Abstract
The covalently bound phytochromobilin (PϕB) prosthetic group is required for the diverse photoregulatory activities of all members of the phytochrome family in vascular plants, whereas by contrast, green algal and cyanobacterial phytochromes use the more reduced linear tetrapyrrole pigment phycocyanobilin (PCB). To assess the functional consequence of the substitution of PϕB with PCB in plants, the phytochrome chromophore-deficient hy2 mutant of Arabidopsis was transformed with a constitutively expressed pcyA gene that encodes the cyanobacterial enzyme, PCB:ferredoxin oxidoreductase. Spectroscopic analyses of extracts from etiolated seedlings revealed that PcyA expression restored photoactive phytochrome to WT levels, albeit with blue-shifted absorption maxima, while also restoring light lability to phytochrome A. Photobiological measurements indicated that PcyA expression rescued phytochrome-mediated red high-irradiance responses, low-fluence red/far-red (FR) photoreversible responses, and very-low-fluence responses, thus confirming that PCB can functionally substitute for PϕB for these photoregulatory activities. Although PcyA expression failed to rescue phytochrome A-mediated FR high-irradiance responsivity to that of WT, our studies indicate that the FR high-irradiance response is fully functional in pcyA-expressing plants but shifted to shorter wavelengths, indicating that PCB can functionally complement this phytochrome-mediated response in vascular plants.
Light, the quality, quantity, direction, and duration of which influences nearly every stage of their growth and development, is a critical environmental factor for photosynthetic organisms. In vascular plants, red (R) and far-red (FR) light are primarily sensed by the phytochrome family of photoreceptors (1–4). The phytochrome family in Arabidopsis consists of five members, termed phyA–E (5), whereas monocotyledonous plant species possess as few as three phytochromes, phyA–C orthologs being found in all angiosperms so far examined (6). Dimeric biliproteins, plant phytochromes all possess the linear tetrapyrrole prosthetic group, phytochromobilin (PϕB), that is thioether-linked to a highly conserved N-terminal domain (7). Because of their inability to produce photoactive holophytochromes, plants that are unable to produce PϕB display aberrant photomorphogenesis with pleiotropic phenotypes that are most pronounced under R and FR illumination (8).
PϕB biosynthesis occurs entirely within the plastid compartment via a pathway that shares common intermediates with those of heme and chlorophyll (9, 10). Two enzymes are committed to PϕB biosynthesis, and their corresponding genes have been identified. Heme is first cleaved to biliverdin (BV) IXα by a ferredoxin-dependent heme oxygenase that is encoded by the HY1 locus in Arabidopsis (11–13). BV is subsequently reduced to PϕB by the enzyme PϕB synthase (EC 1.3.7.4), a ferredoxindependent bilin reductase that is encoded by the HY2 locus in Arabidopsis (14). Mutations in both loci yield plants with elongated hypocotyls, fewer leaves, chlorosis, and early flowering owing to the loss of all phytochrome photoactivity (8, 15).
The discovery of the HY2 gene has shed considerable light on the biosynthetic pathway of the phycobilin prosthetic groups of the light harvesting phycobiliproteins in cyanobacteria (16). These studies identified three new families of HY2-related bilin reductases that mediate the conversion of BV to phycocyanobilin (PCB) and phycoerythrobilin (PEB), the precursors of the chromophores of allophycocyanin, phycocyanin, and phycoerythrin (10, 17). PcyA genes encode PCB:ferredoxin oxidoreductases (EC 1.3.7.5), which catalyze the four electron reduction of BV to PCB, whereas the pebA and pebB genes encode bilin reductases involved in the two-step conversion of BV to PEB (16, 18). The unexpected identification of phytochrome-related genes in cyanobacteria led to the discovery of cyanobacterial phytochrome 1 (Cph1) from Synechocystis sp. PCC6803 in 1997 (19, 20). A R/FR photoreceptor with photochemical properties similar to those of plant phytochromes, Cph1 possesses a thioether-linked PCB prosthetic group instead of PϕB that is found in plant phytochromes (21). These studies established that cyanobacterial phytochromes share phycobilin intermediates with the phycobiliprotein antennae, one of many links between cyanobacterial and plant photosensory networks (22).
