Abstract
The Epstein–Barr virus nuclear leader protein LP (EBNALP) and EBNA2 are expressed first in lymphocyte infection, coordinately regulate cell and viral gene transcription, and are critical for lymphocyte outgrowth into lymphoblastoid cell lines (LCLs). We have now found that EBNALP readily associated with EBNA2 or with the EBNA2 C-terminal acidic activation domain (E2AD) when both components were expressed by bacteria. In lymphoblasts, EBNALP and EBNA2 did not stably associate. However, EBNALP deleted for only 10 C-terminal amino acids stably associated with EBNA2 in lymphoblasts or with EBNA2 acidic activating domain from bacteria. The E2AD was essential for EBNALP coactivation of the latent membrane protein 1 promoter in lymphoblasts; EBNALP could coactivate with a deficient mutant EBNA2, EBNA2W454T, but not with EBNA2 deleted for E2AD. Moreover, EBNALP 31 amino acids (dW2Y1) with 24 C- or N-terminal amino acids was a specific and efficient affinity matrix for EBNA2 or EBNALP. Even an EBNALP 22-aa peptide, dW2, specifically bound EBNALP or EBNA2. These biochemical interactions between EBNALP and EBNA2 enable coordinated transcriptional regulation of cell and viral gene expression in lymphoblasts only when the interaction is unstable; deletion of the EBNALP C-terminal 10 aa stabilized association with EBNA2 and prevented coactivation. Because EBNALPd10 dominantly inhibited EBNALP coactivation with EBNA2, EBNALPd10 expression in LCLs may be useful in assessing the role of EBNALP coactivation in LCL growth or survival.
Keywords: EBV nuclear protein, transcription, association, transformation
Epstein–Barr virus (EBV) infection of lymphocytes usually results in a latency III infection, characterized by limited EBV gene expression, genome persistence as an episome, and EBV-driven continuous lymphocyte proliferation (for reviews, see refs. 1 and 2). EBV nuclear leader protein LP (EBNALP) and EBNA2 are the first viral proteins expressed in lymphocytes. Their RNAs are transcribed by the same promoter and differential splicing determines mRNA and protein levels (3–5). EBNALP and EBNA2 up-regulate EBV and cell gene transcription, including the viral oncogene, latent membrane protein 1 (LMP1), and are required for lymphocyte outgrowth into lymphoblastoid cell lines (LCLs) (6–9). EBNA2 interactions with cellular sequence-specific DNA-binding proteins, such as RBPJkappa, PU.1, and AUF1, are critical for EBNA2 regulation of specific virus and cell promoters (10–17). Another critical component of EBNA2 is an acidic activating domain (E2AD), which can recruit TFIIB, TAF40, TFIIH, p100, and CBP/p300, thereby up-regulating cell and virus gene transcription (8, 18–23).
Less is known about the mechanism by which EBNALP strongly and specifically potentiates EBNA2-mediated transcription (24–28). EBNALP mRNA is comprised of four nearly complete W1 and W2 exons, which encode 22- and 44-aa repeats, and Y1 and Y2 exons, which encode 11 and 34 unique amino acids (refs. 29 and 30 and Fig. 1A). Although the four EBNALP W1W2 repeats have substantial coactivation effect in transient assays with EBNA2 (27, 28), the unique Y1- and Y2-encoded domains are critical for full activity and for long-term effects, including efficient cell growth transformation (9). EBNALP is highly phosphorylated, and phosphorylation of W2 amino acids S14 has a role in coactivation with EBNA2 (31–34). EBNALP can also self-associate. W2-encoded amino acids R23RVRRR28 have been implicated in EBNALP homotypic interaction and in coactivation (31, 35). The mechanism through which EBNALP specifically coactivates transcription from promoters that have recruited EBNA2 may involve direct interaction with EBNA2 or indirect interaction through EBNA2 or EBNALP associated cellular proteins, which include RBP-Jκ, TFIIB, TAF40, TFIIH, p100, p300/CBP, HSP70, HA95, Hax-1, and estrogen-related receptor 1 (10, 11, 20–23, 36–41). The objective of these experiments was to investigate the possibility that EBNALP interacts directly with EBNA2.
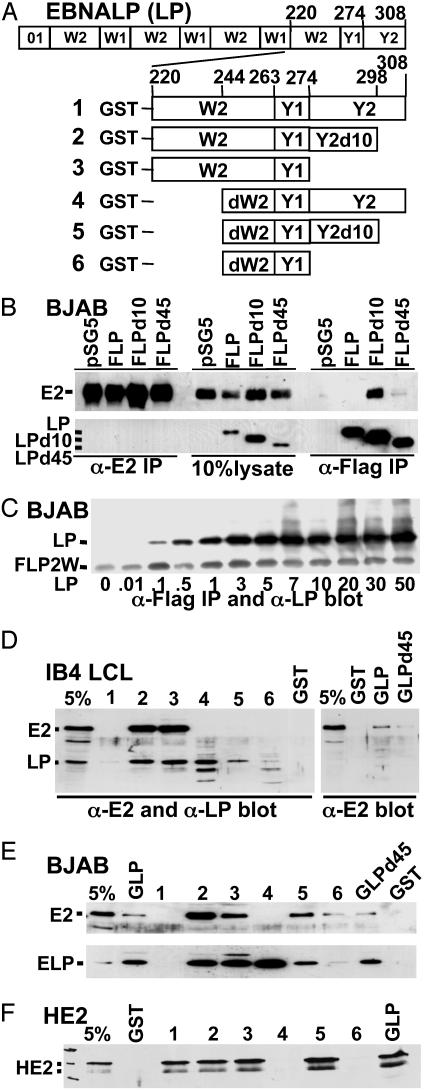
Fig. 1.
