Abstract
B cell chronic lymphocytic leukemia (B-CLL) is characterized by a highly variable clinical course. Recurrent chromosomal imbalances provide significant prognostic markers. Risk-adapted therapy based on genomic alterations has become an option that is currently being tested in clinical trials. To supply a robust tool for such large scale studies, we developed a comprehensive DNA microarray dedicated to the automated analysis of recurrent genomic imbalances in B-CLL by array-based comparative genomic hybridization (matrix–CGH). Validation of this chip in a series of 106 B-CLL cases revealed a high specificity and sensitivity that fulfils the criteria for application in clinical oncology. This chip is immediately applicable within clinical B-CLL treatment trials that evaluate whether B-CLL cases with distinct chromosomal abnormalities should be treated with chemotherapy of different intensities and/or stem cell transplantation. Through the control set of DNA fragments equally distributed over the genome, recurrent genomic imbalances were discovered: trisomy of chromosome 19 and gain of the MYCN oncogene correlating with an elevation of MYCN mRNA expression.
Chronic lymphocytic leukemia of B cell type (B-CLL) is characterized by a highly variable clinical course reaching from rapid progression with fatal outcome to normal life expectancy (1). Characteristic chromosomal imbalances were shown to provide highly significant prognostic markers (2). In particular, patients with deletions within chromosome arms 17p or 11q show advanced disease, significantly shorter treatment free interval, and shorter overall survival times (2). More recently, hypermutation of the variable region of the Ig heavy chain gene (IGV), which is considered as a marker for B cell transit through the germinal centers of the lymph node, was associated with a better prognosis (3–5). The majority of cases carrying loss on 17p or 11q show no IGV hypermutation, however, the IGV mutation status does not identify all patients with a cytogenetic high risk profile (5). Risk-adapted therapy based on such molecular findings has become an option and is currently being evaluated in clinical trials.
High-throughput analysis of genomic alterations for risk-adapted patient stratification and monitoring within treatment trials relies on efficient and automated diagnostic techniques. The potential of array-based comparative genomic hybridization (matrix–CGH) (6) for high resolution screening of small DNA copy number changes was demonstrated (6–8). However, although genomic DNA arrays are considered powerful research tools, their potential to meet specific needs in clinical diagnostics have been intensively discussed. Initially, matrix–CGH has focused on the analysis of cell lines or the investigation of inherited diseases, both characterized by a genetically homogeneous cell population (6–12). More recently, it was also used in research studies addressing issues of tumor classification and correlation with array expression studies (13–16). To obtain a robust tool for large scale clinical diagnostics in B-CLL, we developed a DNA microarray dedicated to the automated analysis of recurrent genomic imbalances in B-CLL by custom-made matrix–CGH hybridization. This chip was validated in a series of 106 B-CLL patients in a blinded fashion comparing microarray profiles to interphase cytogenetic data obtained by fluorescence in situ hybridization (FISH). Surprisingly, previously uncharacterized recurrent genomic imbalances were identified, which were further assessed by means of FISH, chromosomal CGH, or quantitative RNA analysis.
Methods
Patients. Peripheral blood samples, bone marrow aspirates, or lymph node specimens from 106 patients with B-CLL and one patient with a leukemic mantle cell lymphoma (MCL) were obtained after informed consent. The diagnosis was confirmed by standardized clinical, morphological, and immunological criteria. All patient samples were evaluated with a well established FISH probe set for chromosomal regions 3q26, 6q21, 8q24, 11q22, 12q13, 13q14, 17p13, and 18q21, according to standard protocols (2, 17). According to the FISH data, the proportion of tumor cells bearing clonal genomic aberrations was in the range of 7–98% (median, 70.5%; interquartile range, 45–83%). A subset of 20 B-CLL cases was analyzed by CGH to metaphase chromosomes as described elsewhere (18).
