Abstract
Background:
Dengue fever and dengue hemorrhagic fever are caused by dengue virus. Dengue infection remains a burning problem of many countries. To diagnose acute dengue in the early phase we improve the low cost, rapid SYBR green real time assay and compared the sensitivity and specificity with real time Taqman® assay and conventional nested PCR assay.
Aims:
To develop low cost, rapid and reliable real time SYBR green diagnostic dengue assay and compare with Taqman real-time assay and conventional nested PCR (modified Lanciotti).
Materials and Methods:
Eight cultured virus strains were diluted in tenth dilution down to undetectable level by the PCR to optimize the primer, temperature (annealing, and extension and to detect the limit of detection of the assay. Hundred and ninety three ELISA and PCR proved dengue clinical samples were tested with real time SYBR® Green assay, real time Taqman® assay to compare the sensitivity and specificity.
Results:
Sensitivity and specificity of real time SYBR® green dengue assay (84% and 66%, respectively) was almost comparable to those (81% and 74%) of Taqman real time PCR dengue assay. Real time SYBR® green RT-PCR was equally sensitive in primary and secondary infection while real time Taqman was less sensitive in the secondary infection. Sensitivity of real time Taqman on DENV3 (87%) was equal to SYBR green real time PCR dengue assay.
Conclusion:
We developed low cost rapid diagnostic SYBR green dengue assay. Further study is needed to make duplex primer assay for the serotyping of dengue virus.
Keywords: RT-PCR, real time PCR, dengue, sybr green
Introduction
Dengue is a human disease caused by a virus transmitted by Aedes spp. Mosquitoes with four distinct, but closely related serotypes of viruses, referred to as dengue “serotypes” DENV-1, DENV-2, DENV-3, and DENV-4. Annually 50 to 100 million people are affected by dengue, and 500,000 of them progress to Dengue hemorrhagic fever (DHF) with 25,000 deaths[1]. Area of risk for dengue infection is increasing year by year. About 2.5 billion people are at risk of dengue and additional 120 million people visit dengue endemic area every year[2]. The disability adjusted life per year for the dengue is 520 to 580 per million populations[3].
Dengue fever (DF) and dengue hemorrhagic fever (DHF) are the main clinical manifestation of the disease. DHF with shock (dengue shock syndrome or DSS) is the most severe stage of the disease. Clinical presentation of DF and DHF patients at 3-5 days after the onset of illness are somehow similar with the presence of defervescence in the same time[4], so that DF and DHF are difficult to diagnose at this critical period. DF and DHF are classified according to the WHO guideline 1998, the overall sensitivity and the specificity of this clinical classification still remain quite low[5]. Lifelong immunity of dengue virus for the homologous serotype is possible, however partial or lack of immunity after infection with heterologous serotype exacerbate the virus replication in the secondary infection[6]. Immunity based antibody dependent enhancement (ADE) is one of the declare pathogenesis process, for the severe cause of the disease[7]. As the secondary infection progresses with more severe clinical manifestation, it can be correlated with higher viremia and DHF/DSS[8].
Antibody detection by enzyme-linked immunosorbent assay (ELISA) is useful and most popular tool for diagnosis, after 1 week of the fever[9], but it cannot be used earlier than 3-5 days after the fever nor can be used for discriminating the serotypes[10]. Molecular method of diagnosis of dengue is going to replace the traditional method of cell culture as the gold standard test for viral detection[11–13]. SYBR green real time reverse-transcription polymerase chain reaction (RT-PCR) assay has more advantage in epidemiological survey as it is rapid, simple and sensitive than traditional RT-PCR methods[14]. Due to detection of the amplified product and the probe in the same closed vessel, the contamination is greatly reduced compared to other PCR reaction. RT-PCR is a useful tool for the detection and the quantitation of dengue[15].
Taqman probe RT-PCR containing specific primer pair for each serotype and probes is more specific. This is rapid, specific and costly diagnostic method than SYBR green RT-PCR[16]. Developing the specific primers for detection and typing dengue virus by SYBR green RT-PCR assay could be more accurate[17]. The primer pair developed in previous studies cannot amplify the respective sample because these primers could not align with most of the Asian strain due to the variation of the genotypes in different countries. The PCR primers must vary to achieve higher sensitivity. Objective of our study was to develop the specific primer from the most conserved capsid region of the virus and optimise the condition for this primer, to develop a rapid lower cost, real-time SYBR green dengue assay and compare the sensitivity to the slower conventional nested PCR and more expensive Taqman probe real-time PCR assay.
Materials and Methods
The study was conducted in the collaboration of Faculty of Tropical Medicine, Mahidol University and Armed forces Research Institute of Medical Sciences (AFRIMS) Bangkok, Thailand. The research was done after approval by the Ethics Committee of the Faculty of Tropical Medicine, Mahidol University and permission granted by Walter Reed National Military Medical Center, USA.
