Abstract
In olfactory neurons, expression of a single odorant receptor (OR) from a repertoire of >1,000 genes is required for odor coding and axonal targeting. Here, we demonstrate a role for OR protein as an essential regulator in the establishment of monoallelic OR expression. An OR-promoter-driven reporter expresses in a receptor-like pattern but, unlike a native OR, is coexpressed with an additional OR allele. Expression of a functional OR from the identical promoter eliminates expression of other OR alleles. The presence of an untranslatable OR coding sequence in the mRNA is insufficient to exclude expression of a second OR. Together, these data identify the OR protein as a critical element in a feedback pathway that regulates OR selection.
The mammalian olfactory system detects odors with remarkable sensitivity while decoding the complex odorant stimuli present in the environment. Populations of olfactory neurons with similar response profiles generate patterned activity within the olfactory bulb (OB) and lead to the perception of a given odor. These cellular responses are defined by the selective expression of seven-transmembrane odorant receptor (OR) proteins (1).
Each olfactory neuron expresses only one allele of an individual OR gene from >1,000 related genes dispersed throughout the genome, a process termed monoallelic expression (2–4). The neurons expressing a particular OR reside in one of multiple zones defined by their dorsal–medial to ventral–lateral position in the olfactory epithelium (OE) (5, 6). These OR expression zones contribute broad topographic organization for the axons of olfactory neurons as they target the OB (5, 7, 8). ORs also play an important role in olfactory axon pathfinding (9, 10). Neurons that express the same OR, while interspersed with neurons expressing other ORs in the epithelium, project axons that converge to a small number of stereotyped glomeruli in the OB (11, 12). A transgene comprised of proximal OR promoter sequences is in many cases sufficient to recapitulate normal expression (13, 14), although the transgene integration site can significantly influence reporter expression. This variability has confounded the elucidation of the processes that contribute to monoallelic, zonal OR expression.
The monoallelic expression of ORs is critical to the establishment of the functionally distinct neuronal populations required for odor discrimination and organization of axonal projections to the OB (15, 16). This highly regulated expression could be achieved through a probabilistic promoter selection mechanism that is independent of the production of functional OR protein (10). Alternatively, OR protein or mRNA could initiate a feedback signal that ensures monoallelic expression.
To determine whether monoallelic OR expression depends on feedback regulation, we targeted an OR promoter-driven reporter to a defined, reproducible transgene insertion site amid a cluster of ORs. This approach alleviates transgenic variability while facilitating analysis of potential coexpression with adjacent ORs within that cluster. We demonstrate that OR promoter activation in the absence of OR protein expression is insufficient to restrict expression from additional OR promoters in a given neuron. Inclusion of an OR coding region restores monoallelic expression. We conclude that monoallelic OR expression is achieved through a mechanism in which OR protein functions in olfactory neurons to abrogate expression of other OR genes.
Methods
Gene Targeting. Targeting vectors were constructed with 129/Sv mouse genomic DNA fragments isolated from a λ phage library (Stratagene). Homology regions consisted of a 2.3-kb 5′ arm and the 8-kb 3′ sequence that contained the M71 coding sequence. The recombination event results in the simple insertion of the M4 transgenic constructs with no elimination or duplication of the sequences surrounding M71. The M4-driven LacZ transgene was generated by ligation of a 6.7-kb HindIII fragment containing the promoter and intron (13) to a Tau-LacZ reporter (9) and SV40 polyadenylation site (Clontech). The related reporter constructs, M4 IRES-Tau-LacZ and LacZ-M4RNA, were created by replacing a Tau-LacZ reporter with the PCR-generated M4 coding region followed by an internal ribosome entry site (IRES) Tau-LacZ SV40 pA or a Tau-LacZ reporter followed by the M4 coding sequence lacking an initiation codon. Each targeting vector included a Lox-Neomycin-Lox (LNL) cassette (17) for selection of neomycin-resistant embryonic stem cell clones that was subsequently removed by mating chimeric male mice to Cre-transgenic females (18). Sequences and methods for generating these constructs are available from the authors.