DNA Preparation and Spotting. Bacterial artificial chromosome (BAC) and P1-derived artificial chromosome (PAC) clones, which were selected according to their chromosomal localization or their gene content, were identified from the Entrez genome database (www.ncbi.nlm.nih.gov/mapview/map_search.cgi?chrhum_chr.inf&query) or by blast search (www.ncbi.nlm.nih.gov/blast) against finished and unfinished genome sequence data and ordered from the libraries RPCIB753 (same as RP11) and RPCIP704 (same as RP1;3;4;5) (Resource Centre/Primary Database of the German Human Genome Project, Berlin). Furthermore, a set of gene-specific BAC/PAC clones was identified by filter hybridizations of these libraries using cDNA probes. The identity of BACs, which contain the MYCN gene and revealed copy number gain in five B-CLL cases as well as the MCL case, was verified by gene-specific PCR. After DNA isolation according to standard protocols (Qiagen, Hilden, Germany), DNAs from all clones were sonicated to fragments of 500–5,000 bp in size. Sized DNA was spotted at a concentration of 0.75 μg/μl in 3× SSC in replicas of eight onto Corning CMT-Gaps II glass slides by using the Omnigrid microarrayer (Gene Machines, San Carlos, CA). The slides were baked at 80°C for 10 min, and DNA fragments were crosslinked by UV light (254 nm per 2,400 μJ). Finally, the slides were stored at room temperature.
Matrix–CGH. Peripheral blood mononuclear cells were isolated by Ficoll–Hypaque gradient centrifugation, and DNA was isolated by using standard protocols including alkaline lysis and affinity chromatography or TRIzol reagent. Tumor and reference DNA were fluorescently labeled via incorporation of Cy3- or Cy5-conjugated dCTP by either random priming or, in cases were only minimal DNA amounts were available, by linker–adaptor PCR amplification as described (11, 19). Unincorporated nucleotides were removed through Microcon membrane column centrifugation. Labeling efficiency was assessed by photometric measurements. Specimens with low nucleotide incorporation rates were excluded from the study (n = 8). For hybridization, labeled reference DNA (a 1:1 mixture of normal human male and female DNA) and differentially labeled tumor DNA were coprecipitated together with 70 μg of human Cot-1 DNA and resolved in 120 μl of prewarmed Ultrahyb buffer (Ambion, Austin, TX) at 37°C for 30 min. After denaturation (75°C for 10 min) and preannealing (60 min at 37°C), hybridization was allowed for 36 h at 37°C by using a GeneTac hybridization chamber (Genomic Solutions, Cambridgeshire, U.K.). Slides were washed automatically three times for 20 s “flow,” 3 min “hold” in 50% formamide, 2× SSC, and 0.1% Tween 20, pH 7.0 at 45°C in the hybridization chamber followed by additional washing for 2 min in 1× PBS, 0.05% Tween 20 at 25°C. Finally, slides were dried by spinning for 5 min at 2,500 rpm in a clinical centrifuge. Images of fluorescence signals were acquired by a dual laser scanner (GenePix 4000 A, Axon Instruments, Foster City, CA). Assessment of fluorescence signal intensities was achieved by using genepix pro 4.0 imaging software. For final analysis, raw data of fluorescence ratios of two independent hybridization experiments were calculated and averaged applying dedicated matrix–CGH software, developed previously by our group (11).
Fluorescence ratios were normalized by using the median of the fluorescence ratios computed as log2 values from the 211 DNA control fragments spanning the whole genome. The diagnostic cutoff level was determined for each individual experiment, i.e., after averaging the ratios from the two color-switch hybridizations and subsequent normalization, a set of “balanced” DNA fragments was used to calculate the mean and standard deviations to define the cutoff level as mean plus/minus three times the standard deviation. For the present study with a chip dedicated to diagnostics in B-CLL, the balanced control set consisted of those of the 644 DNA fragments, which do not localize on a chromosome involved in recurrent B-CLL aberrations; i.e., DNAs of the following chromosomes were excluded: 3, 6, 8, 10, 11, 12, 13, 14, 17, 18. Although chromosome 19 was previously not described as a frequently aberrant region in B-CLL, we excluded this chromosome from the balanced set because, in the present study, we identified trisomy affecting this chromosome. The final balanced set of clones amounted to n = 198.
Increasing the redundancy of targeted DNA fragments per genomic imbalance resulted in superior diagnostic performance. Each genomic region recurrently altered in B-CLL was covered by 9–33 nonoverlapping DNA fragments. With one exception, an imbalance was scored if ≥50% of fragments within the aberrant region exhibited ratio values beyond the cutoff level. Because a deletion within the chromosomal band 13q14 may be as small as 100 kb (20), this region was covered by a set of overlapping PACs, and deletions were scored even when only the ratio value for PAC RP1–250J10, representing the minimally affected critical region, indicated a deletion (20). Eighteen clones were excluded from the matrix–CGH analysis due to incorrect FISH localization (a list containing these clones is available in Table 3, which is published as supporting information on the PNAS web site).
