Highlights
► Bacterial S-layer proteins have a great potential as nanoscale building blocks. ► By genetic engineering, functional domains were incorporated into S-layers. ► Recombinant fusion proteins were functional and maintained self-assembly capability. ► Nanobiotechnological applications are sensor technology, diagnostics, vaccine development, and biocatalysts.
Abstract
Crystalline bacterial cell surface layers (S-layers) are the outermost cell envelope component of many bacteria and archaea. S-layers are monomolecular arrays composed of a single protein or glycoprotein species and represent the simplest biological membrane developed during evolution. The wealth of information available on the structure, chemistry, genetics and assembly of S-layers revealed a broad spectrum of applications in nanobiotechnology and biomimetics. By genetic engineering techniques, specific functional domains can be incorporated in S-layer proteins while maintaining the self-assembly capability. These techniques have led to new types of affinity structures, microcarriers, enzyme membranes, diagnostic devices, biosensors, vaccines, as well as targeting, delivery and encapsulation systems.
In this article, a survey is given to more recent developments in applied S-layer research.
Introduction
Despite the considerable diversity among the cell envelope structure of prokaryotic organisms, a common feature is the presence of monomolecular arrays of protein or glycoprotein species (Mw: 40–200 kDa) termed S-layers [1]. S-layers have now been identified on organisms of nearly every taxonomic group of walled bacteria and represent an almost universal feature of archaea [2•,3]. They exhibit either oblique (p1, p2), square (p4), or hexagonal (p3, p6) symmetry (Figure 1) with unit cell dimensions in the range of 3–30 nm. Depending on the lattice symmetry, one morphological unit (unit cell) will consist of one, two, three, four or six identical protein or glycoprotein monomers. The crystalline arrays are generally 5–10 nm thick and represent highly porous protein lattices (30–70% porosity) with pores of uniform size and morphology in the 2–8 nm range. Moreover, S-layers are highly anisotropic structures with respect to their topography and physicochemical surface properties (for review see [3]).
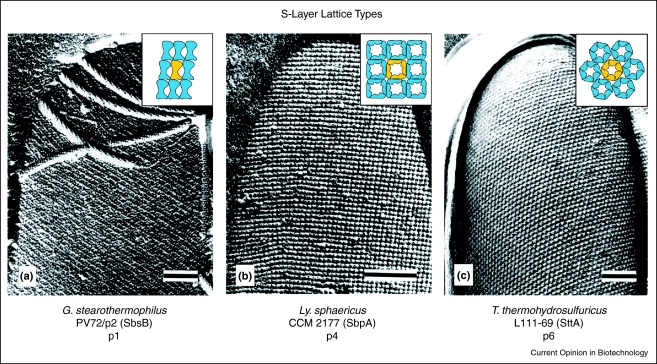
Figure 1.
Electron micrographs of freeze-etched preparations of intact cells from (a)Geobacillus stearothermophilus PV72/p2 exhibiting oblique (p1) lattice symmetry, Bar, 50 nm, (b)Lysinibacillus sphaericus CCM 2177 showing a square (p4) lattice type, Bar, 150 nm, and (c)Thermoanaerobacter thermohydrosulfuricus L111-69 showing hexagonal (p6) lattice symmetry, Bar, 100 nm. Inserts: schematic drawings of the different S-layer lattice types. Depending on the lattice type, one morphological unit (shown in yellow) is composed of (a) one, (b) four, or (c) six identical subunits.
Despite the fact that considerable knowledge has been accumulated on the genetics, morphogenesis, structure–function relationship, and the biochemical and biophysical properties of S-layer (glyco)proteins, no overall tertiary structure of a native protein has been elucidated due to the two-dimensional crystallization behavior of S-layer proteins. Recently, a structural model of an S-layer protein, namely the protein SbsB of Geobacillus stearothermophilus PV72/p2 could be determined by means of molecular dynamic simulations and small angle X-ray scattering [4,5].
Isolated S-layer proteins reassemble into regular arrays upon removal of the disrupting agent (e.g. urea and guanidine hydrochloride) used in the dissociation procedure. The self-assembly products generated in suspension may have the form of flat sheets, open-ended cylinders or closed vesicles (for review see [6•,7]). Most important for many applications of S-layer lattices in nanobiotechnology and biomimetics was the observation that S-layer proteins are capable of reassembly into large coherent monolayers on solid supports (e.g. metals, semiconductors, and polymers) at the air/water interface and on Langmuir lipid films and liposomes (for review see [7–9]).
Studies on a great variety of S-layers from Bacillaceae revealed the existence of specific binding domains on the N-terminal part of S-layer proteins for heteropolysaccharides (secondary cell wall polymers; SCWPs) covalently linked to the peptidoglycan matrix of the cell wall [10••]. Most relevant, functionalizing surfaces with isolated SCWP facilitates recrystallization of S-layers in defined orientation which is of particular importance for nanobiotechnological applications.