Targeting constructs were linearized with XhoI and electroporated into 129/Sv embryonic stem cells (Johns Hopkins Medical Institute Transgenic Core Laboratory), and homologous integration events were identified by Southern blot analysis. Genetically modified embryonic stem cells were injected into C57/BL6 blastocysts and implanted into pseudopregnant females to generate chimeric mice.
Whole-Mount Imaging. 5-Bromo-4-chloro-3-indolyl β-d-galactoside (X-gal) staining on whole-mount preparations was performed essentially as described (9). Tissues were isolated from 4-week-old mice after intracardiac perfusion with 4% paraformaldehyde in PBS. OE and OB were then dissected and imaged directly for GFP and/or stained with X-gal for 2 h or overnight when indicated and subsequently imaged. Images were acquired on a Leica FZ III dissection microscope or a Zeiss Axioplan with a ×5 objective and an Axiocam digital camera.
In Situ Hybridization and Immunohistochemistry. Tissues were isolated as above after perfusion with 4% paraformaldehyde or Bouin's solution (Sigma). Twenty- to 40-μM sections were cut on a cryostat. The M71, K22, and M4 coding regions were used for in situ hybridization probes (M72 is also detected by cross hybridization to M71). In situ hybridization was performed essentially as described (19). NBT/BCIP (Roche Applied Science) was used to visualize the bright field in situ hybridization signal, and HNPP/Fast Red detection (Roche Applied Science) or TSA (NEN) was used to visualize fluorescent signals. Immunohistochemistry was performed by incubating sections with rabbit antibody to β-galactosidase (β-gal) (5′ to 3′) at 1:1,000 dilution in Tris-buffered saline with 0.1% Tween 20 with 1% normal goat serum. After washing, sections were incubated with Alexa 488- or Alexa 546-conjugated anti-rabbit antibody (Molecular Probes) at a 1:1,000 dilution and imaged on a Zeiss Axioplan or LSM 510 confocal microscope. X-gal staining of sections was performed by using the same protocol used for whole-mount staining.
Results
We targeted an OR M4 promoter-driven reporter to a gene cluster comprising ≈100 OR genes (20, 21). The expression construct consisted of 2.4 kb of M4 promoter sequence, the native transcription start site, and a 4-kb intron followed by a Tau-LacZ reporter (13). In this transgene, the reporter replaced the coding sequence of M4 (Fig. 1A). The construct was inserted via homologous recombination between the well characterized OR genes M71 and M72 (20, 21). The M71 and M72 receptor genes are expressed in a more dorsal zone of the OE (zone I) than M4 (zone II) (13), and cells expressing M71 or M72 project axons to glomeruli in a dorsal region of the OB (13, 22–25). If M4 promoter-driven LacZ is coexpressed with either of these genes, LacZ-expressing cells would adopt the zonal expression pattern and glomerular targets of M71 and/or M72.
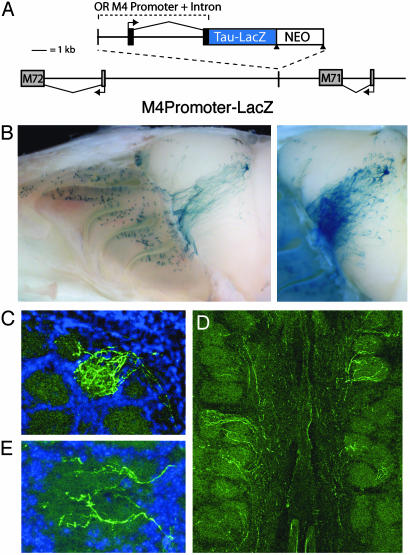
Fig. 1.