Real-Time Quantitative PCR. Real-time quantitative RT-PCR for MYCN was performed as described (21). After RNA extraction using TRIzol reagent and subsequent DNase 1 digestion, cDNA was synthesized with random hexamer primers by using the GeneAmp kit (RNA PCR System, Applied Biosystems). Quantitative assessment of cDNA amounts was detected with SYBR green dye according to the manufacturer's manuals (Applied Biosystems). The following primers were used for amplification: forward, 5′-CTGAGCGATTCAGATGATGAAGAT-3′; reverse, 5′-ATGTGGTGACAGCCTTGGTG-3′. Serially diluted cDNA from the lymphoma cell line Jurkat served for calibration. For normalization, mean cDNA expression levels of the housekeeping genes HPRT and Lamin B were used (21).
Results
Compilation of B-CLL Chip. We here present a disease-specific genomic DNA microarray consisting of a total of 644 immobilized DNA fragments, which was developed and optimized to meet the specific needs of clinical diagnostics in B-CLL (Fig. 1). For genomic regions recurrently imbalanced in B-CLL, namely 3q, 6q21-q27, 8q24, 10q24, 11q22.3-q23.1, 12q13-q15, 13q14, 17p13, and 18q21, sets of 9–33 nonoverlapping DNA fragments (total = 198) were selected, which had been cloned in PAC or BAC vectors. This compilation was extended by a set of DNA fragments, which were mapped to critical regions of other B cell neoplasms by means of cytogenetics (n = 50), e.g., 2p13, 9p24, or 11q13. In addition, 123 clones were included that represent genes that play a putative pathogenic role in B cell non-Hodgkin lymphomas as well as known proto-oncogenes and tumor suppressor genes. For normalization purposes, 211 BAC clones were isolated, which are linearly distributed across the whole human genome with an average genomic distance of ≈15 Mbp. The detailed list of BACs and PACs is available in Table 4, which is published as supporting information on the PNAS web site. This chip was used for genomic profiling in 106 B-CLLs as well as one MCL. One example is shown in Fig. 2 a and b.
Fig. 1.
A total of 644 BACs/PACs were selected to generate the B-CLL diagnostic microarray. Of these, 211 DNA fragments, equally distributed across the genome, were used for normalization purposes: 198 BACs/PACs covering regions recurrently altered in B-CLL with a density of 9–33 nonoverlapping BACs/PACs per region (an overlapping contig for 13q14), and 173 DNA fragments mapping within critical regions of other B cell neoplasias or containing coding information for genes with assumed pathogenetic relevance in lymphomas.
Fig. 2.
(a) Image of a B-CLL chip after hybridization with DNA derived from a patient carrying a 13q14 deletion (labeled in green; Cy3) versus human control DNA (labeled in red; Cy5). (Inset) PAC clone of the 13q14 contig with a dominantly red fluorescence signal after hybridization, indicating the deletion within this region (see arrowhead). The array size is 24 mm × 19 mm containing 644 BACs or PACs spotted in replicas of eight. (b) Example of a profile of signal ratios obtained for all immobilized DNA fragments arranged in ascending order beginning with 1p and ending with the X and Y chromosome. The cluster of fragments detecting deletion within 13q14 is indicated. (c) Assessment of the diagnostic power of the B-CLL chip by comparison with data obtained through targeted FISH. In the analyzed series of 107 patients displaying a total of 27 gains and 95 losses, all recurrently imbalanced regions were correctly identified, if the proportion of cells carrying the respective gains or losses was >53% (determined by FISH, 68 patients), and 77% of the gains as well as 81% of the losses were scored by matrix–CGH if the aberration was present in <53% but >33% (determined by FISH, 19 patients).