Recombinant S-layer proteins
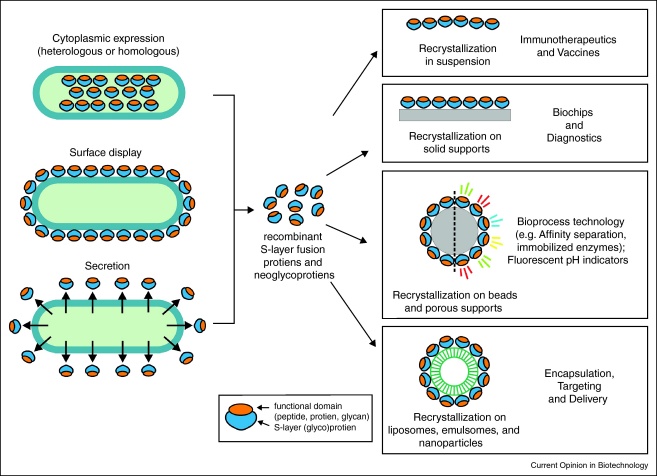
Although native S-layer proteins have already demonstrated their great potential as patterning elements and nanoscale building blocks, genetic approaches opened up the possibility of modifying and changing the natural properties of S-layer proteins thus leading to bio-inspired materials with designed functional properties. However, to be useful in nanobiotechnological applications, S-layer proteins must be capable of tolerating insertions or fusions with foreign proteins or domains, remaining able to assemble into geometrically well-defined layers, and of incorporating a segment that has never participated in lattice formation. To date, two different strategies for the production of nanobiotechnologically relevant S-layer fusion proteins are pursued, namely, homologous expression and secretion by the cells or production inside a host, mostly Escherichia coli (Figure 2).
Figure 2.
Schematic drawing of technologies based on recombinant S-layer fusion proteins and their applications in nanobiotechnology.
The most relevant advantages of the genetically engineered S-layers over less nanostructured approaches are the periodicity of functional domains in the nanometer range on the outermost surface of the S-layer lattice and the general applicability of the ‘S-layer tag’ to any fusion partner. This leads to a high flexibility for variation of the functional groups within a single S-layer array by cocrystallization of different S-layer fusion proteins. On the basis of results of recrystallization studies and surface accessibility screens with various genetically produced N-terminally and/or C-terminally truncated forms, the S-layer proteins of several bacterial strains were chosen as fusion partner for the construction of chimaeric S-layer proteins (Table 1) [11–13].
Table 1.
Nanobiotechnological applications of recombinant S-layer fusion proteins
| Application | S-layer moiety | Genetically fused functionality | Length of function |
|---|---|---|---|
| Sensor technology and diagnostics | SbpA, SbsB | Core-streptavidin | 118 aa |
| SbpA, SbsB | Mimotope of Epstein–Barr virus (EBV) epitope F1 | 20 aa | |
| SbpA | ZZ, IgG-binding domain of Protein A | 116 aa | |
| SbpA | cAb, Heavy chain camel antibody | 117 aa | |
| Vaccine development | SbpA, SbsC | Major birch pollen allergen (Bet v1) | 116 aa |
| SbsA | H. influenzae antigen (Omp26) | 200 aa | |
| SlpA | Antigenic poliovirus epitope (VP1) | 11 aa | |
| SlpA | Human c-myc proto-oncogene | 10 aa | |
| SLH-EA1, SLH-Sap | Levansucrase of B. subtilis | 473 aa | |
| SLH-EA1 | Tetanus toxin fragment C (ToxC) of C. tetani | 451 aa | |
| RsaA | P. aeruginosa strain K pilin | 12 aa | |
| RsaA | IHNV glycoprotein | 184 aa | |
| RsaA | IgG-binding domain of Protein G | GB1xs | |
| RsaA | Domain 1 of HIV receptor CD4 | 81 aa | |
| RsaA | MIP1α ligand for HIV coreceptor CCR5 | 70 aa | |
| Immobilized biocatalysts | SbpA | Hyperthermophilic enzyme laminarinase (LamA) | 263 aa |
| SgsE | Glucose-1-phosphate thymidylyltransferase (RmlA) | 299 aa | |
| RsaA | Beta-1,4-glycanase (Cex) | 485 aa | |
| Fluorescent biomakers | SbpA | Enhanced green fluorescent protein (EGFP) | 238 aa 240 aa |
| SgsE | Enhanced cyan fluorescent protein (ECFP) | 240 aa | |
| SgsE | Enhanced green fluorescent protein (EGFP) | 240 aa | |
| SgsE | Yellow fluorescent protein (YFP) | 225 aa | |
| SgsE | Monomeric red fluorescent protein (RFP1) | ||
| Nanoparticle arrays | SbpA | Incorporated cysteine residues | 3 aa |
| Biosorption of heavy metals | RsaA | His-tag, affinity tag | 6 aa |
S-layer proteins: SbsB of Geobacillus stearothermophilus PV72/p2, SbpA of Lysinibacillus sphaericus CCM 2177, SbsC of Geobacillus stearothermophilus ATCC 12980, SgsE of Geobacillus stearothermophilus NRS 2004/3a, SbsA of Geobacillus stearothermophilus PV72/p6, SlpA of Lactobacillus brevis ATCC 8287, SLH (S-layer homology domain) of EA1 or Sap of Bacillus anthracis, RsaA of Caulobacter crescentus CB15A. For review see [12,13,51].
S-layer based sensor technology and diagnostics
Their intrinsic self-assembly properties as well as their periodicity make S-layers ideal building blocks for all kinds of detection systems (DNA, protein or antibody chips as well as label-free detection systems) (for review see [10••]).