PCB has long been used as a PϕB analog for plant phytochrome reconstitution experiments (23, 24). It is well established that plant apophytochromes autocatalytically bind PCB as well as PϕB via their intrinsic bilin lyase activity (25–27). PCB-apophytochrome adducts are also photochemically active, like native plant phytochromes, and possess the ability to photointerconvert between the R- and FR-light absorbing Pr and Pfr forms. Owing to the reduced number of double bonds in PCB (Fig. 1A), however, reconstituted PCB-apophytochrome adducts are blue-shifted compared with phytochromes isolated from plants that have a PϕB prosthetic group (23). Interestingly, recent studies have established that PCB is the natural prosthetic group of phytochrome from the green alga Mesotaenium caldariorum (28). Taken together, these studies raised the question of whether vascular plant phytochromes would be functional if their PϕB prosthetic groups were substituted with PCB.
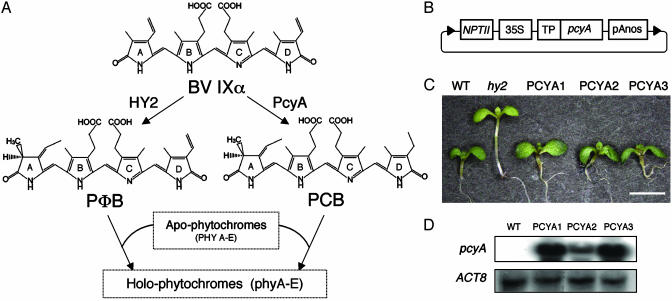
Fig. 1.
Generation of transgenic hy2 lines expressing PCB:ferredoxin oxidoreductase (PcyA). (A) HY2 and cyanobacterial PcyA are ferredoxin-dependent bilin reductases that respectively convert BV IXα to PϕB and PCB. (B) Construct used for PcyA expression in hy2 mutant plants. 35S, cauliflower mosaic viral 35S promoter; NPTII, kanamycin resistance gene; TP, transit peptide from soybean rbcS; pAnos, terminator from nopaline synthase. (C) Ten-day-old seedlings of Ler WT, hy2, and three T3 PcyA-expressing hy2 lines grown under long-day (LD) conditions (16 h of white light at 150 μmol·m-2·s-1 and 8 h of darkness) are shown. (D) RNA blot analysis of total RNA (10 μg) isolated from Ler WT and PCYA transgenic lines by using a Synechocystis pcyA transcript as a probe. The RNA blot was reprobed with an ACT8 transcript to show equal loadings (53).
This question was recently addressed by Hanzawa et al. (29), who fed PCB and other unnatural bilins to chromophore-deficient Arabidopsis plants. Surprisingly, their studies revealed that exogenous PCB and various synthetic PCB analogs rescue most, but not all, of the phytochrome responses in hy1 and hy2 mutant plants. Notable among the responses not restored by PCB feeding was the FR high-irradiance response (FR-HIR), a result implicating the requirement of a PϕB prosthetic group for this phyA-mediated response (29). Owing to the many mitigating factors that complicate pigment feeding experiments (i.e., poor uptake, optical screening, and solvent/adjuvant effects, among others), the present study was undertaken to test the effect of chromophore substitution on the photobiological activities of phytochromes in transgenic plants. Through expression of plastid-targeted PcyA in hy2 mutant plants, we address the hypothesis that the PϕB prosthetic group is required for plant-specific photobiological activities that are not supported by the evolutionarily more ancient PCB prosthetic group.
Materials and Methods
Plant Materials. Arabidopsis thaliana (L.) Heynh. ecotype Landsberg erecta (Ler) was used for all experiments. Mutant strains hy2-1 and phyA-201 were obtained from the Arabidopsis Biological Resource Center at Ohio State University. The phyB-1 line, which lacks the rhg mutation hidden in the original isolate, was kindly provided by M. Tasaka (Nara Institute of Science and Technology). The hy2 host for transformation experiments and for biochemical assays was the null mutant strain, hy2-106 (14). Because of poor seed production of hy2-106, physiological assays for hy2 controls were performed with hy2-1 unless otherwise described.