OR promoter-driven Tau-LacZ displays broad expression and targeting to the OB. (A) Schematic of the genomic structure in targeted transgenic mice. The 2.4-kb OR M4 promoter sequence and native 4-kb intron driving expression of Tau-LacZ were inserted by means of homologous recombination at a location between ORs M71 and M72. The integration site was 3 kb distal to the polyadenylation site of M71, and the insert was oriented to preclude Tau-LacZ transcription originating from the M71 promoter. The neomycin selection marker was removed by Cre-mediated recombination at the LoxP sites (triangles). The orientation and start site of transcription for each receptor is marked with an arrow. (B) Whole-mount image of LacZ expression in homozygous M4 promoter-driven LacZ reporter animals. LacZ+ cells are present in multiple zones of the OE and project to broad regions of the OB (Left). A fraction of labeled axons target a single glomerulus, whereas the remainder target other locations in the OB and can be easily visualized after longer enzymatic reaction (Right). (C) In coronal sections of OB, LacZ+ axons (green) can be seen entering the heavily labeled glomerulus shown in B. (D) Many additional glomeruli on the medial surface of the bulb receive innervation by smaller numbers of LacZ+ axons. (E) Higher magnification reveals extensive branching of these LacZ+ axons consistent with functional glomerular innervation by labeled olfactory neurons.
M4 promoter-driven LacZ was expressed in the OE in a pattern that overlapped with the normal expression zones of both M4 and M71 (Fig. 1B). The axons of LacZ-expressing cells projected to broad regions of the OB. This projection pattern, consisting of axonal targeting to discrete locations in the dorsal bulb and to a large region across the medial surface, was observed in all animals (n > 20), and indicated that LacZ+ axons targeted defined locations. Although LacZ+ projections were broad, a significant fraction of labeled axons converged to a single site (Fig. 1B). In OB sections, these dorsally targeting LacZ+ axons clearly entered the glomerular layer and displayed multiple branches consistent with their functional innervation of this glomerulus (Fig. 1C). Many additional glomeruli were innervated by one or a few LacZ+ axon fibers that displayed a similar branching phenotype (Fig. 1 D and E). Previous studies have shown that the OR is critical for targeting olfactory axons to defined glomeruli in the OB (8–10). Therefore, we infer that each reporter-labeled cell also expresses a native OR that determines its glomerular target.
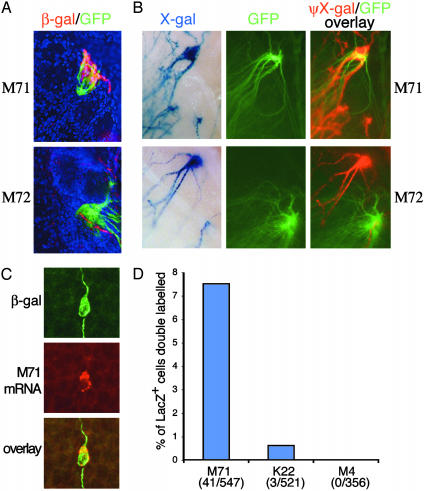
To identify the glomeruli innervated by LacZ+ axons, we compared the projection patterns of LacZ+ axons to those of axons expressing the adjacent genes encoding ORs M71 and M72. Mice expressing M4 promoter-driven LacZ were crossed to animals in which M71 or M72 expressing cells could be visualized with an IRES-Tau-GFP reporter (22, 23). We then examined the axonal projection patterns of these two cell populations. The GFP-labeled M71 glomerulus on the medial surface of the OB received robust innervation from LacZ+ axons (Fig. 2 A and B, n = 10). The medial M72 glomerulus was also innervated by LacZ+ axons but to a far lesser extent (Fig. 2 A and B, n = 10). Targeting of LacZ+ axons to the M71 glomerulus indicates that a fraction of LacZ+ cells express this OR. In double heterozygous mice, the GFP-labeled M71 allele and the allele containing the targeted M4 transgene are always located on the apposing homologs. Interestingly, no neuronal cell bodies in the OE of these double heterozygous animals were labeled with both β-gal and GFP (n = 200 cells; data not shown). We conclude that coexpression of LacZ and M71 occurs frequently when the two genes are present in cis. Coexpression was confirmed by using fluorescent in situ hybridization for M71 combined with an antibody against β-gal. A significant number of cells (41/547 LacZ+ cells, 7.5%) in the OE expressed both the M4 promoterdriven LacZ and M71 (Fig. 2 C and D). Moreover, the intensity of the M71 in situ signal was similar in LacZ+ and LacZ– cells. By the same method, additional LacZ+ cells were found to coexpress another OR gene, K22 (zone I), located in the same genomic region (Fig. 2D). Coexpression of M71, M72, K22, and presumably other OR genes, along with the M4 promoter-driven LacZ, directs the reporter-labeled axons to multiple targets in the OB. These data demonstrate that two OR promoters can be active in a single cell, at least under circumstances where only one promoter directs expression of a receptor protein.