EBNA2 and EBNALP binding to EBNALP. (A) Schematic depiction of EBNALP (LP), which is encoded by four repeats of W01 (21 aa) or W1 (22 aa) and W2 (44 aa) exons and by Y1 (11 aa) and Y2 (34 aa) exons. EBNALPd10 (LPd10) and EBNALPd45 (LPD45) are deleted for the C-terminal 10 or 45 aa. Smaller EBNALP components were expressed as GST fusion proteins 1–6. (B) BJAB lymphoblasts were transfected with 10 μg each of EBNA2 and EBNALP, mutant EBNALP, or vector control (pSG5) expression plasmids. F is Flag epitope. After 48 h, lysates were immunoprecipitated with PE2 or M2 antibody to E2AD or the Flag epitope. Precipitates were separated by SDS/PAGE and immunoblotted with PE2 or JF186 monoclonal antibody to EBNALP. (C) BJAB cells were transfected with 5 μg of pSG5-FLP that has two W1W2 repeats (FLP2W) and with increasing amounts of pSG5 expressing EBNALP. M2 immunoprecipitates were analyzed by SDS/PAGE and immunoblotted with JF186 antibody, which recognizes FLP2W less well than EBNALP. (D and E) Binding of EBNA2 or EBNALP from LCL or EBNA2 or EBNALP transfected BJAB cell lysates to GST, GLP, GST-LPd45 (GLPD45), or GLP polypeptides 1–6 was assayed by SDS/PAGE and immunoblot. (F) Binding of nickel-bead-purified (His)6-tagged EBNA2 from Escherichia coli to GST, GLP, or mutant GST EBNALP fusion proteins as assayed by SDS/PAGE and immunoblot with PE2 antibody.
Materials and Methods
Expression Plamids. GST-EBNALP (GLP) was expressed from the EBNALP cDNA (29) inserted into the BamHI and EcoRI sites of pGEX-2TK (Amersham Pharmacia) and includes PIP-STQ, encoded by 5′ leader cDNA. GLPd45 was expressed from a modified cDNA with an XbaI linker in the SfiI site before Y1, adding PLV to the C terminus (Fig. 1 A). GST-W2Y1Y2, GST-W2d10, GST-W2Y1, GST-dW2Y1Y2, GST-dW2d10, and GST-dW2Y1 were made from EBNALP cDNA by PCR and cloned into BamHI and EcoRI sites of pGEX-2TK vector. W2 constructs start with codon 220 (ggt ccc ctc, GPL) and have all 44 W2 codons, whereas dW2 starts with codon 244 (gtc cgt aga, VRR) and has the C-terminal 20 codons. The GST-LP fusion proteins have three extra amino acids, THM, between GST and W2 or dW2.
GLP W2 mutants used in Fig. 2A were made in pGEX-2TK after cloning and mutagesis by PCR, using 5′ and 3′ primers that included BamHI or EcoRI sites, respectively, and EBNALP-specific sequences. Plasmids that express FLP, FLPd10, FLPd45, GST–E2AD, E2ADm, His-tagged EBNA2 (HE2), EBNA2, EBNA2-Flag, or EBNA2 with the E2AD W454T mutation have been described (18, 21–23, 27). pSG5-EBNA2ΔAD is EBV W91 strain EBNA2 DNA in pSG5 (Stratagene) with codon 401 mutated to a nonsense codon. GAL4 DNA-binding domain–EBNA2 fusion protein expression vectors with W91 EBNA2 codons 19–484, 426–465, 433–465, or 433–484 were generated by PCR using primers with 5′ EcoRI or 3′ XbaI sites and EcoRI- and XbaI-digested pSG424 (42). Recombinant plasmids were sequenced through the manipulated DNA.
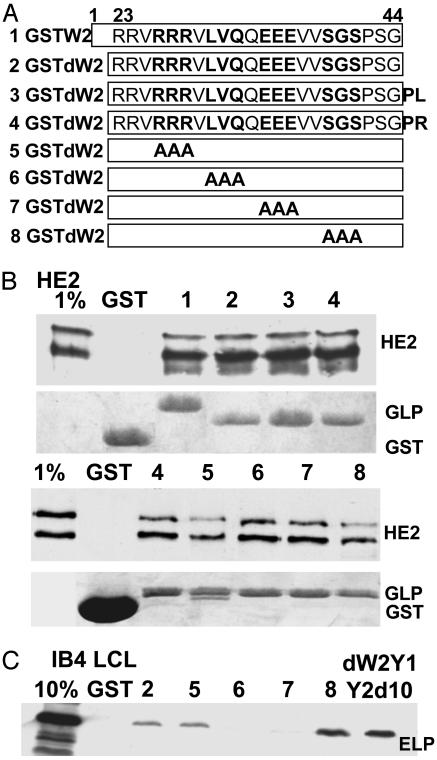
Fig. 2.
Binding of His-EBNA2 from E. coli or of EBNALP from LCL lysates to GST fusions with EBNALP W2-derived peptides. (A) Schematic depiction of the GST-fusions with EBNALP W2 (row 1), dW2 (row 2), dW2 with PL (row 3), which are the next amino acids in EBNALP Y1, dW2 with PR (row 4), which are the next amino acids in W1, and triple alanine mutations in dW2 (rows 5–8). (B) Binding assays with His-6–EBNA2 and PE2 immunoblot. Coomassie blue-stained images of the polyacrylamide gels show the relative amounts of fusion protein. (C) Binding assays with IB4 LCL cell lysate and JF186 blot. dW2Y1Y2d10 is described in Fig. 1.