Plant Transformation. A plant transformation vector, i.e., pKT219, to target any protein of interest to the plastid stroma under regulatory control of the Cauliflower Mosaic Virus (CaMV) 35S promoter containing a dual enhancer region and the tobacco etch viral translational enhancer sequence (30) was generated by introducing a HindIII–SalI fragment from pBS-35S-TP, a plasmid derived from p35S-TPBVR (31), into the corresponding site of the binary plant transformation vector pBIB-Kan (32). The DNA fragment containing the coding region of pcyA from Synechocystis PCC6803 was amplified from Synechocystis pcyA plasmid DNA (16) by PCR with the primers PCYA-F, GGAGATCGATATCGCCGTCACTGAT T TA AG, and PCYA-R, 5′-GGAGATCGAT T TAT TGGATA ACATCAAATA, digested with ClaI, and cloned into the ClaI site of pKT219 to produce plasmid pKT225. Hy2-106 plants were transformed with pKT225 by using the Agrobacterium-mediated vacuum infiltration method (33). Transformants were selected on Murashige and Skoog media containing 30 mg/liter kanamycin. Independent lines carrying the single transgene locus were selected, and homozygous T3 lines were used for these studies.
Phytochrome Spectrophotometric and Protein Assays. Phytochrome spectrophotometric assays were performed essentially as described (34). Partially purified phytochrome extracts were resuspended in 100 mM Tris·HCl (pH 8.3) buffer containing 2 mM EDTA and 25% (vol/vol) ethylene glycol. Phytochrome difference spectra were obtained by using a HP8453 UV-visible spectrophotometer (Agilent Technologies, Palo Alto, CA) on extracts kept at 15°C after saturating R and FR irradiation with an Intralux 6000-1 light source (Volpi AG, Schlieren, Switzerland) filtered with band-pass interference filters (BP-W1 660, Kenko, Tokyo; 660 nm, half bandwidth 7 nm) and a 700-nm cut-off filter (R-72, Kenko), respectively. Total protein concentrations were determined by Bradford assay using the Protein Assay Kit (Bio-Rad) with BSA as a protein standard (35).
Zinc Blot and Immunoblot Analyses. Zinc blot analyses were performed essentially as described (36). For immunoblot analysis, 20 μg of protein extract was separated by SDS/PAGE and electroblotted to poly(vinylidene difluoride) (PVDF) membranes (Bio-Rad). The phyA-specific monoclonal Arabidopsis antibody mAA1 was kindly supplied by A. Nagatani (Kyoto University). The protocol for immunoblot analysis was performed according to the manufacturer's instructions (ECL Plus, Amersham Biosciences).
Germination Assays. Low-fluence (LF) and very-low-fluence (VLF) germination assays were carried out essentially according to Shinomura et al. (37). Monochromatic irradiation was done at the Okazaki large spectrograph (38). Germination frequencies (%) are based on the presence of an emerged radicle. Maximum germination frequency in seed lots was measured by incubating seeds for 4 h under white fluorescent light (50 μmol·m-2·s-1). Light-independent “background” germination frequencies were determined for FR-pretreated seeds incubated in darkness for 5 days.
Hypocotyl Measurements. For measurement of hypocotyl lengths, 80–100 surface sterilized seeds sown on 0.8% agar in MS medium without sucrose were incubated in the dark at 4°C for 2 days and irradiated for 4 h with white fluorescent light (100 μmol·m-2·s-1) to induce uniform germination, and kept in the dark for 24 h. For continuous irradiation experiments, hypocotyl lengths were measured after 5 days' irradiation at 21°C with R light using R-light-emitting diodes (LEDs) (MIL-R18, Sanyo Electric Biomedical, Osaka; peak emission at 657 nm, a half bandwidth in 12 nm, maximum output of 30 μmol·m-2·s-1) or with FR light using FR-LEDs (MIL-F18, Sanyo Electric Biomedical; peak emission at 734 nm, a half bandwidth of 13 nm, maximum output of 50 μmol·m-2·s-1). For intermittent irradiation experiments, seeds were pretreated as described above but were kept in the dark for 48 h, rather than 24 h, before intermittent illumination. Hypocotyl lengths were measured after repeated treatment of preinduced seeds with 3-min cycles composed of 1 min of irradiation with intervals of 2 min of darkness for a 24-h period using the Okazaki large spectrograph followed by 5 days darkness. Hypocotyl length measurements were performed by using a digimatic caliper (CD15C, Mitutoyo, Kawasaki, Japan).
RNA Gel Blot Analysis. Total RNA was isolated from seedlings by using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). RNA blots were performed as described (14). The pcyA DNA fragment from the plasmid construction described above was used as a probe.