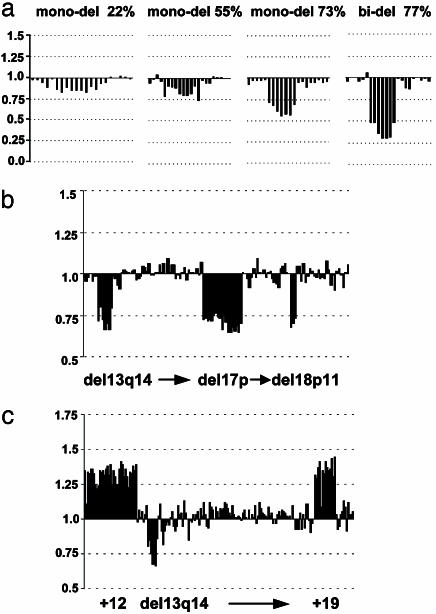
Assessment of the Sensitivity of the B-CLL Chip. The B-CLL chip was validated in comparison to interphase cytogenetic data in a blinded fashion. For all cases, the known recurrent genomic changes characteristic for B-CLL were assessed by FISH: in the series of 107 patients, 27 chromosomal gains and 95 losses were detected. When matrix–CGH was used, a total of 68,908 data points, equivalent to single FISH experiments, were analyzed in this study (a detailed list containing all ratios and individual thresholds for all 107 tumors is available in Table 5, which is published as supporting information on the PNAS web site). Less than 7% of these matrix–CGH data points were excluded from the analysis because of low intensity-to-background ratios, high standard deviations of the replicas, or obvious fluorescent artifacts over the spots. Comparison of matrix–CGH and FISH data revealed an overall sensitivity of the chip of 100% for B-CLL samples with clonal genomic aberrations in >53% of mononuclear cells (determined by FISH, n = 68). This result is illustrated in Fig. 2c. With a decrease of the proportion of aberrant cells, the detection sensitivity was reduced. When this proportion was between 33% and 53%, seven of nine small copy number gains and 9 of 11 losses were detected by using matrix–CGH (Fig. 2c). The validity of the signal ratios allowed reliable differentiation between mono- and biallelic deletions, and genomic imbalances were detectable even when the percentage of cells with clonal aberrations was ≈25% (Fig. 3a). Eighteen tumor samples revealed no aberrations in FISH experiments and were also scored as balanced for the B-CLL relevant chromosomal regions by using array-based CGH.
Fig. 3.
(a) Relationship between tumor cell content and signal ratio. Examples are shown for deletions within 13q14 with various proportions of aberrant cells (indicated). The high validity of the ratio values allows a reliable differentiation between mono- and biallelic deletions. Note that even a genomic imbalance present in <25% of the aberrant cells was scored correctly. (b and c) Previously unknown imbalances in B-CLL detected by matrix–CGH comprise one case with loss on 18p11 (b) and a recurrent trisomy of chromosome 19 (5 of 106 B-CLLs), which coincided in all instances with trisomy 12 (c). Clones are arranged in ascending order; see Table 1 for details.
As compared to chromosomal CGH, the spatial resolution of matrix–CGH is superior, allowing the detection of much smaller gained or lost chromosomal regions: based on the small size of the imbalanced regions, only 20 of 28 deletions or gains identified by matrix–CGH were also detectable by chromosomal CGH (data not shown).
Assessment of the Specificity of the B-CLL Chip. A critical region did not score falsely positive in any of the cases. Of the 68,908 matrix–CGH data points, 0.6% revealed 39 gains and losses (in 25 samples) when applying the thresholds derived from intrachip variability (examples are shown in Fig. 3 b and c). Even if all of these additional aberrations were considered as “false positive,” a specificity of >99% is achieved. However, for a subset of patients, sufficient cell material or DNA was available, allowing assessment of the aberrations by FISH or chromosomal CGH (Table 1). Most interestingly, two of these unknown aberrations occurred recurrently in this series of patients: (i) trisomy 19 was detected in five B-CLL samples, all of which contained a trisomy 12, as well as the MCL carrying a partial gain of chromosome 19, and (ii) a small copy number gain located ≈16,084 kbp on 2p24 in five B-CLLs and the MCL case comprising the MYCN gene (Table 1). Because of the distance between the closest DNA fragments proximal and distal to the MYCN containing BACs (28,600 kbp and 10,500 kbp) on the chip, the critical region of the respective genomic gain can be narrowed down to 18,100 kbp. FISH to interphase nuclei or chromosomal CGH confirmed trisomy 19. In all analyzed cases (five of five) with trisomy of chromosome 19 the IGV region was hypermutated, suggesting an association of this aberration with B cell maturation. In contrast, gain of the MYCN locus was found in IGV mutated and unmutated B-CLL samples (Table 1).