Four laboratory strains and eight standard strains of dengue virus were utilized to detect the limit of detection (LOD) in conventional nested PCR. Eight standard strains were utilized to optimize the new primer for SYBR green real-time assay. To calculate the sensitivity specificity and the confidence interval, the stats calculator from the KT Clearinghouse website was used at: http://ktclearinghouse.ca/cebm/practise/ca/calculators/stats calc.
Serum Samples
Two hundred and twenty three clinical serum samples were utilized in the study was collected since 2006 to 2008 from febrile patients. Diagnostic confirmation of dengue was done by both ELISA and conventional nested PCR. Samples were kept in -70°C in AFRIMS. Out of 223 samples 30 blind samples were utilized for the initial validation of the SYBR green real-time assay. Out of 193 samples 43 were primary, 130 were secondary and 20 were negative by ELISA.
Dilution and RNA Extraction of Wild and Standard Strain
Four laboratory virulent strains: DENV-1 Hawaii strain, DENV-2 NGC strain, DENV-3 H87 Philippines strain, and DENV-4 814669 Dominica strain and eight standard strains of C6/36 isolated dengue virus were utilized to determine the limit of detection. Standard virus strains were propagated in C6/36 cells; PFU titres were determined by plaque assay and stored at -70°C. Wild virus strain was diluted in RPMI solution, while standard strain was diluted in normal human serum. Ten-fold serial dilutions were performed to make the lowest concentration of 0.0001 pfu/mL in both groups of strains. The diluted solution of each concentration of 140 μL was taken as sample of interest and total RNA was extracted by QIAGEN commercial kit (USA) according to the manufacturer's recommendation. 50 μL of AVE buffer was applied for elution.
RT-PCR
Modified Lanciotti protocol was applied for one step RT-PCR and Nested PCR. The primer applied was shown in Table 1.
Table 1.
The primers used for conventional nested PCR assay

Single Step RT-PCR
Single step 1st round RT-PCR was set on 50 μL final volume containing 5 μL of RNA suspension, 5 μL of 10 × PCR buffer II, 1μL of 10 mM dNTPs, 3 μL of 25 mM MgCl2, 1.25 μL of 10pmoL/μl of forward primer D1, 1.25 μL of 10pmoL/μL of reverse primer D2, 0.25 μL of 1M DTT, 0.10 μL of reverse transcriptase from avian myeloblastosis virus (AMV RT, 10 U/μL, Promega, Madison, WI, 0.25 μL of AmpliTaq DNA polymerase (5U/μL, Applied Biosystems), and 32.9 μL of RNase free water was applied in Thermocycler. The RT step was performed at 42°C for 60 min It was followed by 35 cycles of thermo cycling; 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min, using Gene Amp PCR system 9600 (Perkin Elmer, Norwalk, CT, USA).
Nested PCR
5 μL of 10 × PCR buffer II, 4 μL of 10 mM dNTPs, 5 μL of 25 mM MgCl2, 1.25 μL of 10pmoL/μL of forward primer D1, 1.25 μL of 10pmoL/μL of each reverse primer, DNA polymerase (5U/μL, Applied Biosystems), and 24.50 μL of RNase free water. The RT-PCR product (2 μL) was then diluted to 1:100 dilutions in RNase free water. Five micro litter of this dilution was added in 45 μL of nested PCR mixture.
The nested PCR step was performed for 25 cycles of thermo cycling; 94°C for 30 sec, 55°C for 1 min, and 72°C for 2 min, using Gene Amp PCR system 9600 (Perkin Elmer, Norwalk, CT, USA).
Gel Electrophoresis
PCR products of both RT-PCR and Nested PCR steps (9 μL of PCR product with 1 μL of gel loading buffer) were electrophoresis in agarose gel and stained with ethidium bromide. The common band of 511 base pairs (bp.) in size was observed in the RT-PCR step, while in nested PCR step specimen containing DENV-1, 2, 3 or 4 was identified as the DNA band of 482, 119, 290, or 392 bp.
Design of Oligonucleotide Primers for SYBR Green-Based RT-PCR
The primers described by Shu et al.[17] were considered and re-designed by sequence alignment to give maximum sequence homology among Thai DENV strains and other Asia pacific strains and the Dominican strain. The Thai DENV strains sequences were obtained from GenBank (accession numbers shown in parentheses) included 10 DENV-1 (AY732474-AY732483), 10 DENV-2 (DQ181797-DQ181806), 6 DENV-3 (AY676348-AY676353), 6 DENV-4 (AY618988-AY618993) were aligned by using Clustal X software. The sequences of some primers were modified to degenerate to match with the sequences of DENV Thai strains and other strains. Primers were designed avoiding the primer dimer and the secondary structure. Designed primer is shown in Table 2.