To generate a universal affinity matrix for binding of any kind of biotinylated molecules (e.g. proteins, allergens, antibodies or oligonucleotides), S-layer-streptavidin fusion proteins carrying core-streptavidin either at the N-terminal or C-terminal end have been constructed with a great application potential in protein-chip, allergy-chip or DNA-chip technology [14–16].
Another application potential can be seen in the development of label-free detection systems. In the surface plasmon resonance (SPR) or surface acoustic wave (SAW) technique, specific binding of functional molecules (e.g. proteins or antibodies) to the sensor chip functionalized with an oriented chimaeric S-layer can be visualized directly by a mass increase on the chip without the need of any labeled compound. This was exemplified by the S-layer fusion protein rSbpA31–1068/cAb-PSA carrying the camel antibody sequence recognizing the prostate-specific antigen (PSA) which was recrystallized on gold chips precoated with thiolated SCWP [17]. The fused ligands on the S-layer lattice showed a well-defined spatial distribution down to the subnanometer scale, which might reduce diffusion limited reactions [17,18].
Another field of research deals with the production of S-layer fusion proteins between the S-layer proteins of Ly. sphaericus CCM 2177 or G. stearothermophilus PV72/p2 and peptide mimotopes such as F1 that mimics an immunodominant epitope of Epstein–Barr virus (EBV). Diagnostic studies have been performed by screening 83 individual EBV IgM-positive, EBV-negative, and potential cross-reactive sera and resulted in 98.2% specificity and 89.3% sensitivity as well as no cross-reactivity with related viral diseases. This result demonstrates the potential of these S-layer fusion proteins to act as a matrix for site-directed immobilization of small ligands in solid-phase immunoassays [19].
A further line of development is the production of antibody-binding matrices based on chimaeric S-layers. For that purpose, the S-layer fusion protein rSbpA31–1068/ZZ incorporating two copies of the 58 amino acids long Fc-binding Z-domain (a synthetic analogue of the IgG-binding domain of protein A from Staphylococcus aureus) was constructed and the binding capacity for human IgG was determined by SPR measurements [20]. By recrystallization of this chimaeric protein on microbeads, a biocompatible matrix for the microsphere-based detoxification system used for extracorporeal blood purification of patients suffering from autoimmune disease has been generated [20].
Vaccine development
Owing to their intrinsic adjuvant ability as well as their capability to surface display proteins and epitopes, S-layers are excellent candidates to be used as antigen carriers, either as self-assembly products, recrystallized on liposomes or displayed on bacteria.
On the basis of the fact that S-layer fusion proteins comprising allergens combine reduced allergenicity with immunomodulating capacity they can be considered as novel approach to specific immunotherapy (SIT) of allergic diseases [21–23]. This was exemplified by two chimaeric S-layer proteins, rSbpA31–1068/Bet v1 and rSbsC31–920/Bet v1, carrying the major birch allergen Bet v1 at the C-terminus [22,23]. The recombinant S-layer/Bet v1 fusion proteins altered an established Th2-dominated phenotype as well as the de novo cytokine secretion profile toward a more balanced Th1/Th0-like phenotype [21,24•,25]. This strategy may be generally applied to design vaccines for SIT of type I allergy with improved efficacy and safety.
Since up to now, the inability to establish viable Lactobacillus S-layer null mutants has hampered the biotechnological applications of Lactobacillus S-layers, the utilization of Lactobacillus brevis S-layer subunits (SlpA) for the surface display of foreign antigenic epitopes was envisaged [26]. In terms of immunological studies, an inducible L. brevis expression system for the production, secretion and surface display of antigenic epitopes inserted at distinct sites of the S-layer protein SlpA was developed [26]. While maintaining the S-layer lattice structure, the 11-amino acid long epitope c-myc from the human c-myc proto-oncogene was successfully expressed in every S-layer subunit of the L. brevis S-layer SlpA [26]. Delivery of antigens to mucosal surfaces by lactic acid bacteria was considered to offer a safe alternative to live attenuated pathogens because of their food grade status.
With the objective to deliver candidate antigens for the development of vaccines against nontypeable Haemophilus influenzae (NTHi) infections, the antigen Omp26 was introduced at different positions of the S-layer protein SbsA of G. stearothermophilus PV72/p6 [27]. The fusion protein was expressed in empty bacterial cell envelopes, so-called ghosts and induced an Omp26-specific antibody response in BALB/c mice [28].
The gram-positive pathogen Bacillus anthracis, the causative agent of anthrax, which is a fulminant and lethal infection of mammals, requires S-layer proteins for the pathogenesis of infection [29]. Surface localization and abundant expression of the S-layer protein BslA make this polypeptide a candidate antigen for purified subunit vaccines against B. anthracis [29]. In a former study, chimaeric genes encoding the sequence of the SLH-domains of the two most abundant S-layer proteins, Sap or EA1, as well as the levansucrase of Bacillus subtilis or the C fragment of the tetanus toxoid of Clostridium tetani were cloned and expressed in B. anthracis [30,31]. The fusion proteins were secreted and could attach to the bacterial cell surface of B. anthracis and one resulting recombinant strain, RPL-ToxC, was used for veterinary vaccinal purposes [31].