Table 1. With matrix–CGH identified aberrations in B-CLL.
| Imbalanced in matrix–CGH
|
|||
|---|---|---|---|
| B-CLL no. | Chromosome localization | Genes covered by BACs or PACs | IGV mutation status* |
| 56 | +19 | BCL3, Cyclin E, AKT2, BAX, LIG1 | M |
| 40 | +19 | BCL3, Cyclin E, AKT2, BAX, LIG1 | M |
| 29 | +19 | BCL3, Cyclin E, AKT2, BAX, LIG1 | M |
| 28 | +2p24 | MYCN | UM |
| 54 | -18p11 | EMILIN-2 | UM |
| 23 | +2p24 | MYCN | M |
| 25 | +2p24 | MYCN | M |
| 48 | -7p14-p22 | - | UM |
| 47 | -4p15 | - | UM |
| -15q11-q21 | GPR | ||
| 39 | -10q24.2-q24.33 | FLJ10512, C10orf2, SEMA4G, FLJ23209, FKSG28, BA108L72, KIAA1813, MRPL43, FLJ22559, COL17A1, SLK | UM |
| 46 | -6q22.1 | - | UM |
| 66 | +2p24 | MYCN | UM |
| 68 | +19 | BCL3, Cyclin E, AKT2, BAX, LIG1 | M |
| 77 | +2p24 | MYCN | UM |
| -7q31-q33 | MET | ||
| 86 | +2p11-p23 | - | - |
| -20q | BCLX, AIB1, ZNF217, BCAS1, MYBL2, PTPN1, CSTF1, HEFL, STK15 | ||
| 87 | -2p24 | - | M |
| 4 | -11q13 | - | UM |
| 6 | -X† | DMD, ASB11, DKFZP564L0862, FLJ22601, CCNB3, KIAA0902, AR, SRPUL, SYTL4, GRIA3, ATP2B3, FLJ10727, F8A, H2AFB, | M |
| 7 | +19† | BCL3, Cyclin E, AKT2, BAX, LIG1 | - |
| 59 | -1p36.33 | TP73, FLJ20321, | M |
| 91 | -10q23-q24.2‡ | MGC16202, PTEN, LGI1, COL17A1, SLK, HIF1AN, WNT8B, | UM |
| 92 | -9p24† | JAK2 | UM |
| +2q22† | - | ||
| +7q31† | MET | ||
| +11q24-q25† | ETS-1 | ||
| 96 | +1p36.3† | TP73 | UM |
| 104 (MCL) | +2p24‡ | MYCN | M |
| -8p21-p23‡ | TNFRSF 10B | ||
| +8q24 | CMYC | ||
| -10p13-p15‡ | BS69, PFKP, PRKCQ | ||
| +18q21-q23‡ | EMILIN-2, RPL17, LIPG, POLI, MBD2, SCOP, DCC, CLUL1, HSRTSBETA, YES1, BCL2 | ||
| +19q13‡ | LIG1, BCL3, AKT2, BAX | ||
| 107 | +14q32‡ | AKT1 | - |
| -20 | AIB-1, | ||
| -21 | - | ||
| -22 | - | ||
Expression of MYCN Transcripts. The morphology of FISH signals for MYCN did not allow us to discriminate between a disomic and a tandem duplication status. However, in one case, the gain was sufficiently large to be confirmed by chromosomal CGH (Table 1). A comprehensive expression study in B-CLL applying the Affymetrix chip technology and testing for some 12,000 human genes included the five B-CLL cases with copy number gain of MYCN as well as 23 other cases from the present study (C.H., Norbert Schweifer, S.S., H.D., P.L., Norbert Kraut, Christian Stratowa, and Roger Abseher, unpublished data). In agreement with the genomic copy number, the median expression level of the five cases was 2.09 times above the median level of the other cases indicating that the genomic copy number gain is reflected on the RNA level (Table 2). In contrast, for the NCYM gene directly adjacent to MYCN on 2p24, no elevation of the expression level was observed. Similarly, measurement of the MYCN transcript level by quantitative RT-PCR (see Methods) revealed a 2.14-fold increase in the five cases with gain of MYCN in comparison to B-CLL patients without gain on 2p24 (Table 2).
Table 2. Comparison of DNA and transcript copy number levels of MYCN.
| B-CLL cases | n | Ratio matrix-CGH, median | Expression chip, normalized arbitrary values | Ratio quantitative RT-PCR, median* |
|---|---|---|---|---|
| Gain on 2p24 | 5 | 1.34 | 314 | 1.67 |
| 5 | 1.01 | 0.78 | ||
| Balanced on 2p24 | 23 | 150 | ||
| Ratio gain/balanced | 1.33 | 2.09 | 2.14 |
Relative gene expression in relation to the housekeeping genes HPRT and Lamin B.