Table 2.
The primers used for the SYBR green real-time RT-PCR
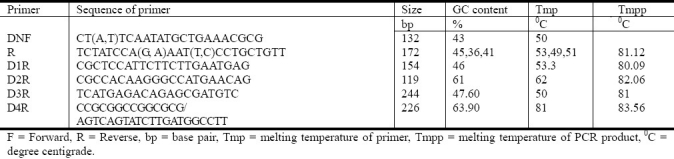
Designed primer from the most conserved capsid region of the virus was optimized in annealing at 54°C for 30 sec and extension at 60°C for 30 sec. For one step RT-PCR, RT master mix was prepared according to the protocol of the Power SYBR Green RNA-to-CT 1-Step Kit (Applied Biosystems, Foster, CA, USA). 20 μL of PCR mix was prepared with RNase-free water to 4.24 μL, 10 moL volume, Power SYBR® Green RT-PCR Mix (2 ×) 0.3 μL, forward primer (10 μmoL) 0.3 μL reverse primer (10 μmoL), RT Enzyme Mix (125 ×), 0.16 μL. The tubes with mix were aliquoted in the micro tube and added 5 μL of RNA template. The reaction optical tubes were sealed with optical caps and subjected in 7300 Applied Bio Systems real time PCR with reverse transcription (48°C, 30 min), Activation of ampliTaq Gold® DNA polymerase, UP (Ultra Pure) (95°C, 10 min), denaturing (95°C, 15 sec) annealing 54°C 30 sec and the extension 60°C 30 sec and dissociation step with 95°C 15 sec, 60°C 1 min, 95°C 15 sec, and 60°C 15 sec.
Taqman® Real Time RT-PCR
The Taqman® real time RT-PCR was performed according to Armed Forces Research Institute of Medical Sciences AFRIMS's protocol using AgPath-ID™. One-Step RT-PCR Kit (Ambion, Austin, TX, USA). Two separate duplex reactions were carried out for each RNA sample, one for simultaneous detection of DENV-1 and DENV-3 and one for DENV-2 and DENV-4. DENV-1/DENV-3 reaction mixture contained 2.5 μL of RNA suspension, 10 pmoL (1 μL) of each DENV-1 and DENV-3 primers, 5 pmol (0.5 μL) of each DENV-1 and DENV-3 probes, 1.83 μL of DNase RNase-free water, 1.67 μL of Detection Enhancer, 20 U of RNase, 12.5 μL of 2 × RT-PCR buffer, and 1 μL of 25 × RT-PCR enzyme mix, in total volume of 25 μL. DENV-2/DENV-4 reaction mixture had the same composition of primers and probes were used. The single-step RT-PCR consisting of a 10 min RT step at 45°C, 15 min of Taq polymerase activation at 95°C, followed by 40 cycles of PCR at 95°C for 15 sec and 60°C for 1 min was performed on an ABI 7300 Real Time PCR system. Accumulation of PCR products was detected by monitoring the fluorescence emitted by probe hydrolysis using SDS software. The amplification cycle (Ct value) at which fluorescence was detected above the threshold was determined. A sample was defined as positive if the Ct value was <40, based on background cross-reactivity of the primers and probes in non-template control reactions.
Results
Limit of detection (LOD) of Conventional Nested PCR for Laboratory and Standard Strains
One sample each of four serotype laboratory strains and two samples each of four standard serotype strains, tenth dilution from 1000 to 0.00001 pfu/mL were subjected as a triplicate to determine the LOD by conventional nested PCR. Consistent results for all triplicate reactions of the same sample were taken as the valid result. For the laboratory strains, DENV-1 and DENV-4 were detected down to 0.1and 10 PFU/mL, whereas DENV-2 were detected down to 0.0001 PFU/mL, DENV-3 to 0.01 PFU/mL. The results using the standard strain showed that the LOD for DENV-2 and DENV-3 (0.1 and 0.01 pfu/mL, respectively) were the same as those of laboratory strains, but LOD for DENV-1 standard strain was only 1 pfu/mL and that for DENV-4 was 10 pfu/mL.
Limit of Detection (LOD) of Standard Strain in Real time SYBR Green Assay
Two samples for each of four standard serotype strains (total eight samples) were subjected in real-time PCR to find the LOD. LOD as pfu/ml for DENV1, DENV2, DENV3 and DENV4 was 0.1, 0.01, 1.0 and 1.0 pfu/mL, respectively. LOD determined by the specific primer pair was the same as those determined by the universal primer except for DENV4 (10 pfu/mL). GC tail was added in the primer to improve the sensitivity as Lo et al.[18] to improve by 1 exponential to 1pfu/mL.
