Abstract
Background:
the occurrence of the different types of Extended spectrum beta Lactamase producing Escherichia coli with the, Sulphurhydryl variable, Temonera and the Cefotaximase have been on the rise
Aim:
The study was to determine the prevalence of extended spectrum beta lactamase gene resistance across the clinical isolates of hospitalized patients.
Materials and Method:
Three hundred and fifty isolates of Escherichia coli were received from different clinical specimens. The susceptibility profile of the isolates against 10 different antibiotics was examined, the MICs (Minimum Inhibitory Concentration) for ceftazidime were also determined using micro-broth dilution assay. Isolates showing MIC ≥ 6 μg/ml for ceftazidime were screened for ESBL (PCT)phenotypic confirmatory test and subjected to PCR (polymerase chain reaction) to further.
Results:
By disk diffusion test, there was resistance to ceftazidime and cefotaxime were 180(51.4%) and 120 (34.2%) respectively. However, all strains were susceptible to imipenem. 250 isolates showed MICs≥ 6 μg/ml for ceftazidime of which 180 (72%) were positive for extended spectrum beta lactamase. The prevalence of Sulphurhydryl variable, Temonera and the Cefotaximase among these isolates were 17.1%, 6.6% and 17%, respectively.
Conclusion:
For the identification of extended spectrum beta lactamase producing isolates it is recommended that clinical laboratories adopt simple test based on Cinical laboratory standard institute recommendation for confirming extended spectrum beta lactamase production in enterobacteriacea species.
Keywords: Extended spectrum beta Lactamase, bla TEM, SHV and CTX-M, Cephalosporins
Introduction
Antimicrobial resistance is a growing problem in many bacterial pathogens and is of particular concern for hospital-acquired nosocomial infections[1]. Escherichia coli is an important pathogen that causes urinary tract infections (UTIs), pneumoniae, and intra-abdominal infections in hospitalized immunocompromised patients with severe underlying diseases[2]. Resistance of Escherichia coli to broad spectrum antibiotics such as extended spectrum cephalosporins due to plasmid mediated enzymes (extended spectrum β-lactamases: ESBLs) results in treatment failure of infections caused by these isolates[3].
Difficult treatment of these infections may allow ESBL producing pathogenes to remain within the environment and patients for the long period of time and to spread easily within and between hospitals. Within a few years of the commercial release of β-lactams, gram negative bacilli (especially Escherichia coli) that harbored mutated versions of the potent TEM and SHV enzymes were detected. These and other newly detected β- lactamases (for example CTX-M) hydrolyze β-lactam antibiotics containing the oxymino side-chain[4]. CTX-M preferentially hydrolyze cefotaxime and based on the changes in amino acids sequences identities is divided into five groups[5]. strong ESBL activity which can efficiently hydrolyzes penicillins and cephalosporins. Because of inappropriate usage of antibiotic in treatment of infection caused by ESBL producing pathogens, it seems that studies about correct detection and antibiotic resistance pattern of these organisms are necessary. In recent years a few studies were done about the E. coli isolates producing ESBLs in our country[7,12].
Also despite of importance of PER type β-lactamase, there is not any information concerning E. coli isolates harboring these bla gene in Yola south Nigeria. Therefore, the present study was carried out to determine the prevalence of the genes encoding SHV, TEM, and CTX-M are responsible for drug resistance.
Materials and Methods
A total of three hundred and fifty (350) patient samples were collected, all from the following biological specimens, Urine(180), Sputum(20) Blood (25), Stool (50) High Vaginal Swabs (20) Wound swabs (30), and Abscess (25). Blood samples were collected into clean blood culture bottles and incubated anaerobicaly, urine samples were also collected into sterile universal bottles, sputum and stool samples were also collected into clean universal sterile containers and sent to the laboratory for microbiological processing.
Standard microbiological methods according to (CLI) specimens were cultured aerobically and aerobically all at 37°C except otherwise stated.
Susceptibility Profile
The standard disk diffusion pour plate method according to Kirby buer was used, the various antibiotics used were, Ceftazidime, Cefotaxime, Ceftriaxome, Cefixime (30 μg), Gentamycine, Ampicillin, Imipenem (10 μg), Tetracycline (10 μg), Aztronam (30 μg), and Augumentin (30 μg), zones were reported as sensitive and resistance according to the width of the zone of inhibition.