Yeast expression vectors for EBNALP were made by inserting GATCCATATGGCACAA between the EBNALP cDNA BamHI and BstXI site, Tth111I cleavage 3′ to the EBNALP ORF, T4 DNA polymerase blunt ending the Tth111I site, NdeI release of the EBNALP ORF, and cloning into NdeI- and SmaI-digested pAS1-CYH2, a yeast GAL4 DNA-binding domain fusion protein expression vector (gift of S. J. Elledge, Harvard Medical School, Boston). EBNALPd45 cDNA was obtained by NdeI and SfiI cleavage and was similarly cloned. EBNALP and EBNALPd45 were also cloned into the BamHI and EcoRI sites of GAL4 acidic domain fusion protein expression vector pACT-2 yeast by PCR amplification using primers with 5′ BamHI and 3′ EcoRI sites.
Cell Lines and Transfections. IB4 LCLs (43) and BJAB EBV-negative Burkitt tumor lymphoblasts (44) were cultured in RMPI medium 1640 (GIBCO/BRL) supplemented with 10% FCS. BJAB cells at 1.0–1.5 × 107 per 400 μl were electroporated with LMP1 promoter (–512/+72)–Luc reporter to assay EBNA2 promoter up-regulation and EBNALP coactivation (45). CMV– β-galactosidase (2 μg) was cotransfected to control for expression efficiency. Pfr-Luc, with five GAL4-binding sites upstream of a promoter and a luciferase reporter was used to assay Gal4 DNA-binding domain–EBNA2 gene fusion enhancement (19).
GST- or His-Tagged Protein Binding, Immunoprecipitation, and Immunoblotting. All GST- or His-tagged proteins were expressed in E. coli BL21 (DE3) (Stratagene) and were purified by glutathione or nickel-nitrilotriacetic acid (NTA) beads (Qiagen, Valencia, CA) (21). For immunoprecipitations or GST-fusion protein pull-downs, 1–1.5 × 107 BJAB cells were cotransfected with 10 μg of pSG5-EBNA2 and 10 μg of pSG5-FEBNALP (–FLP), -FLPd10, or -FLPd45 expression plasmids and harvested after 24 h. Lysates were mixed at 4°C for 30–60 min with M2 beads (Pierce), primary monoclonal antibody, and Protein G Sepharose beads (Amersham Pharmacia) or GST-fusion protein beads. Beads were collected, washed five times with lysis buffer, and analyzed by SDS/PAGE and immunoblot (23, 27).
Results
EBNA2 and EBNALP Are Not Stably Associated in EBV-Transformed LCL Lysates. To assess whether EBNA2 and EBNALP stably associate, EBNA2 or EBNALP were immunoprecipitated from 0.15 or 0.4 M (after adjustment to 0.15 M) salt extracts of an LCL by using JF186 (46, 47) or PE2 (47) monoclonal antibodies to EBNALP W1 or to E2AD, respectively, followed by Western blots for EBNALP and EBNA2. Although the antibodies were efficient in immunoprecipitating as much as 20–30% of their cognate protein and were sensitive in detecting <1% of the cognate protein in immunoblots, EBNALP and EBNA2 did not coprecipitate (data not shown).
Because PE2 reacts with the E2AD and JF186 with EBNALP W1 and EBNALP comprised only of W1W2 repeats (EBNALPd45) can coactivate transcription with EBNA2 (27), these antibodies could fail to coprecipitate an associated protein if EBNA2 and EBNALP associate through the E2AD and EBNALPW1. We therefore attached a Flag epitope to the EBNALP N terminus (FLP) or to the EBNA2 C terminus (FE2) and used Flag-specific antibody for immunoprecipitations. FLP and EBNA2 or FE2 and EBNALP were expressed in BJAB lymphoblasts. Although 10–30% FLP and 5–15% FE2 immunoprecipitated with Flag antibody from BJAB lysates, EBNA2 did not significantly coprecipitate with FLP and EBNALP did not coprecipitate with FE2 (Fig. 1B and data not shown).
EBNA2 Stably Associates with EBNALPd10. To further evaluate whether EBNA2 can associate with a functional component of FLP, EBNA2 was transiently expressed in BJAB cells that stably express FLP, FLPd45, or FLPd10 at similar levels. EBNALPd45 can coactivate transcription with EBNA2, whereas EBNALPd10 does not (ref. 27 and data below). In multiple experiments, Flag monoclonal antibody precipitated ≈30% of FLP, FLPd45, or FLPd10. EBNA2 did not coprecipitate with FLP, <1% coprecipitated with FLPd45, and ≈10% coprecipitated with FLPd10 (Fig. 1B and data not shown). Given the 30% efficiency of FLPd10 immunoprecipitation in these experiments, ≈30% of EBNA2 was stably associated with FLPd10. In contrast, PE2 immunoprecipitated 20–30% of EBNA2 from these same lysates, but neither FLPd10 nor FLPd45 or FLP coprecipitated with EBNA2 (Fig. 1B). These experiments in lymphoblasts indicate that EBNALP deleted for only 10 C-terminal amino acids stably associates with EBNA2, that stable EBNALP and EBNA2 association correlated with null activity, and that the E2AD may mediate association with EBNALPd10.