Validation of Real-Time SYBR® Green Assay by Clinical Samples
Out of 223 samples, thirty blinded serum samples were tested by SYBR green real-time PCR assay using universal primer (DNF+R) set. Out of 24 positive differentiated samples by SYBR green real-time assay, 7 samples were randomly selected and tested by duplex PCR and single-plex specific primers DNF+D1R+D3R, DNF+D2R and DNF+D4R. Sensitivity of the universal primer group was 82.75% while specificity was 100%.
Sensitivity of the specific primer for each serotype tested on 7 samples. Among them only 4 were positive and 3 were negative. We conclude that duplex primer DNF+D1R+D3R is less sensitive than the single-plex primer.
Sensitivity and Specificity of Real-Time Taqman and SYBR Green PCR Assays in Comparison to Conventional Nested PCR
Hundred and ninety three dengue positive human serum samples diagnosed by conventional PCR and ELISA were tested by real-time Taqman and the SYBR green real time PCR assay and analyzed for the sensitivity and specificity as shown in Table 3.
Table 3.
Comparison of sensitivity and specificity of two real-time RT-PCR methods with conventional nested PCR
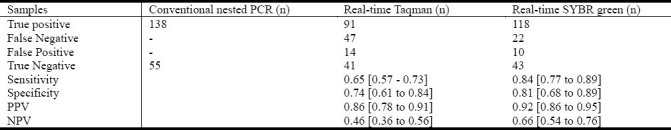
Real-time SYBR green RT-PCR test was more sensitive than real-time Taqman RT-PCR test. The sensitivity/specificity of the SYBR green RT-PCR assay was less than those of conventional RT-PCR but better than Taqman real-time PCR assay.
Sensitivity of SYBR Green Real Time Assay in Primary and Secondary Infections
Out of 193 samples, 109 positive samples in comparison with conventional nested PCR detected by real-time SYBR green analyzed with respect to primary and secondary infection. DENV-1 and DENV-2 serotype were more sensitive in primary infection, but DENV-3 and DENV-4 were sensitive in secondary infection.
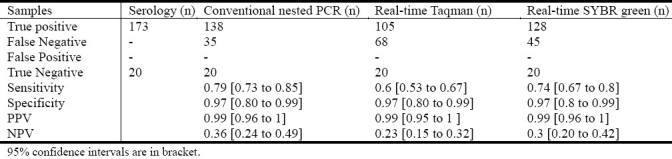
Analysis of the sensitivity of Real Time SYBR green dengue assay in different serotype with respect to primary and secondary infection is shown in Table 4.
Table 4.
Analysis of sensitivity of different PCR methods
Sensitivity of SYBR Green Real-Time Assay in Due Course of Primary and Secondary Infections
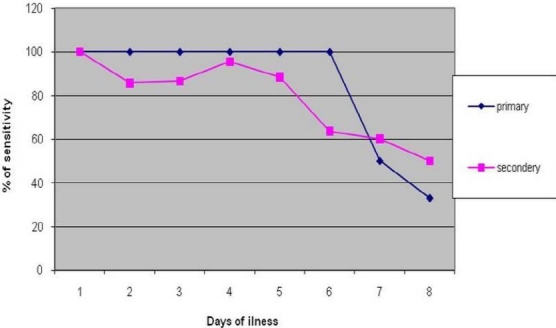
Out of 193 samples, the conventional nested PCR serotype discriminated as positive primary 38 samples and secondary 100 samples were analysed in the progression of the days of illness. Primary infection was detected consistently until the 6th days of infection and then the detection rate went down in the second week. In the secondary dengue infection cases, the detection rate decreased after the 5th days of illness (Figure 1).
Fig. 1.
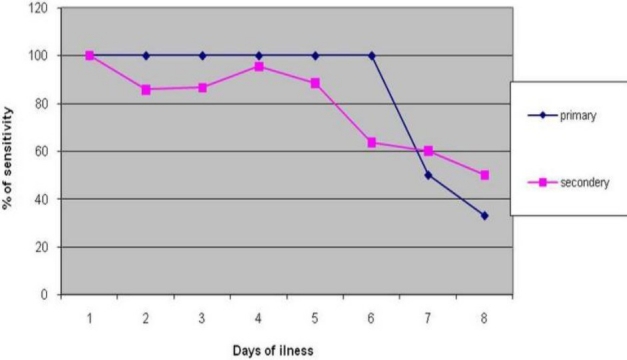
Comparison of the sensitivity of Sybr green dengue assay in primary and secondary infection. Sybr green test% of sensitivity was higher and consistent till day sixth of fever and decrease less than 50% after day seven of fever in primary infection, while sensitivity was higher till day fifth of fever and decreased in the second week.