Screening for ESBL
ESBL production ability of isolates showing MICs ≥ 6 μg/ml for ceftazidime was examined by using phenotypic confirmatory test (PCT). In brief, pairs of discs containing cefotaxime ESBLs in Klebsiella pneumoniae from Iran Iran J Basic Med Sci, Vol. 13, No. 3, Summer 2010 113 (30 μg) and ceftazidime (30 μg) with and without clavulanic acid (10 μg) were placed on opposite sides (at a distance of 20-30 mm) of the same inoculated plate containing Muller Hinton agar (BBL-Becton Dickinson). A positive test result was defined as a≥5 mm increase in zone diameter compared to a disk without clavulanic acid[16].
DNA Extraction And PCR
ESBL positive isolates were cultured in LB (Luria-Berneti) broth at 37 °C overnight and plasmid DNA was extracted according to the published method of Johnson and Woodford[17].
Specific primers and annealing temperature for amplifying the blaSHV, blaTEM, and blaCTX genes by PCR were shown in Table 1. PCR was carried out in solution containing 200 μM concentration of dNTPs, 10 Pmol of each primer, 0.8 mM/μl MgCl2, 0.5 U Taq polymerase and 50 ng DNA template in a final volume of 25 μl Escherichia coli 6681 containing blaSHV, blaCTX-M and blaTEM gene (Kindly provided by Patrice Nordmann) were used as controls.
Table 1.
Showing the Percentage resistance pattern of the isolates across the various antibiotic

Table 2.
showing the primer names across the tables and the annealing temperature
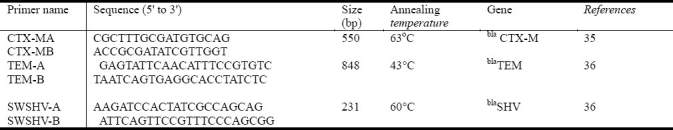
Table 3.
Showing the percentage occurrence of the genes across the biological specimens

PCR-RFLP
The CTX, PER and TEM ESBL amplified genes were characterized by PCR-Restriction Fragment Length Polymorphism (RFLP). This analysis was performed using PstI for TEM and PER and PvuII for CTX-M ESBL amplified genes. Restriction fragments were analyzed using gel electrophoresis in a 1% (W/V) agarose gel.
Statistical Analysis
Parametric methods (t-test) were used for statistical analysis of the data obtained from drug susceptibility testing and also using standard percentage occurrence for the calculations of data received.
Results
The susceptibility pofile showed a remarkable 100% susceptibility of the isolate to Imipenem and there was also the resistance to Ceftazidime and Cefotaxime with (51.4 % and 34.2 %) 180 and 120 respectively, the resistance pattern across board showed a (83%) resistance pattern of the isolates to tetracycline and Ampicillin. The resistance pattern for Ceftriaxome (23%), Cefixime (16%), Gentamycine (32%) Aztronam (18%), and Augumentin (29%). A total of 181 isolates (52%) were seen to be resistant one or more of the third generation Cephalosporin. 250 isolates showed MICs ≥ 6 μg/ml for ceftazidime of which 180 (72%) The MICs of ceftazidime in ESBLs producing isolates ranged from 6 to > 512 μg/ml, 63% of which showed MICs≥ 128 μg/ml. The ESBL-producing isolates were recovered mostly from urine (n = 180), stool (n = 50) and wound (n = 30) specimens. Table 1 showed the results of percentage resistance to antibiotics in ESBL positive isolates.
Discussion
Escherichia coli is a known pathogenic organism that has caused sever nosocomial, urinary tract infection, blood borne disease and gastro enteritis which has led to sever morbidity and mortality. Following the extensive use and constant abuse of the extended spectrum beta lactam agent, the outbreaks of infection caused by extended spectrum beta Lactamase producing Escherichia coli have been widely reported throughout the world[18]. The production of ESBLs is a major threat to the use of new generation of cephalosporins [19,20]. Long hospitalization, diabetes, age over 60 and previous antibiotic treatment have been reported as the risk factors to acquire infections with ESBL strains[21].