EBNALP Associates with EBNALP in Lymphoblasts and in Directed Yeast Two-Hybrid Assays. To assess the extent of EBNALP association with EBNALP in LCLs, FLP was stably expressed at the same level as endogenous EBNALP in IB4 LCLs. In a typical experiment where the FLP immunoprecipitation efficiency was 15–20%, ≈5% of the endogenous EBNALP coprecipitated with FLP (data not shown). Thus, as much as 50% of EBNALP in an LCL can stably associate with EBNALP.
In transient cotransfections of BJAB lymphoblasts with FLP, FLPd10, or FLPd45 and EBNALP, EBNALPd10, or EBNALPd45 expression vectors, immunoprecipitations with Flag antibody immunoprecipitated up to 50% as much non-Flagged protein (data not shown). In another assessment of EBNALP association with EBNALP, FLP with two W1W2 repeats instead of four (FLP2W) was expressed in BJAB lymphoblasts with increasing amounts of WT EBNALP expressed from an identical vector. The amount of WT EBNALP coimmunoprecipitating with FLP reached its maximum at a 1:1 ratio of FLP2W to EBNALP expression plasmids (Fig. 1C). These data are most consistent with a 1:1 association of EBNALP with FLP2W.
The association of EBNALP or EBNALPd45 was also assessed in directed yeast two-hybrid assays. EBNALP or EBNALPd45 fused in frame to the C terminus of the GAL4 DNA-binding domain (GAL4DBD) or the GAL4 acidic transactivating domain (GAL4AD), were expressed in Y190 yeast clones, and were assayed for GAL4DBD and GAL4AD-dependent β-galactosidase expression by clonal 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) conversion. Clones with GAL4DBD-EBNALPd45 and GAL4AD-EBNALPd45 were dark blue after 1-h incubation, whereas GAL4DBD-EBNALP and GAL4AD-EBNALP transfected yeast were only light blue after overnight incubation and clones transfected with GAL4DBD-EBNALPd45 and GAL4AD-EBNALP or GAL4DBD-EBNALP and GAL4AD-EBNALPd45 were dark blue after overnight incubation. Thus, EBNALP and EBNALPd45 self-associate or associate with each other in yeast, and EBNALPd45 self-association is particularly strong.
EBNA2 and EBNALP Bind to EBNALP W2Y1 or dW2Y1Y2d10. To characterize the interaction between EBNA2 or EBNALP and EBNALP, parts of EBNALP were expressed in E. coli as GST fusion proteins (Fig. 1 A) and used to pull down interacting proteins from lysates of LCLs or of BJAB cells that express EBNA2 or EBNALP (Fig. 1 D and E). EBNA2 and EBNALP were the only EBNAs detected when immunoblots were probed with human serum reactive against EBNA proteins or with α-EBNA2 or with α-EBNALP antibody (Fig. 1 D and E and data not shown).
EBNA2 from lymphoblasts did not bind to GST–W2Y1Y2 or -dW2Y1Y2 (Fig. 1 D and E and data not shown). Binding to GLP or GLPd45 was barely above background, whereas binding to GST–dW2Y1d10 or –dW2Y1 was usually substantially above background and to GST–W2Y1 or –W2Y1Y2d10 was at least 10% (Fig. 1 D and E). These in vitro binding data map strong lymphoblast EBNA2 binding to the 55 amino acids of EBNALP W2Y1. Further, deletion of the EBNALP C-terminal 10 aa from W2Y1Y2 substantially enhanced lymphoblast EBNA2 binding to EBNALP, reminiscent of the effect of d10 on EBNA2 association with EBNALP in lymphoblasts.
In contrast to the dissociation of EBNA2 from EBNALP in lymphoblasts, HE2 from E. coli stably associated at a >5% level with GLP from E. coli (Fig. 1F). HE2 also bound well to GST–W2Y1Y2, –W2Y1Y2d10, –W2Y1, and –dW2Y1Y2d10, but not to GST–dW2Y1Y2. Overall, the data in Fig. 1 indicate that lymphoblast proteins or posttranslational modifications down-modulate the intrinsic affinity of EBNA2 for full-length EBNALP.
Consistent with the high-level association of EBNALP or EBNALPd45 in B lymphoblasts or yeast, EBNALP from LCL or transfected BJAB lymphoblast lysates bound at ≈5% level to GLP or GLPd45, and at similar or higher levels to GST-W2Y1d10, -W2Y1, -dW2Y1Y2, or -dW2Y1Y2d10 (Fig. 1 D and E, and data not shown). Thus, EBNALP in LCL or other lymphoblast extracts bound specifically and substantially to EBNALP fusion proteins from E. coli, including GST-W2Y1, which also bound EBNA2 and dW2Y1Y2, which did not bind EBNA2.
EBNALP dW2 Binds EBNA2 or EBNALP with Different Specificities. The high-level binding of EBNA2 or EBNALP to W2Y1 was further investigated by using GST-W2, -dW2, and mutant -dW2 fusion proteins purified from E. coli (Fig. 2). HE2 and a nearly full-length HE2 degradation product bound to GST-W2, -dW2, and most -dW2 fusion proteins with ≈1% efficiency and did not bind to GST, even when GST was in severalfold excess (Fig. 2B). HE2 bound slightly less to GSTdW2 RRR/AAA or SGS/AAA (mutants 5 and 8 in Fig. 2 A and B). Surprisingly, EBNALP bound particularly well to GST-dW2 SGS/AAA and not at all to GST-dW2 LVQ/AAA or -dW2 EEE/AAA (mutants 8, 6, and 7 in Fig. 2 A and C). These results indicate that EBNA2 and EBNALP can both bind specifically to dW2 and that EBNALP differs from EBNA2 in not binding to dW2 LVQ/AAA or dW2 EEE/AAA.