Discussion
Here we report on a robust chip developed by extensive and elaborate optimization procedures that fulfils the criteria for application in clinical diagnostics. Testing of this chip in a series of 106 B-CLL cases revealed a high diagnostic specificity and sensitivity underlining the potential and reliability of matrix–CGH as a diagnostic tool. The sensitivity was shown to be 100% for cases with clonal genomic aberrations in >53% of the test cell population (determined by FISH). Thus, a routine analysis of genomic alterations in B-CLL should include preenrichment of the leukemic B cell population to >50%, e.g., via enrichment by surface antigen binding to microbeads.
Reliable scoring of genomic imbalances by CGH to microarrays depends on the application of adequate diagnostic cutoff levels of the signal ratio values. Different procedures have been used to define such threshold values including fixed cutoff values as well as statistical values derived, e.g., from intrachip variances or from matches to a normal distribution of ratio values (see ref. 22 and references therein). However, an imbalance might not be detectable when solely applying threshold criteria, because the cell clone carrying a given aberration is rare or the quality of the source material is limited. Nevertheless, if the ratio values of all DNA fragments from a given region deviate from the balanced status in the same direction, this suggests the presence of an imbalance. Thus, the unidirectional deviation of ratios from DNA fragments clustering in the genome but hardly reaching the threshold criteria might provide an additional parameter for the evaluation of disease-specific matrix–CGH chips. In such rare situations, the pattern of ratios should trigger additional locus-specific tests (e.g., FISH). As suggested by the present study, this criterion would be a useful supplement when analyzing cell populations with clonal aberrations in less than ≈50% of cells.
Although it had been assumed that the recurrent genomic alterations in B-CLL are known by now and the respective genes are already under investigation, the present study uncovered previously unrecognized recurrent genomic imbalances: copy number gain of chromosome 19 as well as of the MYCN oncogene on 2p24. Interestingly, trisomy of chromosome 19, described here as a recurrent alteration in B-CLL, was strongly associated with trisomy of chromosome 12 and IGV hypermutation. Because mutation of the variable region of the Ig heavy chain gene is associated with good clinical prognosis (3–5), these findings suggest future investigations of the prognostic potential of chromosome 19 trisomies in B-CLL.
Although a recent comprehensive analysis of B-CLL revealed gain of the respective region on chromosome 2 in 2 of 36 cases (23), direct evidence for activation of MYCN in non-Hodgkin lymphoma remained so far anecdotal (24, 25). The coincidence of gene copy number increase and elevated transcript level of MYCN discovered in the present study turns MYCN into a previously undescribed B-CLL candidate gene. The potential of the MYCN transcription factor to down-regulate the gene coding for the leukemia inhibitory factor, LIF (26), might point to a yet unrecognized mechanism involved in B-CLL pathogenesis. Because trisomy of chromosome 19 and 2p24 were detected by analyzing ≈400 DNA fragments not located in known B-CLL aberrant regions, it is possible that higher density genomic scanning, e.g., by matrix–CGH with microarrays consisting of many thousand BAC clones, will still uncover yet unrecognized genomic alterations.
In principle, matrix–CGH with BAC or PAC clones provides a resolution comparable to interphase cytogenetics by FISH using the respective clones. However, analysis of 10 or more genomic regions by interphase FISH, as requested in B-CLL diagnostics, requires multiple hybridization experiments or a complex multicolor FISH approach with demanding evaluation procedures in series of cell nuclei. Although quantitative PCR procedures have the potential to detect genomic imbalances, reliable scoring of alterations with ratios between 1.5 and 0.5 is still a challenge in routine diagnostics.
In B-CLL, chromosomal translocations are rare events and do not appear to identify distinct prognostic subgroups of patients (27). Based on a detailed analysis of the comprehensive compilation of all published cytogenetic aberrations in B cell non-Hodgkin lymphomas, Johansson et al. (28) proposed a model on tumor evolution. In this model, translocations are primary events useful for lymphoma classification, whereas secondary alterations are mainly chromosomal imbalances, many of which are associated with clinical outcome (28). Similarly, in carcinomas, imbalances such as amplifications of MYCN, MYC, or HER2, are well established prognostic markers, which are currently used as basis for therapy decisions. Thus, a robust automated tool for the diagnosis of genomic imbalances is of particular importance for tumor diagnosis, eventually leading to new patient stratification schemes. We here present a prototype of such a tool, which can be expanded or modified to meet the clinical needs in molecular oncology. The information about the required complexity of targeted sequences for a highly reliable diagnosis by matrix–CGH also provides basic information for other disease-specific microarrays in clinical genetics. Such chips can be produced and hybridized within clinical treatment trails in specialized academic centers maintaining chip-facilities. Moreover, biotechnology companies may develop commercially available platforms allowing to perform genomic analysis in specific disease.