A number of vaccine approaches involve the development of vaccination vehicles, serving to potentiate the immune response to an antigen. In this context, the highly ordered S-layer structure [32,33] as well as the C. crescentus RsaA secretion apparatus was exploited to display foreign peptides on the C. crecentus cell surface. The feasibility of the commercially available PurePro TM Caulobacter expression and secretion system (Invitrogen) for the surface display of various functional epitopes (see Table 1) could already be demonstrated [32,34–37]. This presentation system could have many potential applications, such as the development of whole-cell vaccines, tumor suppressors, cellular adsorbents, and peptide display libraries [38–40].
Since Aeromonas hydrophila is an important fish pathogen in aquaculture systems, there is an urgent need for a potent vaccine to fight Aeromonas infections. A possible reason why there is no commercial vaccine available can be seen in the inability of these vaccines to cross-protect against different isolates of A. hydrophila, which is a biochemically as well as serologically heterogeneous bacterium [41]. The results of a recent study demonstrated that the recombinantly produced S-layer protein of A. hydrophila may be an important antigen for conferring protection in common carp against a variety of virulent isolates of this pathogenic bacterium [41].
A further recombinant subunit model vaccine was developed to fight infectious hematopoietic necrosis virus (IHNV) which causes a hemorrhagic disease in young salmonid fish. For that purpose, a 184-amino-acid segment of IHNV glycoprotein was fused to the C-terminal portion of the C. crescentus S-layer protein and expressed by the S-layer secretion system [37]. Investigations by laboratory trials revealed a relative percent survival of 26–34% in rainbow trout fry [37].
Immobilized biocatalysts
Concerning the biotechnological application of S-layer fusion proteins aiming at controllable display of biocatalytic epitopes, storage stability, and reuse, S-layer/enzyme fusion proteins comprising S-layer proteins of Bacillaceae and enzymes from extremophilic organisms (extremozymes) were cloned and expressed in E. coli. For proof of principle, the enzyme β-1,3-endoglucanase LamA from the extremophilic P. furiosus was C-terminally fused to the S-layer protein SbpA31–1068 of Ly. sphaericus CCM 2177 [42]. It is remarkable to note that measured enzyme activities of the recrystallized S-layer/enzyme fusion proteins reach up to 100% compared to the native enzyme. In a further approach, S-layer/enzyme fusion proteins comprising the glucose-1-phosphate thymidylyltransferase RmlA from G. stearothermophilus NRS 2004/3a as well as the SgsE derivatives SgsE31–773 or SgsE31–573 originating from the same organism, were produced [43].
Recent research activities are focused on the production of immobilized biocatalysts based on fusion proteins comprising self-assembling S-layer proteins and multimeric enzymes of extremophiles (e.g. xylose isomerase of Thermoanaerobacterium strain JW/SL-YS 489) (Ferner-Ortner-Bleckmann, unpublished results).
Fluorescent biomarkers
In current studies, S-layer fusion protein technology was used to build periodic biomimetic nanostructures with controlled fluorescent properties [44,45].
The S-layer protein SgsE from G. stearothermophilus NRS 2004/3a was used for the production of four S-layer fusion proteins carrying three different mutants of the green fluorescent protein GFP and the red fluorescent protein mRFP1 [44]. In addition, Kainz and coworkers reported on the construction of a bifunctional S-layer tandem fusion protein (ECFP–SgsE–YFP) which was able to self-assemble on solid supports without losing its functionality [45]. The results indicated that the assembling and the fluorescence properties of the newly designed fluorescent S-layer fusion proteins can be used for building nanopatterned biofunctional surfaces for future applications in nanobiotechnology, such as pH indicators both in vivo and in vitro or as fluorescent markers for drug delivery systems [44,45].
A further promising application potential can be seen in the development of drug and delivery systems based on liposome–DNA complexes coated with functional S-layer fusion protein for transfection of eukaryotic cell lines. In this context, the S-layer fusion protein rSbpA31–1068/EGFP incorporating the sequence of enhanced green fluorescent protein (EGFP) was recrystallized as a monolayer on the surface of positively charged liposomes thus representing a useful tool to visualize the uptake of S-layer coated-liposomes into eukaryotic cells [46].
Nanoparticle arrays
In soluble and recrystallized rSbpA/STII/Cys fusion proteins, the short affinity tag for streptavidin, Strep-tag II, was used for prescreening of the surface accessibility, whereas the thiol group of the end-standing cysteine residue was exploited for site-directed chemical linkage of differently sized preactivated macromolecules via heterobifunctional cross-linkers [47]. Such functionalized two-dimensional S-layer lattices formed by rSbpA/STII/Cys exhibited highly accessible cysteine residues in a well-defined arrangement on the surface and could be utilized for the template-assisted patterning of gold nanoparticles [47].
Bioremediation of heavy metals
A current trend in the field of bioengineered materials is to investigate the role of S-layer-carrying bacteria in the biosorption of heavy metals [e.g. U, Cu, Pd(II), and Au(III)] [48,49]. Recently, the genetically engineered S-layer protein RsaA was employed as a delivery system for displaying hexa-histidine peptides on the Caulobacter cell surface and to construct a recombinant bioremediation agent to remove cadmium from contaminated water samples [50].