EBNALP Binds to E2AD. PE2 antibody precipitation of EBNA2 without coprecipitation of EBNALPd10 from lymphoblast lysates despite high-level FLPd10 association with EBNA2 (Fig. 1B) identified the E2AD as a candidate site for EBNALP association. To evaluate this hypothesis, EBNALPd10 binding to GST–E2AD (GE2AD) was assessed. GE2AD specifically pulled down 3–5% of FLPd10 from transfected BJAB lysates and variably pulled down a trace of FLP or FLPD45 (Fig. 3A). Thus, the d10 mutation markedly increased EBNALP binding to the E2AD, much as d10 increased EBNALP association with EBNA2.
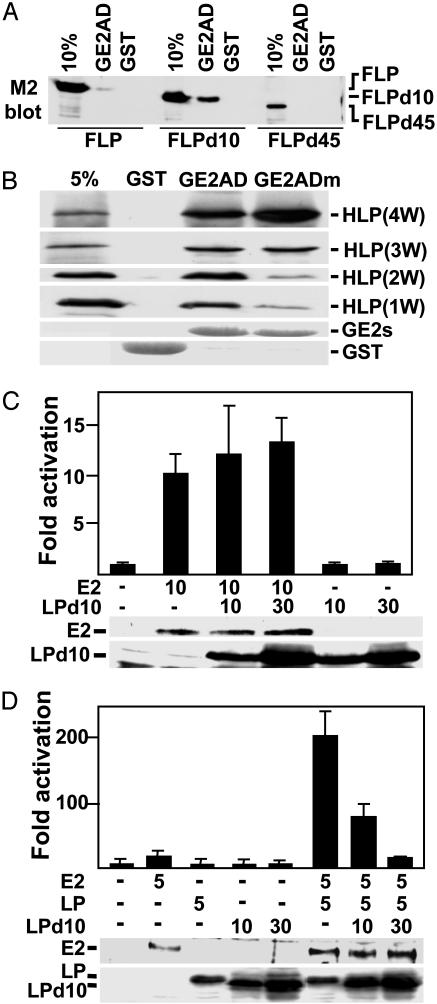
Fig. 3.
EBNALP expressed in E. coli and EBNALPd10 bind to the E2AD; EBNALP coactivates with EBNA2; and EBNALPd10 inhibits coactivation. (A) Lysates of BJAB cells 48 h after transfection with FLP, FLPd10, or FLPd45 expression vectors were incubated with GST or GST–E2AD (GE2AD). Proteins bound to the GST beads were eluted and compared with 10% of the input by SDS/PAGE and immunoblot using M2 antibody. (B) The binding of HLP(4W), which is His-tagged EBNALP or of HLP(4W)-deleted for 1, 2, or 3 W1W2 repeats, HLP(3W), HLP(2W), or HLP(1W) to GST, GE2AD, or the transcriptional null mutant GE2ADW454T (GE2ADm) was compared with 5% of the input by SDS/PAGE and immunoblot with JF186 antibody. (C) BJAB cells were transfected with 10 μg of EBNA2 expression plasmid (E2), 5 μg of LMP1 promoter–luciferase reporter plasmid, 0 or 30 μg of EBNALPd10 expression plasmid, an appropriate amount of control expression vector DNA, and 5 μg of control GK-β-galactosidase expression plasmid. (D) BJAB cells were transfected with 5 μg of EBNA2 (E2) expression plasmid, 5 μg of LMP1 promoter–luciferase reporter plasmid, 5 μg of EBNALP expression plasmid, 10 or 30 μg of EBNALP10 expression plasmid, an appropriate amount of control expression plasmid DNA, and 5 μg of control GK-β-galactosidase expression plasmid. In C and D, immunoblots for EBNA2, EBNALP, or EBNALPd10 are shown below the activation data. Luciferase activities were corrected for β-galactosidase activity.
His-tagged WT EBNALP, which has four W1W2 repeats (HLP4W) and was purified from E. coli, bound specifically at a 10–20% level to GE2AD, despite the absence of the d10 mutation, as was characteristic of HE2 binding to GLP. Although highly specific for GE2AD versus GST, HLP4W bound slightly better to the null point mutant GE2ADm (E2ADW454T; ref. 18) than to GE2AD (Fig. 3B). Deletion of one, two, or three W1W2 repeats from HLP4W progressively lessened binding to GE2ADm, whereas binding to GE2AD remained relatively constant; HLP2W and HLP1W bound GE2AD preferentially over GE2ADm. Thus, HLP bound well to GE2AD, just as HE2 bound well to GLP (Fig. 1E), indicating that unmodified EBNALP readily associates with E2AD or EBNA2. These data support the hypothesis that EBNA2 in lymphoblasts is not stably association with EBNALP because of associated cell proteins or posttranslational modification(s) of EBNALP or EBNA2 that regulate EBNA2 and EBNALP association (20, 21, 23).
EBNALPd10 Has a Dominant Negative Effect on EBNALP Coactivation. Because EBNALPd10 stably associated with EBNA2 in lymphoblasts, without coactivating transcription, we evaluated whether EBNALPd10 could dominantly inhibit EBNA2 activation or EBNALP coactivation. In multiple experiments, EBNA2 activated the LMP1 promoter in BJAB cells ≈10-fold and increasing amounts of EBNALPd10 had no effect (Fig. 3C). However, in a smaller number of experiments, EBNA2 activated the LMP1 promoter only 2- to 6-fold and increasing amounts of EBNALPd10 inhibited EBNA2 as much as 50% (data not shown). Without EBNA2, EBNALPd10 did not affect LMP1 promoter activity (Fig. 3C and multiple experiments not shown).