Supplementary Material
Acknowledgments
We gratefully acknowledge Andrea Wittmann, Kathrin Wildenberger, Sibylle Ohl, Antoaneta Mincheva, Armin Pscherer, Holger Kohlhammer, Felix Kokocinski, Stephan Wolf, Boris Zielinski, Björn Fritz, Daniel Göttel, Cordula Tschuch, André Müller, Martina Enz, Elsbeth Brückle, Annett Habermann, Alexander Kröber, Dirk Kienle, and Sandra Ruf from Heidelberg and Ulm for excellent technical assistance and sharing of material and assays. We thank Christian Stratowa, Roger Abseher, and Norbert Kraut from Boehringer Ingelheim, Austria, for the fruitful collaboration in expression profiling by microarray analyses. PACs and BACs for this study were ordered from libraries provided by theResource Centre/Primary Database of the German Human Genome Project (Berlin). This work was supported by Bundesministerium für Bildung und Forschung (BMBF), Grants 01KW9937 and 01GR0101-P6T7 NGFN, the Land Thüringen, the Tumorzentrum Heidelberg/Mannheim, and the Landesforschungsschwerpunkt Baden-Württemberg Project No. 1423/98101-0.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: B-CLL, B cell chronic lymphocytic leukemia; CGH, comparative genomic hybridization; FISH, fluorescence in situ hybridization; MCL, mantle cell lymphoma.
References
- 1.Rozman, C., Montserrat, E. (1995) N. Engl. J. Med. 333, 1052–1057. [DOI] [PubMed] [Google Scholar]
- 2.Döhner, H., Stilgenbauer, S., Benner, A., Leupolt, E., Krober, A., Bullinger, L., Döhner, K., Bentz, M. & Lichter, P. (2000) N. Engl. J. Med. 343, 1910–1916. [DOI] [PubMed] [Google Scholar]
- 3.Damle, R. N., Wasil, T., Fais, F., Ghiotto, F., Valetto, A., Allen, S. L., Buchbinder, A., Budman, D., Dittmar, K., Kolitz, J., et al. (1999) Blood 94, 1840–1847. [PubMed] [Google Scholar]
- 4.Hamblin, T. J., Davis, Z., Gardiner, A., Oscier, D. G. & Stevenson, F. K. (1999) Blood 94, 1848–1854. [PubMed] [Google Scholar]
- 5.Kröber, A., Seiler, T., Benner, A., Bullinger, L., Brückle, E., Lichter, P., Döhner, H. & Stilgenbauer, S. (2002) Blood 100, 1410–1416. [PubMed] [Google Scholar]
- 6.Solinas-Toldo, S., Lampel, S., Stilgenbauer, S., Nickolenko, J., Benner, A., Döhner, H., Cremer, T. & Lichter, P. (1997) Genes Chromosomes Cancer 20, 399–407. [PubMed] [Google Scholar]
- 7.Pinkel, D., Segraves, R., Sudar, D., Clark, S., Poole, I., Kowbel, D., Collins, C., Kuo, W. L., Chen, C., Zhai, Y., et al. (1998) Nat. Genet. 20, 207–211. [DOI] [PubMed] [Google Scholar]
- 8.Pollack, J. R., Perou, C. M., Alizadeh, A. A., Eisen, M. B., Pergamenschikov, A., Williams, C. F., Jeffrey, S. S., Botstein, D. & Brown, P. O. (1999) Nat. Genet. 23, 41–46. [DOI] [PubMed] [Google Scholar]
- 9.Snijders, A. M., Nowak, N., Segraves, R., Blackwood, S., Brown, N., Conroy, J., Hamilton, G., Hindle, A. K., Huey, B., Kimura, K., et al. (2001) Nat. Genet. 29, 263–264. [DOI] [PubMed] [Google Scholar]
- 10.Bruder, C. E., Hirvela, C., Tapia-Paez, I., Fransson, I., Segraves, R., Hamilton, G., Zhang, X. X., Evans, D. G., Wallace, A. J., Baser, M. E., et al. (2001) Hum. Mol. Genet. 10, 271–282. [DOI] [PubMed] [Google Scholar]
- 11.Wessendorf, S., Fritz, B., Wrobel, G., Nessling, M., Lampel, S., Goettel, D., Kuepper, M., Joos, S., Hopman, T., Kokocinski, F., et al. (2002) Lab. Invest. 82, 47–60. [DOI] [PubMed] [Google Scholar]