Fig. 2.
Coexpression of OR promoter-driven LacZ reporter with endogenous OR genes. (A) Confocal images of OB sections. M4-driven LacZ+ axons detected with anti-β-gal antibody (red) converge with M71 IRES-Tau-GFP axons (Upper) in LacZ/GFP double heterozygous mice (Lower). A small number of LacZ+ axons also innervate the M72 glomerulus visualized in M72 IRES-Tau-GFP heterozygous mice (Lower). (B) Whole-mount analysis of M4-driven LacZ/M71 IRES-Tau-GFP or M72 IRES-Tau-GFP double heterozygous mice. For M71, multiple LacZ+ axons (Upper Left) and GFP labeled axons (Middle) converge to the same glomerulus (Right). Overlay was created by combining the GFP panel with an intensity inverted, red pseudocolored image (photoshop, Adobe Systems, Mountain View, CA) of the X-gal panel. For M72, the analogous overlay (Lower) revealed modest innervation of this glomerulus by LacZ+ axons. Landmarks outside of the displayed field were used to register the images in all panels. (C) LacZ and OR M71 are expressed in the same neuron. Anti-β-gal immunocytochemistry (green) was performed in conjunction with fluorescent in situ hybridization for M71 (red) and analyzed by two-color confocal microscopy. (D) Quantification of endogenous OR mRNA expression by LacZ+ cells. A fraction (41/547, 7.5%) of LacZ+ cells express the M71 gene. A smaller percentage (3/521, 0.6%) of LacZ+ cells express the K22 OR located 14 kb to the right of M71 as depicted in Fig. 1.
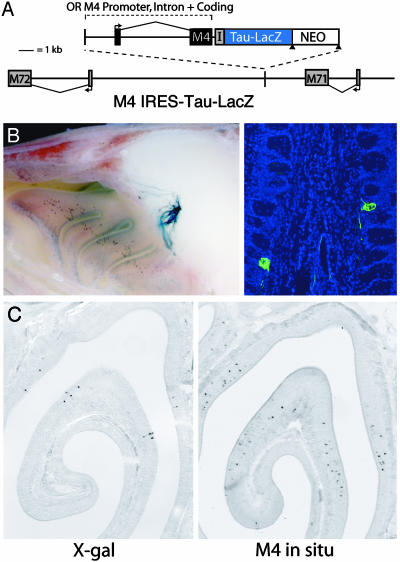
If OR protein serves to restrict expression of other OR alleles, an OR promoter-driven transgene that expresses an intact receptor should preclude expression of other OR genes. We tested this hypothesis by targeting the same location with the identical M4 promoter driving expression of a functional M4 protein. The receptor sequence was followed by an IRES-Tau-LacZ to visualize expression (Fig. 3A). The expression of M4 protein resulted in a dramatic change in the LacZ expression pattern in the OE and targeting of labeled axons to the OB (Fig. 3B). M4 IRES-Tau-LacZ expression was restricted to a zone of the epithelium similar to that of the native M4 OR (Fig. 3C), and LacZ+ axons targeted to a single medial and a single lateral glomerulus in more ventral positions of the OB (Fig. 3B). Importantly, inclusion of M4 coding sequence was sufficient to abolish reporter expression from the more dorsal epithelial expression zone where M71-expressing cells reside and eliminate targeting of labeled axons to the M71 glomerulus. We infer that the M4 receptor protein in the labeled cells prevents coexpression of M71 and other ORs. When the M71 in situ hybridization and β-gal antibody staining described above were performed on this line, no cells in the OE coexpressed these two genes (n = 300 cells; data not shown). These data demonstrate that an OR promoter expressing a functional receptor is able to restrict the expression of other receptor alleles in the same cell.
