only observed in the reactions that contained either M. tuberculosis cells (lane 13) or a plasmid template containing the M. tuberculosis DNA target region (lane 14), demonstrating that the primers used in the assay and the reaction mechanism combine to provide a high degree of specificity.
Discussion
Gene-based testing for infectious diseases offers rapid, specific results with the potential to require fewer resources than standard microbiological testing methods. The World Health Organization's (WHO) Sexually Transmitted Diseases Diagnostics Initiative proposed guidelines for developing rapid tests to diagnose key infectious diseases in developing countries, namely, ASSURED (affordable, sensitive, specific, user friendly, robust and rapid, equipment free and deliverable)11. Most epidemics originate at resource-limited locations, and gene-based ASSURED tests have the potential to provide global benefits through early detection that leads to prevention of the spread of contagious diseases. ASSURED tests may also find use worldwide in settings such as emergency rooms or family medicine clinics where there is a lower volume of patients but a requirement for rapid diagnostics.
Gene-based testing protocols require the ability to amplify a unique sequence region of the DNA from the infectious organism. The most widely accepted method for gene amplification is PCR1. The requirement of instrumentation for thermal cycling and detection, the complexity of procedures and the associated costs decrease the usefulness of PCR in resource-limited locations. While a number of isothermal nucleic acid amplification methods have been developed to overcome these issues, they involve relatively complex protocols requiring the use of multiple enzymes and/or special reagents.
During in vitro amplification of DNA, denaturation is usually achieved by increasing the incubation temperature to 95°C whereas DNA strands are separated during replication in vivo by the action of a DNA helicase12. The current methods for isothermal amplification either require an initial incubation at 95°C (LAMP, SDA) or inclusion of a DNA helicase in the reaction mixture (HDA). In resource poor locations where those carrying out amplification based testing may not be highly trained, a specifically timed incubation at 95°C with a shift to a lower temperature may either be difficult to carry out when the necessary equipment is unavailable, or not be carried out rigorously, compromising the integrity of the diagnostic testing. The CPA protocol documented here proceeds with no demonstrable difference whether an initial denaturation step at 95°C is included or not. As the temperature of a DNA solution is raised, thermal activation can cause the spontaneous formation of local denaturation bubbles in the Watson-Crick double strands, increasing the fraction of single stranded DNA12,13. When denaturation bubbles open up at the chosen priming sites of DNA in a CPA reaction mixture at 63°C, the formation of a primer-template hybrid is favored over re-annealing of the template strands by the high ratio of the concentration of primer to template DNA and the inclusion of betaine in the reaction14,15. Once formed, the primer within the primer-template hybrid is elongated by Bst DNA polymerase. Multiple primers tandemly aligned to the short target sequence enhance the effect. The extension and strand displacement of any upstream primer opens the double stranded target DNA and exposes the binding sites for the downstream primer.
After the initial amplification step, LAMP relies upon the formation of a naked loop structure encoded within the 5′ tail of the primers16. SDA requires enzyme nicking of the DNA to allow for further isothermal amplification cycles, and the 5′ end of the primers used contain a recognition site for the nicking enzyme.
In single priming CPA (Figure 1a), the strands produced by the crossing primer (1s) and non-crossing primer (2a) form structures which are similar to molecular beacon probes. Traditionally, a molecular beacon structure encodes a stem of 5–8 base pairs and a 15–35 nucleotide long loop sequence17,18. However, the hairpin structures formed in single crossing CPA have longer stems consisting of 18–22 base pairs, which is favorable for the formation of intramolecular stem-loop structures over the double stranded products. Formation of the intramolecular stem structures separates the double strands into two hairpin structures and exposes the binding sites for both the crossing primer and the non-crossing primer in the hairpin loops. Extension of the annealed primer opens the stem structure and synthesizes double-stranded products, which in turn form hairpin structures—facilitating the next round of amplification. This allows for the production of size-limited amplification products. However, the mechanism of single crossing CPA (Figure 1) did not predict the large amplification products observed after agarose gel electrophoresis (Figure 2). Subsequent sequencing data indicated that they were composed of tandem repeats of the predicted basic units. These tandem repeats may be the consequence of self-folding extension and phenomena similar to DNA shuffling19,20 and “jumping PCR”21.