Conclusions
Data obtained in studies on the structure, chemistry, genetics, morphogenesis, and function demonstrated that S-layers are the simplest protein membranes developed during evolution. Their structure and physicochemical repetitive uniformity down to the subnanometer scale has already led to numerous applications in nanobiotechnology and biomimetics. Most important, S-layers represent very versatile self-assembly systems which can be exploited as structural basis for a complete supramolecular construction kit, involving all major species of biological macromolecules (e.g. proteins, lipids, glycans, nucleic acids, and combinations of these). Moreover, the possibility to modify the natural properties of S-layer proteins by genetic manipulations and to incorporate single or multifunctional domains of other proteins while maintaining the self-assembly capability has led to a broad spectrum of applications in numerous areas of both, life and material sciences. Another area of future development concerns copying the supramolecular principle of cell envelopes of archaea. The observation that S-layers can be used as supporting and stabilizing structures for lipid membranes incorporating functional molecules will lead to applications in sensor technology, diagnostics, nanobiotechnology, electronic or optical devices, and high-throughput screening for drug discovery (for reviews see [6•,7,10••,12,51,52,53•]).
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
This work was supported by the EU project MemS (Grant agreement no.: 244967), by the Austrian Science Fund (FWF) Project P18510-B12 as well as by the US Air Force Office of Scientific Research (AFOSR) projects FA9550-07-1-0313 and FA9550-10-1-0223.
References
- 1.Sleytr U.B., Messner P., Pum D., Sára M, editors. Crystalline Bacterial Cell Surface Layers. Springer; Berlin: 1988. [DOI] [PubMed] [Google Scholar]
- 2•.Messner P., Schäffer C., Egelseer E.M., Sleytr U.B. Occurrence, structure, chemistry, genetics, morphogenesis, and function of S-layers. In: König H., Claus H., Varma A., editors. Prokaryotic Cell Wall Compounds — Structure and Biochemistry. Springer-Verlag; 2010. pp. 53–109. [Google Scholar]; Most recent survey on basic principles of S-layers
- 3.Sleytr U.B., Sára M., Pum D., Schuster B. Crystalline bacterial cell surface layers (S-layers): a versatile self-assembly system. In: Ciferri A., editor. Supramolecular Polymers. edn 2. 2005. pp. 583–612. [Google Scholar]
- 4.Horejs C., Pum D., Sleytr U.B., Peterlik H., Jungbauer A., Tscheliessnig R. Surface layer protein characterization by small angle X-ray scattering and a fractal mean force concept: from protein structure to nanodisk assemblies. J Chem Phys. 2010;133:175102. doi: 10.1063/1.3489682. [DOI] [PubMed] [Google Scholar]
- 5.Horejs C., Pum D., Sleytr U.B., Tscheliessnig R. Structure prediction of an S-layer protein by the mean force method. J Chem Phys. 2008;128 doi: 10.1063/1.2826375. 065106 (065101–065111) [DOI] [PubMed] [Google Scholar]
- 6•.Sleytr U.B., Egelseer E.M., Ilk N., Messner P., Schäffer C., Pum D., Schuster B. Nanobiotechnological applications of S-layers. In: König H., Claus H., Varma A., editors. Prokaryotic Cell Wall Compounds — Structure and Biochemistry. Springer; 2010. pp. 459–481. [Google Scholar]; Most recent compilation on nanobiotechnological applications of S-layers
- 7.Sleytr U.B., Sára M., Pum D., Schuster B. Characterization and use of crystalline bacterial cell surface layers. Prog Surf Sci. 2001;68:231–278. [Google Scholar]
- 8.Sleytr U.B., Sára M., Pum D., Schuster B. Molecular nanotechnology and nanobiotechnology with two-dimensional protein crystals (S-layers) In: Rosoff M., editor. Nano-Surface Chemistry. Marcel Dekker Inc.; 2001. pp. 333–389. [Google Scholar]
- 9.Sleytr U.B., Sára M., Pum D., Schuster B., Messner P., Schäffer C. Self-assembly protein systems: microbial S-layers. In: Steinbüchel A., Fahnestock S.R., editors. Polyamides and Complex Proteinaceous Materials I. vol 1st Ed. Wiley-VCH; 2002. pp. 285–338. (Biopolymers). [Google Scholar]
- 10••.Egelseer E.M., Ilk N., Pum D., Messner P., Schäffer C., Schuster B., Sleytr U.B. S-layers, microbial, biotechnological applications. In: Flickinger M.C., editor. vol 7. John Wiley & Sons Inc.; 2010. pp. 4424–4448. (The Encyclopedia of Industrial Biotechnology: Bioprocess, Bioseparation, and Cell Technology). [Google Scholar]; Most detailed description of application potentials of S-layers for life and non-life sciences