Fig. 3.
OR promoter driving expression of a functional OR. (A) Schematic of the genomic structure in targeted transgenic mice. The M4 promoter and intron were used to direct expression of a functional M4 protein, and expression was visualized by using an IRES-Tau-LacZ reporter. This construct was integrated at the same location between M71 and M72 as in Fig. 1. (B) LacZ expression in homozygous M4 IRES-Tau-LacZ animals. In whole-mount staining, LacZ+ cells are restricted to one zone of the OE, and labeled axons target to a single glomerulus on the medial surface of the OB (Left). In coronal sections of the OB, all axons labeled with an anti-β-gal antibody (green) enter the glomerular layer and innervate a single medial glomerulus (Right). A corresponding lateral glomerulus was also innervated. (C) LacZ expression in coronal sections of the OE. X-gal staining of M4 IRES-Tau-LacZ mice (Left) reveals a LacZ expression pattern similar to the native zone of M4 as visualized by in situ hybridization (Right). Reporter expression is restricted to the dorsal aspects of the endogenous M4 OR expression zone.
It is formally possible that specific RNA sequences or DNA elements imbedded within the coding region, rather than receptor protein, could serve to restrict the expression of other OR alleles. To discriminate among these possibilities, we targeted to the same M71 locus a transgene in which the OR coding sequence is present in the mRNA but cannot be translated into protein. This construct was generated by inserting the M4 coding sequence into the 3′ UTR of the M4 promoter-driven LacZ replacement construct shown in Fig. 1 (Fig. 4A). The M4 coding sequence differed from wild type only in that it lacked a start codon to ensure that the receptor protein would not be translated. The introduction of premature stop codons into the OR open reading frame of reporter-tagged alleles could disrupt cis elements within the coding region and are reported to destabilize the message (10). We designed this construct, LacZ-M4RNA, to circumvent these issues.
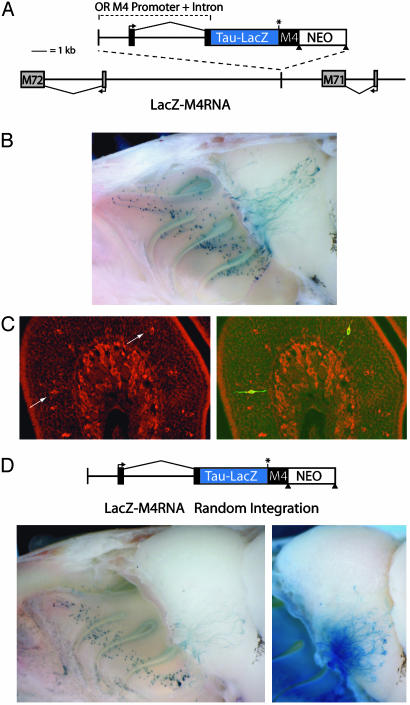
Fig. 4.
Transgenic analysis of an OR promoter-driven Tau-LacZ containing an untranslated M4 coding sequence in the mRNA. (A) Schematic of the genomic structure in targeted transgenic mice. The M4 promoter and intron were used to drive expression of Tau-LacZ. The M4 coding sequence, lacking an initiating methionine (asterisk), was present in the 3′ UTR of the transgene immediately after the stop codon of LacZ. This construct (LacZ-M4RNA) was integrated between M71 and M72. (B) Whole-mount image of LacZ expression in heterozygous LacZ-M4RNA animals. LacZ+ cells are present in multiple zones of the OE and project to broad regions of the OB. A fraction of labeled axons target in the region of the M71 glomerulus, whereas the remainder project to other locations in the OB. (C) Fluorescent in situ hybridization in LacZ-M4RNA animals for M4 (red) along with anti-β-gal immunocytochemistry (green) shows that M4 sequence is retained in the transgene mRNA present in β-gallabeled cells (arrows). (D) Random integration of the LacZ-M4RNA construct. Whole-mount imaging (Lower Left) reveals LacZ is expressed in a single zone of the OE. LacZ+ axons target project broadly across the ventral OB. Multiple glomerular targets of LacZ+ axons can be visualized after longer enzymatic reaction (Lower Right).