Diagnostic Sensitivity of All Three Assays in Comparison to ELISA
ELISA is important to confirm the diagnosis even in convalescent sera; we analyzed the diagnostic values of three PCR methods using acute and convalescent phase serum samples. Out of 193 samples, 173 positive samples were compared with the results of different PCR assays and the percentage of sensitivity was calculated.
SYBR green RT-PCR assay was equally sensitive as the conventional nested PCR. Taqman real-time PCR was less sensitive than other two PCR assays. In terms of the specificity, all three assay methods were nearly 100% specific as shown in Table 4.
Sensitivity of Different Assay on Progression of the Days of Illness
Conventional nested PCR is more sensitive in the early days of infection. All PCR dengue assay was less sensitive after the 7th days of fever.
In comparison of the sensitivity of different PCR assays. All assays are sensitive (100%) in 0 days and the sensitivity decreases after 3 days. Only 50% are detectable at five days after fever by Taqman RT-PCR. The trend of decreasing sensitivity in the SYBR green is similar to the conventional nested PCR but sensitivity of real time Taqman fall less than 50% after three days of fever. Sensitivity is nearly similar on day 7 and 8 for all three assays. Sensitivity of different assays is shown in Figure 2.
Fig. 2.
Sensitivity of different assay in progression of days. All three PCR assays were highly sensitive early days while sensitivity of SYBR green assay was highest in the first week. Sensitivity of all PCR assay decreased after fifth day.
Agreement Statistic
Out of 193 samples tested by different PCR assays were analyzed in 2 × 2 tables to test the agreement of sensitivity with conventional nested PCR test. Statistical agreement of conventional nested PCR is moderate (kappa=.566) with SYBR green but slightly (kappa= 0.279) in real time Taqman assay.
Discussion
Although viral isolation is the gold standard method in dengue diagnosis, it is not common in practice due to the long time consuming, higher cost and laborious with the risk of examiners. Serology methods are in practice but they can diagnose only after the seroconversion in the second week of fever. PCR is the only practical way to diagnose dengue in acute phase of infection.
For conventional PCR, different regions of the virus RNA have been targeted[11,19,20]. To improve specificity/sensitivity, conventional nested PCR was developed[20] and modified[11] in one step RT-PCR. Among the comparison of four RT-PCR method of Dengue diagnosis, modified Lanciotti method was the most sensitive and reliable[21]. Modified Lanciotti method is common conventional method used nowadays in the laboratory. The wild virus (natural infected strain) were more sensitive (Pfu/mL) than standard laboratory strain because of the high mutating nature. Generally conventional nested PCR assay could detect less than 1 pfu/mL in all serotype.
The limit of detection by real-time SYBR green assay was far lower than the conventional PCR except in DENV3 serotype, because of the well matched primer from the capsid region to form the amplicon of the interest[22]. The limit of detection of one step SYBR green real-time PCR was higher than real time Taqman assay and substantially agrees with conventional nested PCR in our data. Less sensitivity of Taqman real time assay was previously reported[23] that only single point mutation in Taqman probe could decrease the sensitivity by 47%. Conventional nested PCR require PCR amplification step to obtain first step product (RT-PCR) using the enzyme and the primer, real time SYBR green RT-PCR did not need any supplementary step to amplify the PCR product. Real time SYBR green could detect more than conventional nested PCR in detection of DENV-1 and DENV-2 and DENV4. Relatively low sensitivity against DENV-3 may be due to repeated freezing/thawing of the samples[24].
The sensitivity of SYBR green dengue RT-PCR assay was equal or even lower than real-time Taqman RT-PCR assay in Poersch's and Lai's studies[16,25]. Over all sensitivity and specificity of universal primer in the test of 193 clinical dengue samples in our study was 84.85% and 74 % which was higher than real-time Taqman 74.54%.
In the test of 193 clinical samples SYBR green real-time assay was highly sensitive. The specificity was low compared to the conventional nested PCR as our gold standard. In this study, serology positive but conventional nested PCR negative samples were taken as false positive. SYBR green was even sensitive after the Seroconversion in the secondary dengue infection when conventional nested PCR became negative. It could be due to higher sensitivity of the assay to detect less copy number of RNA in the convalescent phase of dengue infection[16,26]. Real-time SYBR green assay was more sensitive in DENV-1, DENV-2 (100%) in primary dengue infection while DENV-3 and DENV-4 were more sensitive in secondary infection. It may be due to the unequal distribution of these serotypes in primary and secondary infection. Real-time SYBR green assay was highly sensitive in primary infection until the 6th day of the illness, while in secondary infection it got low sensitivity from the earlier day and least sensitive after the 5th day of illness. It may be due to the persistent viremia in the primary infection[27]. For the diagnostic approach, ELISA could be the choice of diagnostic test[28]. We compared the sensitivity and specificity with ELISA, the specificity was 100% and sensitivity was about same as conventional nested PCR, while real time Taqman assay was less sensitive than other two PCR assays.