In our study antimicrobial susceptibility testing showed that 250 isolates showed MICs≥ 6 μg/ml for ceftazidime of which 180 (72%) were positive for ESBL in PCT. ESBLs in other countries in our region such as India (97.1%), Turkey (57%) and South Korea (30%) (22-24). However, previous studies from Iran about ESBL positive strains of Escherichia coli is rare; Feizabadi et al in 2006 detected 44.5% ESBL positive rate among clinical Escherichia coli isolated from clinical specimens in Tehran[8]. The rates of ESBL producing Escherichia coli isolated from Tehran reported by Aminzadeh et al was 52.5% in 2008[9]. Ramazanzadeh et al reported 34.8% of ESBL producing strains of Gram-negative bacteria isolated from Kurdistan[10]. In 2009 Bazzaz et al also indicated that the prevalence of ESBL producing strains of E. coli and K. pneumonia was 59.2%[11]. As 180 of 250 ceftazidime resistant isolates were ESBL positive in this study, it appears that ESBL production has a significant role in resistance to cephalosporines rather than other mechanisms of resistance such as loss of porins and efflux pumps in our research[25–26]. The MICs of ceftazidime for the majority of ESBL positive isolates which 180 (72%) was >32μg/ml. However, these isolates were susceptible to imipenem. So, the best coverage against ESBL-producing isolates was obtained with imipenem (0% resistance), Since ESBL genes are generally plasmids mediated, many of the organisms that harbor ESBLs also are resistant to other classes of antibiotics such as aminoglicosides, flouroquinolons, tetracyclines, chloramphenicol and sulfonamides[27]. These results obtained from past issues on this confirms the prevalence of occurrence in our results and our study: the rate of resistance to cephalosporines, aminoglicosides and flouroquinolons in ESBL positive strains is higher compared to all of studied isolates. In accordance with our results, 41% of ESBL positive strains were resistant to ciprofloxacin which is lower than Lautenbach's report (60%)[28].
The prevalence of blaSHV and blaTEM genes in this study was 17.1% and 17% respectively which are different from the results of the multi-national study group (67% and 16%, respectively)[30]. In Iran, Feizabadi et al in 2009 showed that 69.7% of Escherichia coli isolated from Tehran were ESBL positive and the prevalence of blaTEM, blaSHV, blaCTX-M-I and blaCTX-M-III among these isolates was 54%, 67.4%, 46.51% and 29%, respectively[12].
Conclusion
The emergence and increased spread of ESBL-producing Escherichia coli strains is worrisome and usage of Cephalosporins against these isolates is ineffective. As imipenem is the drug of choice for serious infection disease these days, prolong and extensive use of this drug in treatment of infection caused by resistant isolates will enhance. Because of this problem prudent use of β-lactam antibiotics containing an oxyimino group and consistent application of basic infection control procedures in treatment centers is necessary. Due to importance of ESBL producing organisms and difficult treatment of infections caused by these bacteria, for rapid identification of ESBL producing isolates clinical laboratories should adopt simple test based on CLSI recommendation for confirming ESBL production in enterobacteracea species. Laboratory services should be available to support every infection control program. Unfortunately there is rare co-operation between clinical settings and laboratories in Nigeria. We should always remember that effective treatment of serious infections will only be achieved by close cooperation between clinical and laboratory staff.
Acknowledgments
All thanks and gratitude goes to the staff and nurses of the Federal Medical Centre Yola and also my appreciation goes to the National Academy for the Advancement of Science for the financial assistance in form of grants and tutelage in carrying out this research.
There was no competing interest in this study.