Results
PcyA Expression in Transgenic hy2 Mutants Rescues Photomorphogenesis. To substitute PϕB with PCB in planta, the PϕB synthase locus (HY2) must be inactivated and a gene for PCB:ferredoxin oxidoreductase (pcyA) must be incorporated into the plant genome. To accomplish this objective, we expressed Synechocystis PcyA in transgenic hy2 mutant plants that were unable to produce PϕB (Fig. 1 A). Because PcyA is a ferredoxindependent enzyme, our expression plasmid was designed to target the cyanobacterial PcyA protein to the plastid compartment where ferredoxin is primarily localized in plant cells (Fig. 1B). Multiple independent T1 transgenic lines were obtained (13 total), all of which exhibited short hypocotyl and expanded green leaves when grown under white light, phenotypes fully consistent with functional complementation of their phytochrome deficiency (data not shown). For further biochemical and physiological analyses, we isolated several homozygous single insertion lines that we named PCYA1–3. Fig. 1C illustrates the light-grown seedling phenotypes of these 35S:pcyA transgenic lines, the hy2 parent and Ler WT. This comparison indicates that, except for slightly larger cotyledons, seedling photomorphogenesis of all three transgenic lines was basically restored to that of WT. The early flowering phenotype of hy2 under either long- or short-day conditions was also rescued in these lines (data not shown). RNA blot analysis confirms that the complementation of these hy2 mutant phenotype correlates with the expression of the pcyA transgene (Fig. 1D). Although all PCYA transgenic lines were further characterized, the following discussion focuses on PCYA1, which is representative of all three homozygous lines.
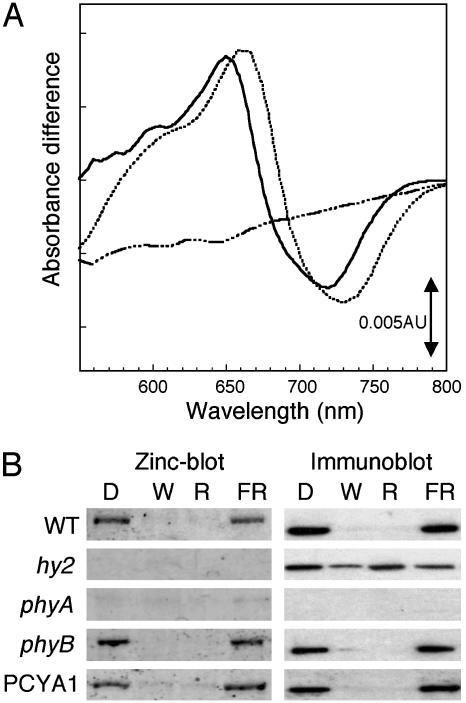
Holophytochrome Levels Are Restored to WT Levels in PCYA1 Transgenic Plants, but Its Spectrum Is Blue-Shifted. To assess whether PcyA expression rescues the phytochrome chromophore deficiency of hy2 mutant plants, extracts from dark grown Ler WT, hy2, and PCYA1 seedlings were analyzed for phytochrome photoactivity (Fig. 2A). Phytochrome difference spectra of dark grown plant extracts primarily reflect phyA, the species of phytochrome that accumulates to high levels in etiolated seedlings (39). As has been well documented in the literature (40), Ler WT extracts exhibited difference maxima and minima at 660 and 730 nm, which correspond to the respective Pr and Pfr forms, whereas no photoreversible phytochrome was detected in hy2 (14). PCYA1 seedling extracts revealed a strong phytochrome difference spectrum with maxima and minima at 650 and 715 nm. Although strikingly blue-shifted from WT, these results indicate that the expression of PcyA restores phytochrome photoactivity close to that of WT seedlings. These observations were fully consistent with earlier in vitro reconstitution experiments with oat PHYA apoprotein that showed that PCB can functionally substitute for PϕB for phytochrome photoactivity (23, 24). To confirm the assumption that PCB was covalently attached to PHYA, we analyzed phytochrome A levels in dark-grown Ler WT, hy2, and PCYA1 plants by using both zinc blot and immunoblot analysis (Fig. 2B). Because zinc blots are selective for bilin-linked proteins (36), the combination of these two methods allows assessment of whether PHYA contains a covalently bound bilin. As positive and negative controls for these analyses, phyB and phyA null mutants were similarly examined. Immunoblot analysis using a PHYA-specific monoclonal antibody showed that PHYA protein was present in dark-grown extracts of all plant lines, except for the phyA mutant (Fig. 2B, lane D). Zinc blot analysis of the same protein extracts showed that PcyA expression fully restored bilin binding to PHYA (compare hy2 and PCYA1 extracts in lane D of Fig. 2B). Taken together, these results indicate that PCYA1 plants synthesize PCB, which becomes covalently attached to PHYA to produce a photochemically active, albeit blue-shifted, holoprotein.