Serology is still commonly used in the general laboratories. We analyzed the diagnostic sensitivity of all three PCR assays in comparison with ELISA. Real time SYBR green assay was more sensitive (73%) than real time Taqman assay (60%) and comparable to the conventional nested PCR (78%). Specificity of all assays was 100%.
By analyzing statistic agreement, the real-time SYBR green assay has moderate agreement in sensitivity with conventional nested PCR while Taqman real-time PCR has just fair agreement.
Real-time PCR dengue assay (real-time Taqman and real-time SYBR green) was more rapid[29–31] to perform the reaction while conventional nested PCR take more time as it has one more step (reverse transcription and then nested) to get the final result. The exact time required to perform different PCR assays were recorded. Real-time Taqman and real-time SYBR green dengue assay were rapid as they could be performed in 1 step in less time. The time to perform SYBR green required 45 minutes more than real-time Taqman assay.
Real-time SYBR green assay was cheaper than real-time Taqman and conventional nested PCR. ABI Biosystems recommended decreasing the volume of the reaction to economize the assay system. SYBR green assay can reduce the PCR reaction cost by half times than probe and other assays[25]. Here, we calculated the price of qualitative real-time from the 48 well plate's reaction and for the conventional nested PCR. For the single reaction of all components, the price was calculated per reaction. Real-time SYBR green assay costs about 2 times less (137 Thai Baht or 4.5 US dollars) than real-time Taqman (260 Thai Baht or 8.6 US dollars) and conventional nested PCR (204 Thai Baht or 7 US dollars).
It was easy to perform but it was not easy to optimize due to the nature of reaction (intercalating dye with ds DNA) and easily contaminated[32].
Further study in clinical sera for real-time SYBR green RT-PCR was needed to discriminate the serotype by single multiplex primer pair[33,34] and the duplex primer to find out the dual serotype infection, which is common nowadays in endemic areas. Furthermore decreasing the volume of the reaction can lower the price of the reaction. Real time SYBR green assay is useful in early diagnosis of dengue before production of immunity by the infected host[35]. Real-time SYBR green assay had many properties i.e. assay can detect many samples once, less handling, low chance of contamination and highly sensitive. These properties support to use this assay as a screening test in the epidemiological survey in almost all human infective flaviviruses[36].
We improved SYBR green real-time assay for dengue to obtain better sensitivity and specificity which would be useful to for the public health setting.
Conclusion
The limit of detection of dengue virus in real time SYBR green was nearly same as conventional nested PCR. Sensitivity and specificity of SYBR green assay was 84% and 81%. It was better than real-time Taqman assay. SYBR green was moderately agreed in the sensitivity with conventional nested PCR. In both primary and secondary infection except DENV-3, all serotype were more sensitive in SYBR green real-time PCR assay than real-time Taqman assay. Real-time Taqman dengue assay is less sensitive than real-time SYBR green. Time to perform the SYBR green assay was less in comparison with conventional nested PCR assay. The reaction cost of the SYBR green assay was also cheapest among three PCR dengue assay. In future we need further more study to find the duplex primer assay to diagnose the dual infection in dengue endemic areas.
High sensitive, specific, low time consuming and low price SYBR green assay can be concluded as the proper assay and recommended as a useful tool for the epidemiological dengue virus survey in the endemic zones.
Acknowledgments
The study was supported by Thailand international cooperation agency (TICA), the Office of the Higher Education Commission and Mahidol University under the National Research Universities Initiative and the Armed Forces Research Institute of Medical Sciences (USAMC-AFRIMS). We would like to express our gratitude to the staff of virology department of AFRIMS for their kind cooperation and advice. There is no potential conflict of interest among the authors. W Chokejindachai is principal investigator, participated in design of the study and approved the final manuscript. D Paudel carried out the all assays and drafted the manuscript. W Chokejindachai, D Paudel, R Jarman and C Klungthong carried out the analysis and interpretation of data. All authors read and approved the conception of the study.