References
- 1.Monnet DL, Archibald LK, Phillips L, Tenover FC, McGowan JE, JR, Gaynes RP. Antimicrobial use and resistance in eight US hospitals: complexities of analysis and modeling. Intensive Care Antimicrobial Resistance Epidemiology Project and National Nosocomial Infections Surveillance System Hospitals. Infect Control Hosp Epidemiol. 1998;19:388–394. doi: 10.1086/647837. [DOI] [PubMed] [Google Scholar]
- 2.Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11:589–603. doi: 10.1128/cmr.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iroha IR, Egwu OA, Ngozi AT, Chidiebube NA, Chika EP. Extended spectrum beta–lactamase (ESBL) mediated resistance to antibiotics among Klebsiella pneumoniae in enugu Metropolis. Maced J Med Sci. 2009;2:196–199. [Google Scholar]
- 4.Paterson DL, Ko WC, Von Gottberg A, et al. Implications of extendedspectrum beta-lactamase production in nosocomial infections international prospective study of Klebsiella pneumoniae bacteremia. Ann Intern Med. 2004;140:26–32. doi: 10.7326/0003-4819-140-1-200401060-00008. [DOI] [PubMed] [Google Scholar]
- 5.Pitout JD, Hossain A, Hanson ND. Phenotypic and molecular detection of CTX-M β-Lactamases produced by Escherchia coli and Klebsiella spp. J Clin Microbiol. 2004;42:5715–5721. doi: 10.1128/JCM.42.12.5715-5721.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases.: a clinical update. Clin Microbiol Rev. 2005;18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shahcheraghi F, Moezi H, Feizabadi MM. Distribution of TEM and SHV Beta-lactamase genes among Klebsiella pneumoniae strains isolated from patients in Tehran. Med Sci Monit. 2007;13:247–250. [PubMed] [Google Scholar]
- 8.Feizabadi MM, Etemadi G, Yadegarinia D, Rahmati M, Shabanpoor S, Bokaei S. Antibiotic-resistance patterns and frequency of extended-spectrum beta-lactamase-producing isolates of Klebsiella pneumoniae in Tehran. Med Sci Monit. 2006;12:BR362–5. [PubMed] [Google Scholar]
- 9.Aminzadeh Z, Sadat Kashi M, Sha’bani M. Bacteriuria by extended-spectrum Beta-lactamase producing Escherichia coli and Klebsiella pneumoniae: isolates in a governmental hospital in South of Tehran, Iran. Iran J Kidney Dis. 2008;2:197–200. [PubMed] [Google Scholar]
- 10.Ramazanzadeh R, Chitsaz M, Bahmani N. Prevalence and antimicrobial susceptibility of extended-spectrum beta-lactamase-producing bacteria in intensive care units of Sanandaj general hospitals (Kurdistan, Iran) Chemothe. 2009;55:287–292. doi: 10.1159/000224656. [DOI] [PubMed] [Google Scholar]
- 11.Bazzaz BS, Naderinasab M, Mohamadpoor AH, Farshadzadeh Z, Ahmadi S, Yousefi F. The prevalence of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae among clinical isolates from a general hospital in Iran. Acta Microbiol Immunol Hung. 2009;56:89–99. doi: 10.1556/AMicr.56.2009.1.7. [DOI] [PubMed] [Google Scholar]
- 12.Feizabadi MM, Delfani S, Raji N, et al. Distribution of bla(TEM) bla(SHV), bla(CTX-M) genes among clinical isolates of Klebsiella pneumoniae at Labbafinejad hospital, Tehran, Iran. Microb Drug Resist. 2010;16:49–53. doi: 10.1089/mdr.2009.0096. [DOI] [PubMed] [Google Scholar]
- 13.Podschun R, Ullmann U. Klebsiella spp as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11:589–603. doi: 10.1128/cmr.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Approved Standard M2-A8. 5th ed. PA, USA: NCCLS, Villanova; 2004. National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests. [Google Scholar]
- 15.Approved document M7-A5. 5thed. Wayne, PA, USA: 2000. National Committee for Clinical Laboratory Standards (NCCLS). Methods for dilution antimicrobial nsusceptibility tests for bacteria that grow aerobically. [Google Scholar]