The results presented here demonstrate that single crossing CPA amplifies target sequences from double stranded templates at an incubation temperature held constant at 63°C. A 60 minute single crossing CPA reaction generates visible amplification products from as few as four bacterial cells, yet produces no measurable amount of spurious products in the absence of a template. In testing 13 different Mycobacterium species, single crossing CPA products were observed only with the single species carrying the target sequence in its template DNA.
Unlike previous isothermal mechanisms CPA is not defined by only one mechanism but is defined by the use of one or more cross primers with a 3′ end complementary to the target template sequence and a non-complementary 5′ tail that encodes for an additional sequence. In this report we use one cross primer with a 5′ tail that encodes for the complementary strand primer. However, multiple cross primers could be used and could incorporate a 5′ tail that encodes for a probe, a separate genomic sequence or an artificial sequence.
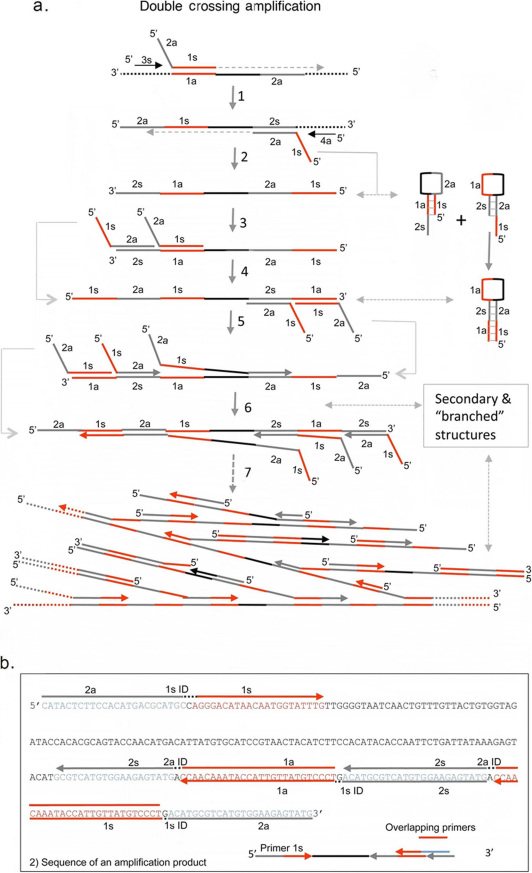
We have previously used an alternative mechanism entitled double crossing CPA for the diagnosis of Mycobacterium tuberculosis infection9, and a sensitive CPA-based test for Enterobacter sakazakii in polluted powdered infant milk formula was recently described22. In the double crossing assay, illustrated in Figure 4a, two cross primers, a forward sense and reverse antisense cross primer, are used (i.e. double crossing CPA). Each cross primer contains 5′ tail sequences identical to each other's priming site and thereby introduces additional priming sites in each round of extension. The addition of identical priming sites in the products is repetitive and complementary, leading to the formation of secondary structures and self-extension. The final product at the end of amplification is a mix of single strands with various secondary structures, branched DNA molecules and double stranded DNA. In single crossing CPA, only one cross primer is used. This helps limit the size of amplification products because additional priming sites are not added in each round of extension. Instead single priming CPA relies on the production of intermediary products that open up the priming sites. We have found that this method is more suitable for probe-based detection methods. In the double crossing CPA reaction, probe binding sites can become “hidden” in the secondary structures produced, thereby lowering the detection efficiency. However double crossing CPA reaction rates are extremely rapid due to simultaneous binding and elongation at multiple sites on the same strand and would therefore be suitable for detection methods that due not rely on nucleotide probes.
Figure 4. Double crossing CPA mechanism and primer design.