References
- 1.Osman O, Fong MY, Devi S. Complete genome sequence analysis of dengue virus type 2 isolated in Brunei. Virus Res. 2008;135:48–52. doi: 10.1016/j.virusres.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Wilder-Smith A, Ooi E-E, Vasudevan S, Gubler D. Update on dengue: epidemiology, virus evolution, antiviral drugs, and vaccine development. Curr Infect Dis Rep. 2010;12:157–64. doi: 10.1007/s11908-010-0102-7. [DOI] [PubMed] [Google Scholar]
- 3.Dengue guidelines for diagnosis, treatment, prevention and control - New edition. Geneva: TDR/World Health Organization Press; 2009. WHO. [PubMed] [Google Scholar]
- 4.Thomas L, Brouste Y, Najioullah F, et al. Predictors of severe manifestations in a cohort of adult dengue patients. J Clin Virol. 2010;48:96–99. doi: 10.1016/j.jcv.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Srikiatkhachorn A, Gibbons Robert V, Green S, et al. Dengue hemorrhagic fever: The sensitivity and specificity of the world health organization definition for identification of severe cases of Dengue in Thailand, 1994–2005. Clin Infect Dis. 2010;50:1135–1143. doi: 10.1086/651268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munoz-Jordan JL, Collins CS, Vergne E, et al. Highly sensitive detection of dengue virus nucleic acid in samples from clinically ill patients. J Clin Microbiol. 2009;47:927–931. doi: 10.1128/JCM.01564-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halsted SB, editor. Dengue Tropical Medicine: Science and Practice. London: Imperial College press; 2008. [Google Scholar]
- 8.Cordeiro MT, Braga-Neto U, Nogueira RM, Marques ET., Jr Reliable classifier to differentiate primary and secondary acute dengue infection based on IgG ELISA. PLoS ONE. 2009;4:e4945. doi: 10.1371/journal.pone.0004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cuzzubbo AJ, Vaughn DW, Nisalak A, et al. Comparison of PanBio dengue duo enzyme-linked immunosorbent assay (ELISA) and MRL dengue fever virus immunoglobulin M capture ELISA for diagnosis of dengue virus infections in Southeast Asia. Clin Diagn Lab Immunol. 1999;6:705–712. doi: 10.1128/cdli.6.5.705-712.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saxena P, Dash PK, Santhosh SR, Shrivastava A, Parida M, Rao PL. Development and evaluation of one step single tube multiplex RT-PCR for rapid detection and typing of dengue viruses. Virol J. 2008;5:20. doi: 10.1186/1743-422X-5-20. doi: 10.1186/1743-422X-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris E, Roberts TG, Smith L, et al. Typing of dengue viruses in clinical specimens and mosquitoes by single-tube multiplex reverse transcriptase PCR. J Clin Microbiol. 1998;36:2634–2639. doi: 10.1128/jcm.36.9.2634-2639.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henchal EA, Polo SL, Vorndam V, Yaemsiri C, Innis BL, Hoke CH. Sensitivity and specificity of a universal primer set for the rapid diagnosis of dengue virus infections by polymerase chain reaction and nucleic acid hybridization. Am J Trop Med Hyg. 1991;45:418–428. doi: 10.4269/ajtmh.1991.45.418. [DOI] [PubMed] [Google Scholar]
- 13.Shu PY, Huang JH. Current advances in dengue diagnosis. Clin Diagn Lab Immunol. 2004;11:642–650. doi: 10.1128/CDLI.11.4.642-650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen CF, Shu PY, Teng HJ, et al. Screening of dengue virus in field-caught Aedes aegypti and Aedes albopictus (Diptera: Culicidae) by one-step SYBR green-based reverse transcriptase-polymerase chain reaction assay during 2004-2007 in Southern Taiwan. Vector Borne Zoonotic Dis. 2010;10:1017–1025. doi: 10.1089/vbz.2008.0069. [DOI] [PubMed] [Google Scholar]
- 15.Espy MJ, Uhl JR, Sloan LM, et al. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin Microbiol Rev. 2006;19:165–256. doi: 10.1128/CMR.19.1.165-256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poersch CdO, Pavoni DP, Queiroz MH, et al. Dengue virus infections: comparison of methods for diagnosing the acute disease. J ClinVirol. 2005;32:272–277. doi: 10.1016/j.jcv.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 17.Shu PY, Chang SF, Kuo YC, et al. Development of group- and serotype-specific one-step SYBR green I-based real-time reverse transcription-PCR assay for dengue virus. J Clin Microbiol. 2003;41:2408–2416. doi: 10.1128/JCM.41.6.2408-2416.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lo CLH, Yip SP, Cheng PKC, To TSS, Lim WWL, Leung PHM. One-step rapid reverse transcription-PCR assay for detecting and typing dengue viruses with GC tail and induced fluorescence resonance energy transfer techniques for melting temperature and color multiplexing. Clin Chem. 2007;53:594–599. doi: 10.1373/clinchem.2006.077446. [DOI] [PubMed] [Google Scholar]