The presence of the M4 coding region in the LacZ-M4RNA construct is insufficient to restrict expression of additional OR alleles in the labeled cells. LacZ+ cells were detected throughout the dorsal OE, and labeled axons targeted to broad regions of the OB, including the M71 glomerulus (Fig. 4B, n = 7). This pattern is indistinguishable from that observed for the M4 promoterdriven LacZ-only construct (Fig. 1B). In situ hybridization with an M4 coding region probe along with antibody staining against β-gal confirmed that all LacZ+ neurons examined also contained M4 RNA (Fig. 4C, n = 75 cells), implying that the transcript was not spliced in a way that removed the M4 coding sequence from the mature mRNA. Moreover, these data confirm that β-gal immunoreactive cells are actively expressing the LacZ-M4RNA transcript, and antibody staining is not a result of reporter protein persisting after expression has been silenced. These observations strongly imply that OR protein rather than OR coding sequences present in the DNA or mRNA directs the selective expression of a single OR allele in each olfactory neuron.
The integration of the transgene in proximity of the M71 gene resulted in the preferential coexpression of LacZ with this particular OR. To demonstrate that coexpression of an OR promoter-driven reporter with additional OR alleles occurs independent of the transgene integration site, we examined a line derived from ES cells that contained a low copy number, random integration of the LacZ-M4RNA construct (Fig. 4C). In these mice, expression in the OE was similar to the native M4 zone, and labeled axons entered many glomeruli in the more ventral regions of the OB, implying that these reporter-labeled cells also express a functional OR (Fig. 4D). Together, these data suggest that monoallelic expression derives from OR protein-mediated feedback regulation rather than a mechanism that permits only one promoter to be active in a given cell.
Discussion
These studies define OR protein as an important factor in the monoallelic expression of OR genes. It is unlikely that a feedback mechanism would, in itself, be responsible for excluding expression from all other receptor loci. Rather, feedback involving the receptor would serve as an additional, modulatory process to refine a stochastic selection mechanism. A simple model for receptor regulation that invoked a purely stochastic mechanism to activate, on average, one OR promoter in each cell would result in the existence of a large number of receptor-null cells in the OE. The model proposed here, in which a negative feedback mechanism regulates a stochastic selection process tuned to activate a few OR loci, results in a far more efficient scheme to ensure that each cell expresses exactly one OR allele. Each cell will, in a temporal, stochastic fashion, activate an OR promoter. If that locus encodes a functional receptor, negative feedback through the OR protein prevents expression from additional OR loci. However, when the activated promoter does not drive expression of functional OR protein, as in the case of the reporter-only constructs and OR pseudogenes, additional promoter regions are subsequently chosen for expression. In this scenario, previously selected OR promoters would remain active.
How is the presence of OR protein sensed by the cell? In the immune system, a feedback pathway exists in which signals originating from the surface expression of antigen receptors contribute to their monoallelic expression (26, 27). The olfactory signal transduction components could play a similar role in signaling from OR protein to OR genes in the nucleus. Genetic disruption of the CNGA2-channel subunit responsible for OR-mediated Ca2+ ion influx does not lead to miswiring that might be expected if multiple receptors are expressed in each cell (28), although second messengers further upstream in the signaling pathway could mediate the feedback signal. Interestingly, glomerular disorganization is reported in mice deficient for the olfactory-specific adenylyl cyclase, ACIII (29). In an alternate scenario, expression of a functional receptor could be a requisite for cells to pass beyond a transient developmental stage when they are competent to select among a large receptor repertoire. Acceleration of neuronal development on receptor expression would then serve to limit the number of receptor genes expressed in a given cell. In either case, one would predict that ubiquitous OR expression that occurs before OR promoter activation would preclude the expression of other receptors while similar expression after the selection process would not be subject to this feedback.