Fig. 2.
Comparative spectroscopic and biochemical properties of phyA-containing extracts from Ler WT, hy2, phyA, phyB, and PCYA1 seedlings. (A) Phytochrome difference spectra of 7-day-old dark-grown Ler WT (dotted line), PCYA1 (solid line), and hy2-106 (dotted–dashed line) seedling extracts. (B) Zinc blot (Left) and phyA immunoblot (Right) analysis of partially purified protein extracts (2 μg per lane) from 5-day-old etiolated seedlings that had been treated for 24 h with darkness (D), white fluorescence light (W; 150μmol·m-2·s-1), R-LED light (657 nm; 20 μmol·m-2·s-1), or FR-LED light (734 nm; 10 μmol·m-2·s-1) before extraction. See Materials and Methods for details.
The PCB Adduct of Arabidopsis PHYA Is Light-Labile. It has long been known that plants possess light-labile and -stable phytochromes, termed type I and type II (41). More recent molecular analysis has shown that the light-labile (type I) phytochrome species can be attributed to phyA, whereas phyB–E are considerably more light-stable (42). The light lability of phyA requires bound chromophore, because the PHYA apoprotein that accumulates in PϕB-deficient plants is considerably more light-stable (8). To assess the stability of PHYA protein in PcyA-expressing hy2 plants, immunoblot analyses were performed by using a PHYA-specific monoclonal antibody (Fig. 2B). These studies revealed that PHYA was markedly destabilized in continuously W- and R-irradiated hy2 mutants expressing PcyA by comparison with hy2 mutant plants (compare hy2 and PCYA1 immunoblots under W and R in Fig. 2B). Our studies show that PcyA expression restored the light lability of PHYA similar to that of Ler WT and phyB mutant controls (Fig. 2B). By comparison with W and R treatments, FR irradiation had little effect on PHYA stability in PCYA1 plants; however, FR irradiation also failed to destabilize phyA holoprotein in WT and phyB plants. These results support the conclusion that PCB binding to PHYA is responsible for its light lability in the hy2 mutant background.
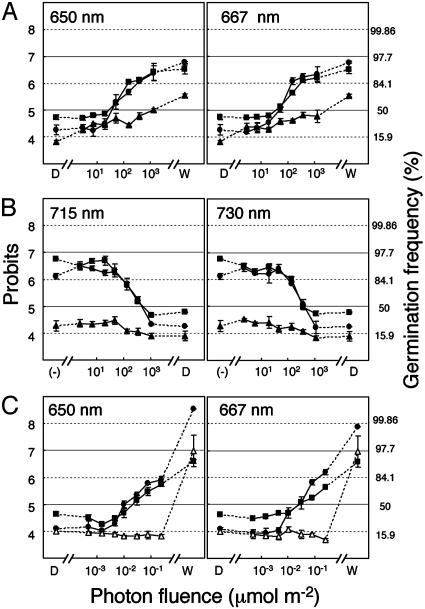
PhyB-Mediated LF and phyA-Mediated VLF Seed Germination Responses Are both Normal in PcyA-Expressing hy2 Mutant Plants. Physiological responses mediated by phytochromes can be classified into three groups according to their light fluence requirements, i.e., VLF response (VLFR), LF response (LFR), and HIR (43). To address whether PcyA expression could restore these responses in the hy2 mutant background, we compared the influence of monochromatic R and FR light on seed germination for Ler WT and PCYA1 transgenics by using protocols that can distinguish between the two phytochrome-mediated responses (44, 45). LF-dependent seed germination was first monitored by measuring the frequency of radicle emergence after an immediate postinhibition R pulse and 5 additional days in darkness (Fig. 3A). Inhibition of R-induced germination by subsequent FR irradiation was similarly evaluated (Fig. 3B). For these studies, we used monochromatic light pulses corresponding to the absorption maxima of the Pr and Pfr forms, i.e., 667 and 730 nm for native phytochromes; 650 and 715 nm for PCB-apophytochrome adducts. Fluence-response measurements showed that the LFR inductive response at both R wavelengths (Fig. 3A) and the FR reversal at both FR wavelengths (Fig. 3B) were identical for PCYA1 and Ler WT plants. As controls, we similarly analyzed a phyB mutant that is strongly impaired in both responses. This comparison clearly showed that PCYA plants possessed fully functional LF-dependent seed-germination responses. VLF-dependent germination responses were next analyzed. For these measurements, preimbibed seeds were first incubated in darkness to accumulate large amounts of phyA (44). As shown in Fig. 3C, VLFR curves were superimposable for Ler WT and PCYA1 plants grown under R, at both 650 and 667 nm. PhyA mutants were similarly analyzed to show that this response is phyA-specific. Taken together, these results support the conclusion that PCB-adducts of both PHYA and PHYB possess “nearly full” biological activity with respect to their respective VLF- and LF-dependent seed-germination responses.