- 19.Chow VT, Seah CL, Chan YC. Use of NS3 consensus primers for the polymerase chain reaction amplification and sequencing of dengue viruses and other flaviviruses. Arch Virol. 1993;133:157–170. doi: 10.1007/BF01309751. [DOI] [PubMed] [Google Scholar]
- 20.Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol. 1992;30:545–551. doi: 10.1128/jcm.30.3.545-551.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raengsakulrach B, Nisalak A, Maneekarn N, et al. Comparison of four reverse transcription-polymerase chain reaction procedures for the detection of dengue virus in clinical specimens. J Virol Methods. 2002;105:219–232. doi: 10.1016/s0166-0934(02)00104-0. [DOI] [PubMed] [Google Scholar]
- 22.Chien LJ, Liao TL, Shu PY, Huang JH, Gubler DJ, Chang GJ. Development of real-time reverse transcriptase PCR assays to detect and serotype dengue viruses. J Clin Microbiol. 2006;44:1295–1304. doi: 10.1128/JCM.44.4.1295-1304.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richardson J, Molina-Cruz A, Salazar MI, Black Wt. Quantitative analysis of dengue-2 virus RNA during the extrinsic incubation period in individual Aedes aegypti. Am J Trop Med Hyg. 2006;74:132–141. [PubMed] [Google Scholar]
- 24.Sa-ngasang A, Wibulwattanakij S, Chanama S, et al. Evaluation of RT-PCR as a tool for diagnosis of secondary dengue virus infection. Jpn J Infect Dis. 2003;56:205–209. [PubMed] [Google Scholar]
- 25.Lai YL, Chung YK, Tan HC, et al. Cost-effective real-time reverse transcriptase PCR (RT-PCR) to screen for Dengue virus followed by rapid single-tube multiplex RT-PCR for serotyping of the virus. J Clin Microbiol. 2007;45:935–941. doi: 10.1128/JCM.01258-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laue T, Emmerich P, Schmitz H. Detection of dengue virus RNA in patients after primary or secondary dengue infection by using the Taqman automated amplification system. J Clin Microbiol. 1999;37:2543–2547. doi: 10.1128/jcm.37.8.2543-2547.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaughn DW, Green S, Kalayanarooj S, et al. Dengue in the early febrile phase: viremia and antibody responses. J Infect Dis. 1997;176:322–330. doi: 10.1086/514048. [DOI] [PubMed] [Google Scholar]
- 28.Yong YK, Thayan R, Chong HT, Tan CT, Sekaran SD. Rapid detection and serotyping of dengue virus by multiplex RT-PCR and real-time SYBR green RT-PCR. Singapore Med J. 2007;48:662–668. [PubMed] [Google Scholar]
- 29.Shu PY, Huang JH. Current advances in dengue diagnosis. Clin Diagn Lab Immunol. 2004;11:642–650. doi: 10.1128/CDLI.11.4.642-650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomes-Ruiz AC, Nascimento RT, de Paula SO, da Fonseca BAL. SYBR green and TaqMan real-time PCR assays are equivalent for the diagnosis of dengue virus type 3 infections. J Med Virol. 2006;78:760–763. doi: 10.1002/jmv.20620. [DOI] [PubMed] [Google Scholar]
- 31.Parida MM. Rapid and real-time detection technologies for emerging viruses of biomedical importance. J Biosci. 2008;33:617–628. doi: 10.1007/s12038-008-0079-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arya M, Shergill IS, Williamson M, Gommersall L, Arya N, Patel HR. Basic principles of real-time quantitative PCR. Expert Rev Mol Diagn. 2005;5:209–219. doi: 10.1586/14737159.5.2.209. [DOI] [PubMed] [Google Scholar]
- 33.Chutinimitkul S, Payungporn S, Theamboonlers A, Poovorawan Y. Dengue typing assay based on real-time PCR using SYBR Green I. J Virol Methods. 2005;129:8–15. doi: 10.1016/j.jviromet.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 34.Santos HWGd, Poloni TRRS, Souza KP, et al. A simple one-step real-time RT-PCR for diagnosis of dengue virus infection. J Med Virol. 2008;80:1426–1433. doi: 10.1002/jmv.21203. [DOI] [PubMed] [Google Scholar]
- 35.Houng H-SH, Hritz D, Kanesa-thasan N. Quantitative detection of dengue 2 virus using fluorogenic RT-PCR based on 3’-noncoding sequence. J Virol Methods. 2000;86:1–11. doi: 10.1016/s0166-0934(99)00166-4. [DOI] [PubMed] [Google Scholar]
- 36.Yang CF, Chen CF, Su CL, et al. Screening of mosquitoes using SYBR Green I-based real-time RT-PCR with group-specific primers for detection of Flaviviruses and Alphaviruses in Taiwan. J Virol Methods. 2010;168:147–151. doi: 10.1016/j.jviromet.2010.05.006. [DOI] [PubMed] [Google Scholar]