In multiple experiments, EBNALP coactivated the LMP1 promoter with EBNA2 10- to 20-fold, resulting in as much as 200-fold increased reporter (Fig. 3D). Cotransfection with two to six times as much EBNALPd10 as EBNALP expression vector resulted in 2- to 6-fold higher EBNALPd10 expression and dose-dependent complete inhibition of EBNALP coactivation (Fig. 3D). Expression of EBNALP at similar levels did not inhibit coactivation (data not shown). EBNALPd34 neither coactivated nor inhibited (ref. 27 and data not shown). Thus, EBNALPd10 has a strong dominant negative effect on EBNALP coactivation with EBNA2 and a variable partial effect on EBNA2 activation. The strong dominant negative effect on EBNALP coactivation could be useful for investigating the role of EBNALP in maintaining LCL growth.
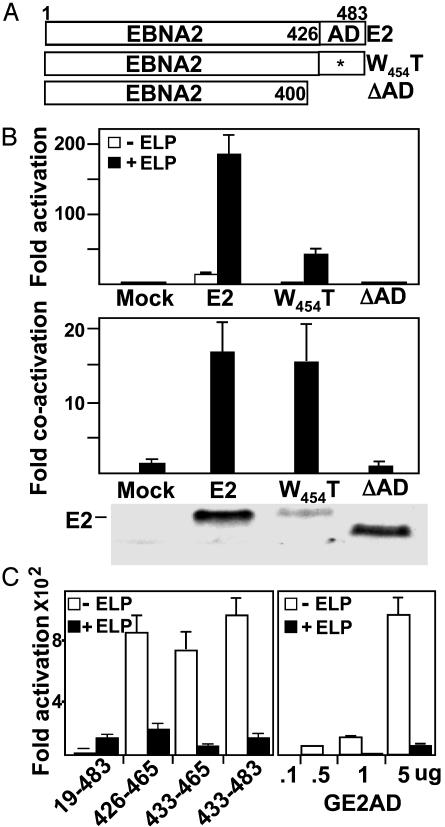
EBNALP Coactivation with EBNA2 Requires the E2AD, but EBNALP Cannot Coactivate the Acidic Domain Alone Fused to the Gal4 DNA-Binding Domain. The direct association EBNALP with the E2AD led us to investigate the role of the E2AD in promoter activation and EBNALP coactivation. Whereas EBNA2 activated the LMP1 promoter ≈10-fold and EBNALP further coactivated ≈17-fold, giving ≈170-fold total activation, EBNA2 deleted for the acidic domain did not activate transcription and was not coactivated by EBNALP (Fig. 4 A and B). EBNA2 with the nearly null E2AD W454T point mutation (18) activated transcription only ≈1- to 2-fold, but was surprisingly almost 15-fold coactivated by EBNALP (Fig. 4 A and B). Thus, EBNA2 activation is highly dependent on E2AD W454, but EBNALP coactivation is largely independent of E2AD W454, similar to the binding of HLP to the E2AD in Fig. 3B.
Fig. 4.
E2AD is critical for EBNA2 activation and for LP coactivation. (A) Schematic diagram of WT and mutant EBNA2, indicating the transactivation domain (amino acid 426–465; ref. 19), defective acidic domain point mutant, EBNA2W454T, and EBNA2 with a stop codon after codon 400, E2ΔAD. (B) BJAB cells were transfected with 10 μg of LMP1 promoter–luciferase reporter plasmid, 5 μg of WT or mutant EBNA2, 10 μg of EBNALP, or an appropriate amount of control expression vector DNA and 5 μg of GK-β-galactosidase. Fold EBNA2 activation and EBNALP coactivation are shown. An EBV-immune human serum was used to detect WT and mutant EBNA2. (C)(Left) BJAB cells were transfected with 5 μg of GAL4DBD-E2AD expression vector and 5 μgof control pSG5 DNA or pSG5-ELP. (Right) BJAB cells were transfected with increasing amounts of GALDBD–E2AD expression vector and 5 μg of pSG5 DNA or pSG5-ELP DNA. (B–C) Luciferase activities were corrected for cotransfected β-galactosidase activity.
To further investigate the E2AD sequence requirements for EBNALP coactivation, GAL4DBD-E2AD amino acids 433–465 was compared with GAL4DBD amino acids 426–465, GAL4DBD amino acids 433–483, and GAL4DBD-EBNA2 amino acids 19–483 in activation of a 5× GAL4 DNA-binding-site-dependent promoter and in coactivation by EBNALP (19, 27). In contrast to our previous report (27), GAL4-E2AD amino acids 426–465, 433–465, or 433–483 strongly activated transcription and EBNALP or EBNALPd10 repressed GAL4E2AD enhanced transcription (Fig. 4C Left and data not shown). This was not an artifact of maximal promoter activation by GAL4E2AD, because transfection with much less expression vector resulted in proportionately less activation and EBNALP inhibited even low levels of activation (Fig. 4C Right and data not shown). The DNA sequence of GAL4DBD-E2AD amino acids 426–462 from the previous study (27) was that of GAL4DBD-E2 amino acids 19–483 (19). GAL4-E2 amino acids 19–483 had ≈1% of GAL4E2AD transcriptional activity (ref. 19 and Fig. 4C) and was coactivated by EBNALP to ≈20% that of E2AD (Fig. 4C). Thus, EBNALP coactivates with the E2AD only in the context of other EBNA2 domains.