- 11.Sára M., Pum D., Schuster B., Sleytr U.B. S-layers as patterning elements for application in nanobiotechnology. J Nanosci Nanotechnol. 2005;5:1939–1953. doi: 10.1166/jnn.2005.502. [DOI] [PubMed] [Google Scholar]
- 12.Sleytr U.B., Egelseer E.M., Ilk N., Pum D., Schuster B. S-layers as a basic building block in a molecular construction kit. FEBS J. 2007;274:323–334. doi: 10.1111/j.1742-4658.2006.05606.x. [DOI] [PubMed] [Google Scholar]
- 13.Sleytr U.B., Huber C., Ilk N., Pum D., Schuster B., Egelseer E. S-Layers as a tool kit for nanobiotechnological applications. FEMS Microbiol Lett. 2007;267:131–144. doi: 10.1111/j.1574-6968.2006.00573.x. [DOI] [PubMed] [Google Scholar]
- 14.Huber C., Egelseer E.M., Ilk N., Sleytr U.B., Sára M. S-layer-streptavidin fusion proteins and S-layer-specific heteropolysaccharides as part of a biomolecular construction kit for application in nanobiotechnology. Microelectron Eng. 2006;83:1589–1593. [Google Scholar]
- 15.Huber C., Liu J., Egelseer E.M., Moll D., Knoll W., Sleytr U.B., Sára M. Heterotetramers formed by an S-layer-streptavidin fusion protein and core-streptavidin as nanoarrayed template for biochip development. Small. 2006;2:142–150. doi: 10.1002/smll.200500147. [DOI] [PubMed] [Google Scholar]
- 16.Moll D., Huber C., Schlegel B., Pum D., Sleytr U.B., Sára M. S-layer-streptavidin fusion proteins as template for nanopatterned molecular arrays. Proc Natl Acad Sci U S A. 2002;99:14646–14651. doi: 10.1073/pnas.232299399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pleschberger M., Saerens D., Weigert S., Sleytr U.B., Muyldermans S., Sára M., Egelseer E.M. An S-layer heavy chain camel antibody fusion protein for generation of a nanopatterned sensing layer to detect the prostate-specific antigen by surface plasmon resonance technology. Bioconjug Chem. 2004;15:664–671. doi: 10.1021/bc049964w. [DOI] [PubMed] [Google Scholar]
- 18.Pleschberger M., Neubauer A., Egelseer E.M., Weigert S., Lindner B., Sleytr U.B., Muyldermans S., Sára M. Generation of a functional monomolecular protein lattice consisting of an S-layer fusion protein comprising the variable domain of a camel heavy chain antibody. Bioconjug Chem. 2003;14:440–448. doi: 10.1021/bc025603+. [DOI] [PubMed] [Google Scholar]
- 19.Tschiggerl H., Casey J.L., Parisi K., Foley M., Sleytr U.B. Display of a peptide mimotope on a crystalline bacterial cell surface layer (S-layer) lattice for diagnosis of Epstein–Barr virus infection. Bioconjug Chem. 2008;19:860–865. doi: 10.1021/bc7003523. [DOI] [PubMed] [Google Scholar]
- 20.Völlenkle C., Weigert S., Ilk N., Egelseer E., Weber V., Loth F., Falkenhagen D., Sleytr U.B., Sára M. Construction of a functional S-layer fusion protein comprising an immunoglobulin G-binding domain for development of specific adsorbents for extracorporeal blood purification. Appl Environ Microbiol. 2004;70:1514–1521. doi: 10.1128/AEM.70.3.1514-1521.2004. Highlight in Nature Reviews Microbiology 1512(1515), 1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bohle B., Breitwieser A., Zwölfer B., Jahn-Schmid B., Sára M., Sleytr U.B., Ebner C. A novel approach to specific allergy treatment: the recombinant fusion protein of a bacterial cell surface (S-layer) protein and the major birch pollen allergen Bet v 1 (rSbsC-Bet v 1) combines reduced allergenicity with immunomodulating capacity. J Immunol. 2004;172:6642–6648. doi: 10.4049/jimmunol.172.11.6642. [DOI] [PubMed] [Google Scholar]
- 22.Breitwieser A., Egelseer E.M., Moll D., Ilk N., Hotzy C., Bohle B., Ebner C., Sleytr U.B., Sára M. A recombinant bacterial cell surface (S-layer)-major birch pollen allergen-fusion protein (rSbsC/Bet v1) maintains the ability to self-assemble into regularly structured monomolecular lattices and the functionality of the allergen. Protein Eng. 2002;15:243–249. doi: 10.1093/protein/15.3.243. [DOI] [PubMed] [Google Scholar]
- 23.Ilk N., Völlenkle C., Egelseer E.M., Breitwieser A., Sleytr U.B., Sára M. Molecular characterization of the S-layer gene, sbpA, of Bacillus sphaericus CCM 2177 and production of a functional S-layer fusion protein with the ability to recrystallize in a defined orientation while presenting the fused allergen. Appl Environ Microbiol. 2002;68:3251–3260. doi: 10.1128/AEM.68.7.3251-3260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24•.Gerstmayr M., Ilk N., Jahn-Schmid B., Sleytr U.B., Bohle B. Natural self-assembly of allergen-S-layer fusion proteins is no prerequisite for reduced allergenicity and T cell stimulatory capacity. Int Arch Allergy Immunol. 2009;149:231–238. doi: 10.1159/000199718. [DOI] [PubMed] [Google Scholar]; Analysis of immunological effects of S-layer fusion proteins