Fig. 3.
Comparative LF- and VLF-dependent seed-germination responses. (A) LF R-mediated induction of seed germination. (B) LF FR-mediated reversal of R-mediated seed germination. Germination frequency by R treatment without FR-mediated reversal is indicated by (-). (C) VLF-mediated seedgermination responses. Ler WT (filled circles), PCYA1 (filled squares), phyA (open triangles), and phyB (filled triangles). Maximum germination frequency under the white light condition and light-independent background germination frequency in the dark are indicated by W and D, respectively. Values are means ± SE of three replicates. The percentage of germinating seeds was transformed by using probits (54). See Materials and Methods for details.
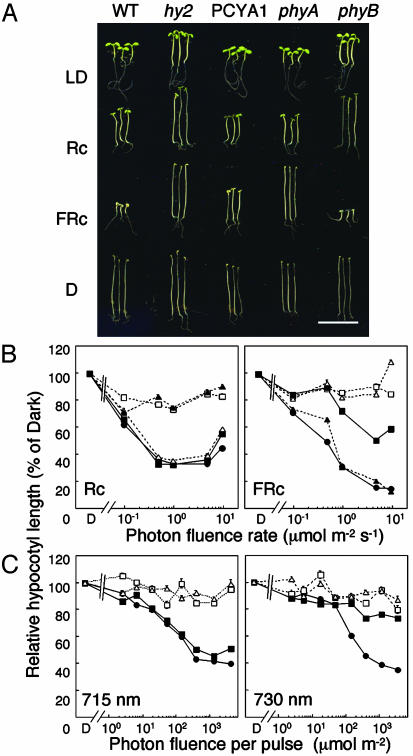
R and FR-HIR-Dependent Inhibition of Hypocotyl Elongation Are both Rescued by PcyA Expression, but the FR-HIR Is Altered. HIRs are best typified by the light-dependent inhibition of hypocotyl elongation under continuous illumination (46). Two types of HIRs have been described in plants: FR-HIR, which is phyA-mediated, and phyB-mediated R-HIR (47). Hypocotyl growth responses were compared in Ler WT and PCYA1 plants, and as controls in hy2, phyA, and phyB mutants. Photographs of seedlings grown under various light regimes and in darkness are shown in Fig. 4A. These measurements show that dark-grown PCYA1 seedlings had elongated hypocotyls, similar to Ler WT, indicating that cell elongation was not affected by expression of PcyA. PCYA1 seedlings also exhibited WT hypocotyl growth inhibition when grown under LD conditions or Rc, suggesting that phyB functioned normally with a PCB chromophore. Hy2 and phyB mutants, as expected, were insensitive to Rc and possessed elongated hypocotyls under LD (Fig. 4A). In contrast with the results of LD and Rc experiments, PCYA1 seedlings were partially elongated under “standard” FRc illumination when compared with FRc-grown Ler WT seedlings. Because PCYA1 seedlings were not as elongated as hy2 or phyA mutant seedlings, these results implicated incomplete complementation of this phyA-mediated response. To more thoroughly assess both HIRs, the effect of continuous irradiation was monitored under different fluence rate conditions (Fig. 4B). Under Rc, we observed no differences in fluence rate-response curves for hypocotyl elongation between Ler WT and PCYA1. By contrast, PCYA1 seedlings were clearly less sensitive to FRc. In this regard, hypocotyl lengths of PCYA1 seedlings were longer than Ler WT even under maximum fluence rates tested (10 μmol·m-2·s-1). Because the PCB-PHYA adduct had blue-shifted Pr and Pfr forms as described above, we hypothesized that this apparent reduced sensitivity to FRc might reflect a blue-shifted action spectrum. We therefore examined inhibition of hypocotyl elongation under monochromatic FR at both 715 and 730 nm wavelengths. For these measurements, intermittent FR pulses for 24 h were used because Shinomura et al. (47) established that the Bunsen–Roscoe law of reciprocity was held for each FR pulse. Fig. 4C illustrates the fluence-response curves when the 3-min cycles of 1-min FR irradiation were followed by 2 min of darkness for 24 h. These results show that hypocotyl elongation of PCYA1 was strongly inhibited by a 715-nm irradiation but not by a 730-nm irradiation. By comparison, Ler WT seedlings strongly responded to both 715- and 730-nm irradiation. Taken together with the responses of phyA and phyB mutant controls shown in Fig. 4, these results indicate that the FR-HIR is functional in PCYA1 plants but is shifted to shorter wavelengths. In contrast to phyB R-HIR that appears normal in PCYA1 plants, chromophore substitution of PCB for PϕB profoundly influences the wavelength selectivity of the phyA-mediated FR-HIR.