Discussion
These experiments use genetic and biochemical approaches to identify domains of EBNALP and EBNA2 that mediate association and cooperative transcriptional regulation. EBNA2 from lymphoblasts and E. coli bound well to the 55 amino acids of EBNALP W2Y1, and EBNA2 from E. coli also bound well to the 55 amino acids of EBNALP dW2Y1Y2d10. EBNALP W2Y1 and dW2Y1Y2d10 both have dW2, which specifically bound EBNA2 at a lower level. Interestingly, EBNA2 binding to dW2 was not substantially increased by adding 22 N-terminal W2 amino acids and was inhibited by adding 11 C-terminal Y1 amino acids. The addition of both sequences or of 24 C-terminal Y2 amino acids enabled high-level EBNA2 binding; interactions among components was required. In the context of the complete EBNALP C terminus, deletion of the C-terminal 10 amino acids was necessary to observe high-level lymphoblast EBNA2 association. These last 10 amino acids are VYIEEEEDED. These acidic residues may directly or indirectly inhibit E2AD interactions with EBNALP dW2 RRVRRR, which has been implicated in EBNA2 coactivation (31, 35). The specific importance of the EBNALP VYIEEEEDED deletion for lymphoblast as opposed to E. coli derived EBNA2 binding strongly suggests that EBNA2 interaction with EBNALP is regulated by posttranslational modifications or cell protein interactions that are specific to eukaryotic cells.
EBNALP coactivation with EBNA2 required the E2AD. Furthermore, the EBNA2 null point mutant in E2AD, EBNA2W454T, was coactivated by EBNALP, consistent with the minimal affect of EBNA2W454T on EBNALP association. EBNALP coactivation with EBNA2 was also linked to the E2AD by the EBNALP d10 mutation, which stabilized E2AD association and abrogated coactivation. A requirement for unstable EBNALP interaction with the E2AD is not surprising, given the role of the E2AD in recruiting p300/CBP and other transcription factors to EBNA2-regulated promoters. In contrast to cellular transcription factors, EBNALP bound more strongly to the transcriptional null point mutant E2ADW454T than to WT E2AD. EBNALP binding to the E2AD with a different specificity than cellular transcription factors may enable EBNALP and cellular transcription factors to interact nearly simultaneously with the E2AD.
These data support a model in which EBNALP coactivation with the E2AD is mediated by direct transient association. Unstable interaction with the E2AD, likely enables EBNALP to recruit transcriptional enhancing or positive regulatory factors to the E2AD at EBNA2-specified promoters. Interestingly, EBNALP coactivation with the E2AD required more of EBNA2 than promoter localization because EBNALP did not coactivate with GE2AD. Current experiments are delineating the essential components of EBNA2 for EBNALP coactivation; the E2AD is the only EBNA2 high-affinity EBNALP-binding site.
The EBNALP dW2 domain also enables homotypic EBNALP association. Mutation of dW2 RRVRRR to AAVAAA abrogated intermolecular interaction in yeast (35) and mutation of dW2 LVQ to AAA or of EEE to AAA abrogated EBNALP binding without affecting EBNA2 binding (Fig. 2). In lymphoblasts and directed yeast two-hybrid assays, EBNALP or EBNALPd45 homo-typically associated, as was observed for EBNALP with fewer repeats (35). Thus, EBNALP is a self-assembling scaffold for integrating factors with EBNA2. Overall, these EBNALP and EBNA2 intermolecular interactions are complex and potentially subject to inhibition. An inhibitor of coactivation would likely abrogate LCL proliferation or survival.
Acknowledgments
Ellen Cahir-McFarland contributed helpful suggestions. This research was supported by National Cancer Institute/U.S. Public Health Service Grant CA47006 and by Japan Ministry of Science and Technology Grant 15019117.
Abbreviations: EBV, Epstein–Barr virus; EBNALP, EBV nuclear leader protein; LMP1, latent membrane protein 1; LCL, lymphoblastoid cell line; E2AD, EBNA2 acidic activating domain; GLP, GST-EBNALP; HE2, His-tagged EBNA2.
References
- 1.Kieff, E. & Rickinson, A. B. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott, Williams & Wilkins, Philadelphia), Vol. 2, pp. 2511–2574. [Google Scholar]
- 2.Rickinson, A. B. & Kieff, E. (2001) in Fields Virology, ed. Knipe, D. & Howley, P. M. (Lippincott, Williams & Wilkins, Philadelphia), Vol. 2, pp. 2575–2628. [Google Scholar]
- 3.Hurley, E. A. & Thorley-Lawson, D. A. (1988) J. Exp. Med. 168, 2059–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rooney, C., Howe, J. G., Speck, S. H. & Miller, G. (1989) J. Virol. 63, 1531–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alfieri, C., Birkenbach, M. & Kieff, E. (1991) Virology 181, 595–608. [DOI] [PubMed] [Google Scholar]
- 6.Hammerschmidt, W. & Sugden, B. (1989) Nature 340, 393–397. [DOI] [PubMed] [Google Scholar]
- 7.Cohen, J. I., Wang, F., Mannick, J. & Kieff, E. (1989) Proc. Natl. Acad. Sci. USA 86, 9558–9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen, J. I., Wang, F. & Kieff, E. (1991) J. Virol. 65, 2545–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mannick, J. B., Cohen, J. I., Birkenbach, M., Marchini, A. & Kieff, E. (1991) J. Virol. 65, 6826–6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henkel, T., Ling, P. D., Hayward, S. D. & Peterson, M. G. (1994) Science 265, 92–95. [DOI] [PubMed] [Google Scholar]