- 25.Gerstmayr M., Ilk N., Schabussova I., Jahn-Schmid B., Egelseer E.M., Sleytr U.B., Ebner C., Bohle B. A novel approach to specific allergy treatment: the recombinant allergen-S-layer fusion protein rSbsC-Bet v 1 matures dendritic cells that prime Th0/Th1 and IL-10-producing regulatory T cells. J Immunol. 2007;179:7270–7275. doi: 10.4049/jimmunol.179.11.7270. [DOI] [PubMed] [Google Scholar]
- 26.Avall-Jääskeläinen S., Kylä-Nikkilä K., Kahala M., Miikkulainen-Lahti T., Palva A. Surface display of foreign epitopes on the Lactobacillus brevis S-layer. Appl Environ Microbiol. 2002;68:5943–5951. doi: 10.1128/AEM.68.12.5943-5951.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuen B., Sleytr U.B., Lubitz W. Sequence analysis of the sbsA gene encoding the 130-kDa surface-layer protein of Bacillus stearothermophilus strain PV72. Gene. 1994;145:115–120. doi: 10.1016/0378-1119(94)90332-8. [DOI] [PubMed] [Google Scholar]
- 28.Riedmann E.M., Kyd J.M., Smith A.M., Gomez-Gallego S., Jalava K., Cripps A.W., Lubitz W. Construction of recombinant S-layer proteins (rSbsA) and their expression in bacterial ghosts — a delivery system for the nontypeable Haemophilus influenzae antigen Omp26. FEMS Immunol Med Microbiol. 2003;37:185–192. doi: 10.1016/S0928-8244(03)00070-1. [DOI] [PubMed] [Google Scholar]
- 29.Kern J.W., Schneewind O. BslA, a pXO1-encoded adhesin of Bacillus anthracis. Mol Microbiol. 2008;68:504–515. doi: 10.1111/j.1365-2958.2008.06169.x. [DOI] [PubMed] [Google Scholar]
- 30.Mesnage S., Tosi-Couture E., Fouet A. Production and cell surface anchoring of functional fusions between the SLH motifs of the Bacillus anthracis S-layer proteins and the Bacillus subtilis levansucrase. Mol Microbiol. 1999;31:927–936. doi: 10.1046/j.1365-2958.1999.01232.x. [DOI] [PubMed] [Google Scholar]
- 31.Mesnage S., Weber-Levy M., Haustant M., Mock M., Fouet A. Cell surface-exposed tetanus toxin fragment C produced by recombinant Bacillus anthracis protects against tetanus toxin. Infect Immun. 1999;67:4847–4850. doi: 10.1128/iai.67.9.4847-4850.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bingle W.H., Nomellini J.F., Smit J. Cell-surface display of a Pseudomonas aeruginosa strain K pilin peptide within the paracrystalline S-layer of Caulobacter crescentus. Mol Microbiol. 1997;26:277–288. doi: 10.1046/j.1365-2958.1997.5711932.x. [DOI] [PubMed] [Google Scholar]
- 33.Umelo-Njaka E., Nomellini J.F., Bingle W.H., Glasier L.G., Irvin R.T., Smit J. Expression and testing of Pseudomonas aeruginosa vaccine candidate proteins prepared with the Caulobacter crescentus S-layer protein expression system. Vaccine. 2001;19:1406–1415. doi: 10.1016/s0264-410x(00)00362-5. [DOI] [PubMed] [Google Scholar]
- 34.Duncan G., Tarling C.A., Bingle W.H., Nomellini J.F., Yamage M., Dorocicz I.R., Withers S.G., Smit J. Evaluation of a new system for developing particulate enzymes based on the surface (S)-layer protein (RsaA) of Caulobacter crescentus: fusion with the beta-1,4-glycanase (Cex) from the cellulolytic bacterium Cellulomonas fimi yields a robust, catalytically active product. Appl Biochem Biotechnol. 2005;127:95–110. doi: 10.1385/abab:127:2:095. [DOI] [PubMed] [Google Scholar]
- 35.Nomellini J.F., Duncan G., Dorocicz I.R., Smit J. S-layer-mediated display of the immunoglobulin G-binding domain of streptococcal protein G on the surface of Caulobacter crescentus: development of an immunoactive reagent. Appl Environ Microbiol. 2007;73:3245–3253. doi: 10.1128/AEM.02900-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nomellini J.F., Li C., Lavallee D., Shanina I., Cavacini L.A., Horwitz M.S., Smit J. Development of an HIV-1 specific microbicide using Caulobacter crescentus S-layer mediated display of CD4 and MIP1alpha. PLoS One. 2010;5:e10366. doi: 10.1371/journal.pone.0010366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simon B., Nomellini J.F., Chiou P., Binale W., Thornton J., Smit J., Leong J.A. Recombinant vaccines against infectious hematopoietic necrosis virus: production line by the Caulobacter crescentus S-layer protein secretion system and evaluation in laboratory trials. Dis Aquat Org. 2001;44:400–402. doi: 10.3354/dao044017. [DOI] [PubMed] [Google Scholar]
- 38.Bhatnagar P.K., Awasthi A., Nomellini J.F., Smit J., Suresh M.R. Anti-tumor effects of the bacterium Caulobacter crescentus in murine tumor models. Cancer Biol Ther. 2006;5:485–491. doi: 10.4161/cbt.5.5.2553. [DOI] [PubMed] [Google Scholar]