The hierarchical processes underlying OR selection and activation are not understood. Our observation that OR promoterdriven reporters are preferentially coexpressed with adjacent receptors may reflect an aspect of the normal mechanism for promoter selection and activation. However, the projection of labeled axons into a large number of glomeruli indicates that many additional OR loci can be selected for coexpression with the reporter. The frequency of coexpression of the reporter with a single endogenous receptor gene is very low and likely accounts for our failure to detect endogenous M4 among the reporter-labeled cells.
Our experimental data makes it unlikely that low-level expression of the introduced transgene allows expression of additional receptors. In situ hybridization with the M4 coding sequence demonstrates that the M4RNA transgene is expressed at levels comparable to the endogenous M4 gene. Additionally, the levels of M71 mRNA in cells that coexpress M71 and M4-driven Tau-lacZ were similar to M71 mRNA levels in cells expressing only M71. The precise overlap in M4 in situ signal and β-gal immunoreactivity suggests that OR-driven reporter expression is not extinguished on activation of an endogenous OR.
Previous studies inferred the absence of OR protein in reporter-expressing cells on the basis of broad axonal projections and accelerated loss of labeled cells from the OE (10). The observed axon projection pattern may have instead reflected the projection of axons to their appropriate targets based on the expression of additional ORs in these reporter-expressing cells. In whole-mount preparations and in antibody staining of tissue sections reported here, most labeled axons appear to enter glomeruli, implying expression of OR protein in LacZ+ cells. Cells expressing M4-driven LacZ are relatively stable in the OE, with a modest reduction in the number of labeled cells at postnatal day 90 (data not shown). The presence of the reporter protein or the precise nature of the reporter construct may contribute to the diminished cell number in both cases.
Targeted transgenesis provides reproducible expression patterns and permits systematic analysis of OR regulation. These studies demonstrate that proximal promoter regions driving an OR are sufficient to fully recapitulate proper monoallelic OR expression, even when imbedded among OR loci with different zonal expression patterns. Moreover, we demonstrate the importance of OR protein in the establishment of selective OR expression. This remarkable protein therefore participates in three diverse and complex aspects of olfactory function: odorant recognition, axon pathfinding, and the regulation of receptor expression.
Acknowledgments
We thank Peter Mombaerts for providing the M71 and M72 Tau-GFP animals, Mitra Cohen and the Johns Hopkins Medical Institute Transgenic Core Laboratory for blastocyst injections, and Sherry Ralls and Heather Kulaga for technical support. This work was supported by the Howard Hughes Medical Institute and a grant from the National Institutes of Health (to R.R.R.).
Abbreviations: β-gal, β-galactosidase; IRES, internal ribosome entry site; OB, olfactory bulb; OE, olfactory epithelium; OR, odorant receptor; X-gal, 5-bromo-4-chloro-3-indolylβ-d-galactoside.