(a) Schematic of Double Crossing CPA. The sequential series of primer extensions and products leading to amplification of the target HPV DNA. (b) Sequence of a representative amplification product with overlaid primer locations.
CPA offers a great deal of flexibility through different primer design strategies and by controlling the ratio of the various primers. We are currently designing mechanisms for multiplex amplification, asymmetric amplification, and allele-specific amplification. Additionally, we are investigating methods to interrogate the possible DNA shuffling that occurred for the detection of separated target areas.
We have succeeded in optimizing conditions of heat, salt and detergent to utilize natural DNA denaturation and rapid primer binding and elongation for true isothermal amplification of target DNA sequences. In CPA, DNA amplification increases with incubation time analogous to the doubling of the DNA sample in each round of traditional PCR. The result is continuous primer binding, elongation and displacement of extension products leading to rapid exponential amplification.
Methods
Enzymes and Oligonucleotides
Bst DNA polymerase large fragment was purchased from New England Biolabs (Beijing, China). All oligonucleotides were synthesized by Shanghai Sangon Biological Engineering Technology and Services Co. Ltd (Shanghai, China). The general features of the primers used in single crossing CPA reactions are provided in Figure 1.
Amplification Conditions
Mycobacterium tuberculosis (TB) was used as template for the development and optimization of single crossing CPA. TB IS6110 clone was constructed by extracting genomic DNA of M. tuberculosis H37Rv using the QIAamp DNA Mini Kit (QIAGEN) according to the manufacturer's protocol. The IS 6110 region of the genomic DNA was amplified with primers TBF3 (5-AGGACCACGATCGCTGATC-3) and TBB3 (5-TGGCCATCGTGGAAGCGA-3) by PCR. The resulting DNA fragment was isolated from an agarose gel and cloned into vector pGEM-T Easy (Promega, Madison, WI). The insert was analyzed by DNA sequencing. Bacterial genomic DNA was used for sensitivity and specificity analysis.
The basic single crossing CPA reaction mixture contains five primers (Figure 1A). The upstream cross primer 1s contains a sequence (1s) at its 3′ end complementary to the upstream priming site (shown in red) and the sequence (2a) of the downstream priming site (shown in blue) at its 5′ end. Primers 3a and 2a for the opposite strand are complementary to two adjacent sequences 3s and 2s. The displacement primers 4s and 5a are complementary to sequences just upstream of the core primers. The two sequences of the cross primer are separated by a 5 nucleotide ID tag (Figure 1B).
Amplification conditions were optimized for assay temperature, concentrations of primers, nucleotides, Mg2+, betaine and reaction time. The optimized reaction was carried out in a total volume of 20 µl containing 0.5 µM of cross primer 1s, 0.3 µM each of primers 3a and 2a, 0.05 µM each of displacement primers 4s and 5a, 0.8 mM of dNTP, 1 M betaine (Sigma), 20 mM Tris–HCl(pH 8.8), 10 mMKCl, 10 mM(NH4)2SO4, 6 mM MgSO4, 0.1% Triton X-100, 8 U Bst DNA polymerase large fragment (New England Biolabs) and the specified amounts of double-stranded target DNA or bacterial cells. The mixture was incubated at 63°C for 1 h, and then heated at 95°C to stop the reaction. The amplified products were detected by electrophoresis on an agarose gel.
Author Contributions
G.X., L.H. and Q.Y. designed studies and analyzed data. G.X., H.Z., H.W., S.Y., and T.W. performed experiments. P.R. and S.P. wrote manuscript and provided guidance on study design. Q.Y. managed project.
Acknowledgments
This study was supported by the 11th Five-year Plan of the State Government of People's Republic of China (2008ZX10301, 2008ZX10002-012), The Ministry of Science and Technology of People's Republic of China (09C26213301184), and a Science and Technology Grant from Zhejiang Provincial Government, People's Republic of China (2008C13002). This research was also funded in part by a grant from the Natural Sciences and Engineering Research Council of Canada to PJR. The authors wish to thank Carl F. Perez (Metaara Medical Technologies, Canada), for proof reading.