- 39.Georgiou G., Stathopoulos C., Daugherty P.S., Nayak A.R., Iverson B.L., Curtiss R., 3rd Display of heterologous proteins on the surface of microorganisms: from the screening of combinatorial libraries to live recombinant vaccines. Nat Biotechnol. 1997;15:29–34. doi: 10.1038/nbt0197-29. [DOI] [PubMed] [Google Scholar]
- 40.Nomellini J.F., Toporowski M.C., Smit J. Secretion or presentation of recombinant proteins and peptides mediated by the S-layer of Caulobacter crescentus. In: Banex F., editor. Expression Technologies: Current Status and Future Trends. Horizon Scientific Press; 2004. pp. 477–524. [Google Scholar]
- 41.Poobalane S., Thompson K.D., Ardo L., Verjan N., Han H.J., Jeney G., Hirono I., Aoki T., Adams A. Production and efficacy of an Aeromonas hydrophila recombinant S-layer protein vaccine for fish. Vaccine. 2010;28:3540–3547. doi: 10.1016/j.vaccine.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 42.Tschiggerl H., Breitwieser A., de Roo G., Verwoerd T., Schäffer C., Sleytr U.B. Exploitation of the S-layer self-assembly system for site directed immobilization of enzymes demonstrated for an extremophilic laminarinase from Pyrococcus furiosus. J Biotechnol. 2008;133:403–411. doi: 10.1016/j.jbiotec.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schäffer C., Novotny R., Küpcü S., Zayni S., Scheberl A., Friedmann J., Sleytr U.B., Messner P. Novel biocatalysts based on S-layer self-assembly of Geobacillus stearothermophilus NRS 2004/3a: a nanobiotechnological approach. Small. 2007;3:1549–1559. doi: 10.1002/smll.200700200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kainz B., Steiner K., Moller M., Pum D., Schäffer C., Sleytr U.B., Toca-Herrera J.L. Absorption, steady-state fluorescence, fluorescence lifetime, and 2D self-assembly properties of engineered fluorescent S-layer fusion proteins of Geobacillus stearothermophilus NRS 2004/3a. Biomacromolecules. 2010;11:207–214. doi: 10.1021/bm901071b. [DOI] [PubMed] [Google Scholar]
- 45.Kainz B., Steiner K., Sleytr U.B., Pum D., Toca-Herrera J.L. Fluorescence energy transfer in the bi-fluorescent S-layer tandem fusion protein ECFP–SgsE–YFP. J Struct Biol. 2010;172:276–283. doi: 10.1016/j.jsb.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 46.Ilk N., Küpcü S., Moncayo G., Klimt S., Ecker R.C., Hofer-Warbinek R., Egelseer E.M., Sleytr U.B., Sára M. A functional chimaeric S-layer-enhanced green fluorescent protein to follow the uptake of S-layer-coated liposomes into eukaryotic cells. Biochem J. 2004;379:441–448. doi: 10.1042/BJ20031900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Badelt-Lichtblau H., Kainz B., Völlenkle C., Egelseer E.M., Sleytr U.B., Pum D., Ilk N. Genetic engineering of the S-layer protein SbpA of Lysinibacillus sphaericus CCM 2177 for the generation of functionalized nanoarrays. Bioconjug Chem. 2009;20:895–903. doi: 10.1021/bc800445r. [DOI] [PubMed] [Google Scholar]
- 48.Pollmann K., Raff J., Merroun M., Fahmy K., Selenska-Pobell S. Metal binding by bacteria from uranium mining waste piles and its technological applications. Biotechnol Adv. 2006;24:58–68. doi: 10.1016/j.biotechadv.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 49.Velasquez L., Dussan J. Biosorption and bioaccumulation of heavy metals on dead and living biomass of Bacillus sphaericus. J Hazard Mater. 2009;167:713–716. doi: 10.1016/j.jhazmat.2009.01.044. [DOI] [PubMed] [Google Scholar]
- 50.Patel J., Zhang Q., McKay R.M., Vincent R., Xu Z. Genetic engineering of Caulobacter crescentus for removal of cadmium from water. Appl Biochem Biotechnol. 2010;160:232–243. doi: 10.1007/s12010-009-8540-0. [DOI] [PubMed] [Google Scholar]
- 51.Ilk N., Egelseer E.M., Ferner-Ortner J., Küpcü S., Pum D., Schuster B., Sleytr U.B. Surfaces functionalized with self-assembling S-layer fusion proteins for nanobiotechnological applications. Colloids Surf A: Physicochem Eng Aspects. 2008;321:163–167. [Google Scholar]
- 52.Pum D, Sleytr UB: Protein-based nanobioelectronics. In Nanobioelectronics for Electronics, Biology, and Medicine Edited by Offenhäusser A, Rinaldi R: Springer; 2009:167–180. [Lockwood D (Series Editor): Nanostructure Science and Technology.]
- 53•.Schuster B., Sleytr U.B. Composite S-layer lipid structures. J Struct Biol. 2009;168:207–216. doi: 10.1016/j.jsb.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; Use of S-layers as stabilizing structures for functional lipid membranes. A biomimetic approach copying the supramolecular structure of archaea