References
- 1.Buck, L. & Axel, R. (1991) Cell 65, 175–187. [DOI] [PubMed] [Google Scholar]
- 2.Zhang, X. & Firestein, S. (2002) Nat. Neurosci. 5, 124–133. [DOI] [PubMed] [Google Scholar]
- 3.Malnic, B., Hirono, J., Sato, T. & Buck, L. B. (1999) Cell 96, 713–723. [DOI] [PubMed] [Google Scholar]
- 4.Chess, A., Simon, I., Cedar, H. & Axel, R. (1994) Cell 78, 823–834. [DOI] [PubMed] [Google Scholar]
- 5.Ressler, K. J., Sullivan, S. L. & Buck, L. B. (1993) Cell 73, 597–609. [DOI] [PubMed] [Google Scholar]
- 6.Vassar, R., Ngai, J. & Axel, R. (1993) Cell 74, 309–318. [DOI] [PubMed] [Google Scholar]
- 7.Yoshihara, Y., Kawasaki, M., Tamada, A., Fujita, H., Hayashi, H., Kagamiyama, H. & Mori, K. (1997) J. Neurosci. 17, 5830–5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bozza, T., Feinstein, P., Zheng, C. & Mombaerts, P. (2002) J. Neurosci. 22, 3033–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mombaerts, P., Wang, F., Dulac, C., Chao, S. K., Nemes, A., Mendelsohn, M., Edmondson, J. & Axel, R. (1996) Cell 87, 675–686. [DOI] [PubMed] [Google Scholar]
- 10.Wang, F., Nemes, A., Mendelsohn, M. & Axel, R. (1998) Cell 93, 47–60. [DOI] [PubMed] [Google Scholar]
- 11.Ressler, K. J., Sullivan, S. L. & Buck, L. B. (1994) Cell 79, 1245–1255. [DOI] [PubMed] [Google Scholar]
- 12.Vassar, R., Chao, S. K., Sitcheran, R., Nunez, J. M., Vosshall, L. B. & Axel, R. (1994) Cell 79, 981–991. [DOI] [PubMed] [Google Scholar]
- 13.Qasba, P. & Reed, R. R. (1998) J. Neurosci. 18, 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vassalli, A., Rothman, A., Feinstein, P., Zapotocky, M. & Mombaerts, P. (2002) Neuron 35, 681–696. [DOI] [PubMed] [Google Scholar]
- 15.Mombaerts, P. (2001) Nat. Neurosci. 4, Suppl., 1192–1198. [DOI] [PubMed] [Google Scholar]
- 16.Buck, L. B. (2000) Cell 100, 611–618. [DOI] [PubMed] [Google Scholar]
- 17.Zhao, H. & Reed, R. R. (2001) Cell 104, 651–660. [DOI] [PubMed] [Google Scholar]
- 18.Schwenk, F., Baron, U. & Rajewsky, K. (1995) Nucleic Acids Res. 23, 5080–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaeren-Wiemers, N. & Gerfin-Moser, A. (1993) Histochemistry 100, 431–440. [DOI] [PubMed] [Google Scholar]
- 20.Sullivan, S. L., Adamson, M. C., Ressler, K. J., Kozak, C. A. & Buck, L. B. (1996) Proc. Natl. Acad. Sci. USA 93, 884–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie, S. Y., Feinstein, P. & Mombaerts, P. (2000) Mamm. Genome 11, 1070–1078. [DOI] [PubMed] [Google Scholar]
- 22.Treloar, H. B., Feinstein, P., Mombaerts, P. & Greer, C. A. (2002) J. Neurosci. 22, 2469–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Potter, S. M., Zheng, C., Koos, D. S., Feinstein, P., Fraser, S. E. & Mombaerts, P. (2001) J. Neurosci. 21, 9713–9723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng, C., Feinstein, P., Bozza, T., Rodriguez, I. & Mombaerts, P. (2000) Neuron 26, 81–91. [DOI] [PubMed] [Google Scholar]
- 25.Ressler, K. J., Sullivan, S. L. & Buck, L. B. (1994) Curr. Opin. Neurobiol. 4, 588–596. [DOI] [PubMed] [Google Scholar]
- 26.Khor, B. & Sleckman, B. P. (2002) Curr. Opin. Immunol. 14, 230–234. [DOI] [PubMed] [Google Scholar]
- 27.Martensson, I. L., Rolink, A., Melchers, F., Mundt, C., Licence, S. & Shimizu, T. (2002) Semin. Immunol. 14, 335–342. [DOI] [PubMed] [Google Scholar]
- 28.Lin, D. M., Wang, F., Lowe, G., Gold, G. H., Axel, R., Ngai, J. & Brunet, L. (2000) Neuron 26, 69–80. [DOI] [PubMed] [Google Scholar]
- 29.Trinh, K. & Storm, D. R. (2003) Nat. Neurosci. 6, 519–525. [DOI] [PubMed] [Google Scholar]