References
- Saiki R. K. et al. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 230, 1350–4 (1985). [DOI] [PubMed] [Google Scholar]
- Walker G. T. et al. Strand displacement amplification--an isothermal, in vitro DNA amplification technique. Nucleic Acids Res 20, 1691–6 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. T., Little M. C., Nadeau J. G. & Shank D. D. Isothermal in vitro amplification of DNA by a restriction enzyme/DNA polymerase system. Proc Natl Acad Sci U S A 89, 392–6 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sooknanan R. & Malek L. T. NASBA. Nat Biotechnol 13, 563–564 (1995). [Google Scholar]
- Fahy E., Kwoh D. Y. & Gingeras T. R. Self-sustained sequence replication (3SR): an isothermal transcription-based amplification system alternative to PCR. Genome Res 1, 25–33 (1991). [DOI] [PubMed] [Google Scholar]
- Fire a. & Xu S. Q. Rolling replication of short DNA circles. Proc Natl Acad Sci U S A 92, 4641–5 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T. et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28, e63 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent M., Xu Y. & Kong H. Helicase-dependent isothermal DNA amplification. EMBO Rep 5, 795–800 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang R. et al. Cross-priming amplification for rapid detection of Mycobacterium tuberculosis in sputum specimens. J Clin Microbiol 47, 845–7 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. T. et al. Multiplex strand displacement amplification (SDA) and detection of DNA sequences from Mycobacterium tuberculosis and other mycobacteria. Nucleic Acids Res 22, 2670–7 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabey D., Peeling R. W., Ustianowski A. & Perkins M. D. Diagnostics for the developing world. Nat Rev Microbiol 2, 231–40 (2004). [DOI] [PubMed] [Google Scholar]
- Kornberg A., Scott J. F. & Bertsch L. L. ATP utilization by rep protein in the catalytic separation of DNA strands at a replicating fork. J Biol Chem 253, 3298–304 (1978). [PubMed] [Google Scholar]
- Metzler R., Ambjörnsson T., Hanke A. & Fogedby H. C. Single DNA denaturation and bubble dynamics. J Phys: Condens Matter 21, 034111 (2009). [DOI] [PubMed] [Google Scholar]
- Wartell R. M., Benight A. S. Thermal denaturation of DNA molecules: A comparison of theory with experiment. Phys Rep 126, 67–107 (1985). [Google Scholar]
- Baskaran N. et al. Uniform amplification of a mixture of deoxyribonucleic acids with varying GC content. Genome Res 6, 633–638 (1996). [DOI] [PubMed] [Google Scholar]
- Nagamine K., Watanabe K., Ohtsuka K., Hase T. & Notomi T. Loop-mediated isothermal amplification reaction using a nondenatured template. Clin Chem 47, 1742–3 (2001). [PubMed] [Google Scholar]
- Tyagi S., Bratu D. P. & Kramer F. R. Multicolor molecular beacons for allele discrimination. Nat Biotechnol 16, 49–53 (1998). [DOI] [PubMed] [Google Scholar]
- Tyagi S. & Kramer F. R. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol 14, 303–8 (1996). [DOI] [PubMed] [Google Scholar]
- Crameri A., Whitehorn E. A., Tate E. & Stemmer W. P. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat Biotechnol 14, 315–9 (1996). [DOI] [PubMed] [Google Scholar]
- Stemmer W. P. Rapid evolution of a protein in vitro by DNA shuffling. Nature 370, 389–91 (1994). [DOI] [PubMed] [Google Scholar]
- Pääbo S., Irwin D. M. & Wilson A. C. DNA damage promotes jumping between templates during enzymatic amplification. J Biol Chem 265, 4718–21 (1990). [PubMed] [Google Scholar]
- Yulong Z. et al. Rapid and sensitive detection of Enterobacter sakazakii by cross-priming amplification combined with immuno-blotting analysis. Mol Cell Probes 24, 396–400 (2010). [DOI] [PubMed] [Google Scholar]