- 16.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing. 15th informational supplement (M100-s15) NCCLS [Google Scholar]
- 17.Johnson A, Woodford N. Plasmid analysis. In: Johnson A, Woodford N, editors. Molecular bacteriology protocols and clinical application. London: Human Press; 1998. pp. 24–28. [Google Scholar]
- 18.Branger C, Lesimple AL, Bruneau B, Berry P, Lambert-Zechovsky N. Long-term investigation of the clonal dissemination of Klebsiella pneumoniae isolates producing extended-spectrum β-lactamases in a university hospital. J Med Microbiol. 1998;47:201–209. doi: 10.1099/00222615-47-3-201. [DOI] [PubMed] [Google Scholar]
- 19.Mendes C, Kiffer C, Segura A, Ribeiro J, Turner P. Klebsiella pneumoniae with multiple antimicrobial resistances. Braz J Infect Dis. 2004;8:109–111. doi: 10.1590/s1413-86702004000100008. [DOI] [PubMed] [Google Scholar]
- 20.Putman M, VanVeen HW, Konings WN. Molecular properties of bacterial multidrug transporters. Microbial Mol Biol Rev. 2000;64:672–693. doi: 10.1128/mmbr.64.4.672-693.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silva1 N, Oliveira M, Bandeira1 AC, Brites C. Risk factors for infection by extended-spectrum beta-lactamase producing Klebsiella pneumoniae in a Tertiary hospital in Salvador, Brazil. Braz J Infect Dis. 2006;10:191–193. doi: 10.1590/s1413-86702006000300007. [DOI] [PubMed] [Google Scholar]
- 22.Jeong SH, Bae IK, Lee JH, et al. Molecular characterization of extended spectrum beta-lactamases produced by clinical isolates of Klebsiella pneumoniae and Escherichia coli from a Korean nationwide survey. J Clin Microbiol. 2004;42:2902–2906. doi: 10.1128/JCM.42.7.2902-2906.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lal P, Kapil A, Das BK, Sood S. Occurrence of TEM and SHV gene in extended spectrum beta-lactamases (ESBLs) producing Klebsiella sp. isolated from a tertiary care hospital. Indian J Med Res. 2007;125:173–8. [PubMed] [Google Scholar]
- 24.Taşli H, Bahar IH. Molecular characterization of TEM- and SHV-derived extended-spectrum beta-lactamases in hospital-based enterobacteriaceae in Turkey. Jpn J Infect Dis. 2005;58:162–167. [PubMed] [Google Scholar]
- 25.Pages JM, Lavigne JP, Leflon-Guibout V, et al. Efflux pump, the masked side of ß-Lactam resistance in Klebsiella pneumoniae clinical isolates. PLoS One. 2009;4:e4817. doi: 10.1371/journal.pone.0004817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ananthan S, Subha A. Cefoxitin resistance mediated by loss of a porin in clinical strains of Klebsiella pneumoniae and Escherichia coli. Indian J Med Microbiol. 2005;23:20–23. doi: 10.4103/0255-0857.13867. [DOI] [PubMed] [Google Scholar]
- 27.Nathisuwan S, Burgess DS. Extended-spectrum beta-lactamases: epidemiology, detection, and treatment. Pharmacother. 2001;21:920–928. doi: 10.1592/phco.21.11.920.34529. [DOI] [PubMed] [Google Scholar]
- 28.Lautenbach E, Strom BL, Bilker WB, Patel JB, Edelstein PH, Fishman NO. Epidemiological investigation of fluoroquinolone resistance in infections due to extended-spectrum beta lactamase-producing Escherichia coli and Klebsiella pneumoniae. Clin Infect Dis. 2001;33:1288–1294. doi: 10.1086/322667. [DOI] [PubMed] [Google Scholar]
- 29.Olson AB, Silverman M, Boyd DA, et al. Identification of a progenitorof the CTX-M-9 group of extended-spectrum beta-lactamases from Kluyvera georgiana isolated in Guyana. Antimicrob Agents Chemother. 2005;49:2112–2115. doi: 10.1128/AAC.49.5.2112-2115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernard H, Tancrede C, Livrelli V, Morand A, Barthelemy M, Labia R. A novel plasmid-mediated extended-spectrum beta-lactamase not derived from TEM- or SHV-type enzymes. J Antimicrob Chemother. 1992;29:590–592. doi: 10.1093/jac/29.5.590. [DOI] [PubMed] [Google Scholar]