- 12.Veltman, J. A., Schoenmakers, E. F., Eussen, B. H., Janssen, I., Merkx, G., van Cleef, B., van Ravenswaaij, C. M., Brunner, H. G., Smeets, D. & van Kessel, A. G. (2002) Am. J. Hum. Genet. 70, 1269–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fritz, B., Schubert, F., Wrobel, G., Schwaenen, C., Wessendorf, S., Nessling, M., Korz, C., Rieker, R. J., Montgomery, K., Kucherlapati, R., et al. (2002) Cancer Res. 62, 2993–2998. [PubMed] [Google Scholar]
- 14.Wilhelm, M., Veltman, J. A., Olshen, A. B., Jain, A. N., Moore, D. H., Presti, J. C., Jr., Kovacs, G. & Waldman, F. M. (2002) Cancer Res. 62, 957–960. [PubMed] [Google Scholar]
- 15.Sanchez-Izquierdo, D., Buchonnet, G., Siebert, R., Gascoyne, R. D., Climent, J., Karran, L., Marin, M., Blesa, D., Horsman, D., Rosenwald, A., et al. (2003) Blood 101, 4539–4546. [DOI] [PubMed] [Google Scholar]
- 16.Martinez-Climent, J. A., Alizadeh, A. A., Segraves, R., Blesa, D., RubioMoscardo, F., Albertson, D. G., Garcia-Conde, J., Dyer, M. J., Levy, R., Pinkel, D., et al. (2003) Blood 101, 3109–3117. [DOI] [PubMed] [Google Scholar]
- 17.Döhner, H., Fischer, K., Bentz, M., Hansen, K., Benner, A., Cabot, G., Diehl, D., Schlenk, R., Coy, J. & Stilgenbauer, S. (1995) Blood 85, 1580–1589. [PubMed] [Google Scholar]
- 18.Bentz, M., Huck, K., du Manoir, S., Joos, S., Werner, C. A., Fischer, K., Döhner, H. & Lichter, P. (1995) Blood 85, 3610–3618. [PubMed] [Google Scholar]
- 19.Wessendorf, S., Schwaenen, C., Kohlhammer, H., Kienle, D., Wrobel, G., Barth, T. F., Nessling, M., Moller, P., Döhner, H., Lichter, P., et al. (2003) Oncogene 22, 1425–1429. [DOI] [PubMed] [Google Scholar]
- 20.Stilgenbauer, S., Nickolenko, J., Wilhelm, J., Wolf, S., Weitz, S., Döhner, K., Boehm, T., Döhner, H. & Lichter, P. (1998) Oncogene 16, 1891–1897. [DOI] [PubMed] [Google Scholar]
- 21.Korz, C., Pscherer, A., Benner, A., Mertens, D., Schaffner, C., Leupolt, E., Döhner, H., Stilgenbauer, S. & Lichter, P. (2002) Blood 99, 4554–4561. [DOI] [PubMed] [Google Scholar]
- 22.Carter, N. P., Fiegler, H. & Piper, J. (2002) Cytometry 49, 43–48. [DOI] [PubMed] [Google Scholar]
- 23.Bea, S., Lopez-Guillermo, A., Ribas, M., Puig, X., Pinyol, M., Carrio, A., Zamora, L., Soler, F., Bosch, F., Stilgenbauer, S. et al. (2002) Am. J. Pathol. 161, 957–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finnegan, M. C., Hammond, D. W., Hancock, B. W. & Goyns, M. H. (1995) Leuk. Lymphoma 18, 511–514. [DOI] [PubMed] [Google Scholar]
- 25.Werner, C. A., Dohner, H., Joos, S., Trümper, L. H., Baudis, M., Barth, T. F., Ott, G., Moeller, P., Lichter, P. & Bentz, M. (1997) Am. J. Pathol. 151, 335–342. [PMC free article] [PubMed] [Google Scholar]
- 26.Hatzi, E., Murphy, C., Zoephel, A., Ahorn, H., Tontsch, U., Bamberger, A. M., Yamauchi-Takihara, K., Schweigerer, L. & Fotsis, T. (2002) Eur. J. Biochem. 269, 3732–3741. [DOI] [PubMed] [Google Scholar]
- 27.Stilgenbauer, S., Bullinger, L., Lichter, P. & Döhner, H. (2002) Leukemia 16, 993–1007. [DOI] [PubMed] [Google Scholar]
- 28.Johansson, B., Mertens, F. & Mitelman, F. (1995) Blood 86, 3905–3914. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.