- 11.Grossman, S. R., Johannsen, E., Tong, X., Yalamanchili, R. & Kieff, E. (1994) Proc. Natl. Acad. Sci. USA 91, 7568–7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ling, P. D., Rawlins, D. R. & Hayward, S. D. (1993) Proc. Natl. Acad. Sci. USA 90, 9237–9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ling, P. D., Hsieh, J. J., Ruf, I. K., Rawlins, D. R. & Hayward, S. D. (1994) J. Virol. 68, 5375–5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laux, G., Adam, B., Strobl, L. J. & Moreau-Gachelin, F. (1994) EMBO J. 13, 5624–5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johannsen, E., Koh, E., Mosialos, G., Tong, X., Kieff, E. & Grossman, S. R. (1995) J. Virol. 69, 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zimber-Strobl, U., Strobl, L. J., Meitinger, C., Hinrichs, R., Sakai, T., Furukawa, T., Honjo, T. & Bornkamm, G. W. (1994) EMBO J. 13, 4973–4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuentes-Panana, E. M., Peng, R., Brewer, G., Tan, J. & Ling, P. D. (2000) J. Virol. 74, 8166–8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen, J. I. (1992) Proc. Natl. Acad. Sci. USA 89, 8030–8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen, J. I. & Kieff, E. (1991) J. Virol. 65, 5880–5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tong, X., Drapkin, R., Reinberg, D. & Kieff, E. (1995) Proc. Natl. Acad. Sci. USA 92, 3259–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tong, X., Drapkin, R., Yalamanchili, R., Mosialos, G. & Kieff, E. (1995) Mol. Cell. Biol. 15, 4735–4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tong, X., Wang, F., Thut, C. J. & Kieff, E. (1995) J. Virol. 69, 585–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang, L., Grossman, S. R. & Kieff, E. (2000) Proc. Natl. Acad. Sci. USA 97, 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang, F., Tsang, S. F., Kurilla, M. G., Cohen, J. I. & Kieff, E. (1990) J. Virol. 64, 3407–3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang, F., Gregory, C., Sample, C., Rowe, M., Liebowitz, D., Murray, R., Rickinson, A. & Kieff, E. (1990) J. Virol. 64, 2309–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abbot, S. D., Rowe, M., Cadwallader, K., Ricksten, A., Gordon, J., Wang, F., Rymo, L. & Rickinson, A. B. (1990) J. Virol. 64, 2126–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harada, S. & Kieff, E. (1997) J. Virol. 71, 6611–6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nitsche, F., Bell, A. & Rickinson, A. (1997) J. Virol. 71, 6619–6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, F., Petti, L., Braun, D., Seung, S. & Kieff, E. (1987) J. Virol. 61, 945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sample, J., Hummel, M., Braun, D., Birkenbach, M. & Kieff, E. (1986) Proc. Natl. Acad. Sci. USA 83, 5096–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng, R., Tan, J. & Ling, P. D. (2000) J. Virol. 74, 9953–9963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yokoyama, A., Tanaka, M., Matsuda, G., Kato, K., Kanamori, M., Kawasaki, H., Hirano, H., Kitabayashi, I., Ohki, M., Hirai, K. & Kawaguchi, Y. (2001) J. Virol. 75, 5119–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kitay, M. K. & Rowe, D. T. (1996) J. Virol. 70, 7885–7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petti, L., Sample, C. & Kieff, E. (1990) Virology 176, 563–574. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka, M., Yokoyama, A., Igarashi, M., Matsuda, G., Kato, K., Kanamori, M., Hirai, K., Kawaguchi, Y. & Yamanashi, Y. (2002) J. Virol. 76, 1025–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mannick, J. B., Tong, X., Hemnes, A. & Kieff, E. (1995) J. Virol. 69, 8169–8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kitay, M. K. & Rowe, D. T. (1996) Virology 220, 91–99. [DOI] [PubMed] [Google Scholar]
- 38.Kawaguchi, Y., Nakajima, K., Igarashi, M., Morita, T., Tanaka, M., Suzuki, M., Yokoyama, A., Matsuda, G., Kato, K., Kanamori, M. & Hirai, K. (2000) J. Virol. 74, 10104–10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han, I., Harada, S., Weaver, D., Xue, Y., Lane, W., Orstavik, S., Skalhegg, B. & Kieff, E. (2001) J. Virol. 75, 2475–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han, I., Xue, Y., Harada, S., Orstavik, S., Skalhegg, B. & Kieff, E. (2002) Mol. Cell. Biol. 22, 2136–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Igarashi, M., Kawaguchi, Y., Hirai, K. & Mizuno, F. (2003) J. Gen. Virol. 84, 319–327. [DOI] [PubMed] [Google Scholar]
- 42.Sadowski, I. & Ptashne, M. (1989) Nucleic Acids Res. 17, 7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.King, W., Thomas-Powell, A. L., Raab-Traub, N., Hawke, M. & Kieff, E. (1980) J. Virol. 36, 506–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Menezes, J., Leibold, W., Klein, G. & Clements, G. (1975) Biomedicine 22, 276–284. [PubMed] [Google Scholar]
- 45.Lin, J., Johannsen, E., Robertson, E. & Kieff, E. (2002) J. Virol. 76, 232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Finke, J., Rowe, M., Kallin, B., Ernberg, I., Rosen, A., Dillner, J. & Klein, G. (1987) J. Virol. 61, 3870–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young, L., Alfieri, C., Hennessy, K., Evans, H., O'Hara, C., Anderson, K. C., Ritz, J., Shapiro, R. S., Rickinson, A., Kieff, E., et al. (1989) N. Engl. J. Med. 321, 1080–1085. [DOI] [PubMed] [Google Scholar]