Fig. 4.
R- and FR-dependent high-irradiance inhibition of hypocotyl elongation responses. (A) Comparative seedling growth phenotypes of 6-day-old seedlings grown under LD conditions (16 h of white light at 150 μmol·m-2·s-1; 8 h dark), Rc (continuous R irradiation at 20 μmol·m-2·s-1), FRc (continuous FR irradiation at 10 μmol·m-2·s-1), or darkness (D). (Scale bar, 10 mm.) (B) Comparative photon fluence rate-response curves for hypocotyl growth of 6-day-old Ler WT (filled circle), PCYA1 (filled squares), hy2 (open squares), phyA (open triangles), and phyB (filled triangles) seedlings grown under Rc (continuous irradiation of R-LED light at 657 nm) and FRc (continuous irradiation with FR-LED light at 734 nm). (C) Photon fluence rate-response curves for intermittent irradiation with monochromatic FR at 715 and 730 nm. Relative hypocotyl lengths for B and C were calculated as the percent of dark-grown seedling lengths. See Materials and Methods for details.
Discussion
In this investigation, we show that the phytochrome chromophore substitution of PCB for PϕB was achieved by plastidtargeted expression of cyanobacterial PcyA in the hy2 mutant of Arabidopsis. The observed complementation of low-fluency, very-low-fluency, and red high-irradiance responses indicates that the PcyA enzyme is active, yielding a PCB chromophore precursor that attaches to all apophytochromes present. The blue-shifted difference spectrum of phyA in dark-grown PCYA1 transgenic plants is in good agreement with PCB adducts of apophytochromes produced in vitro (24, 26). Our studies show that it is this blue shift that is responsible for the altered sensitivity of PCYA plants to high-irradiance FRc, rather than an intrinsic difference in the biological activity of the PCB-PHYA adduct compared with native phyA. Indeed, the reduced FRc sensitivity of PCB-treated and PCB analog-treated hy1 and hy2 plants (29) may reflect the same phenomenon.
The above line of reasoning also predicts that phyA loss-offunction screens using long-wavelength FR light sources would recover novel alleles of phyA with blue-shifted spectrophotometric properties. The phyA Lm-2 allele that has a single point mutation, i.e., M548T, in a region adjacent to the chromophore binding site may be one such allele (48). In the same study, spectroscopic analysis supporting this hypothesis was provided by the observation that introduction of this M548T mutation into oat PHYA shifts the absorption spectrum of the PϕB adduct to shorter wavelengths. The occurrence of the Lm-2 allele of phyA in natural populations of Arabidopsis implicates spectral tuning of phytochromes to be of ecological significance, especially for seedlings grown under FR-enriched canopy shade (49). PCB substitution for PϕB also enabled us to distinguish between phyA-mediated VLFR and FR-HIR, as has also been reported for another mutant allele of phyA (50). It is therefore likely that spectral tuning of phytochromes has been going on for millions of years, caused by mutations in apoprotein and various chromophore substitutions, and that many novel spectrally shifted mutant alleles of phytochromes will be uncovered in years to come. Based on the occurrence of PCB as the chromophore of phytochrome from the green alga M. caldariorum (28), it appears that PCB was replaced by PϕB in the plant lineage sometime after green algae diverged from vascular plants. Although we can speculate that the reason for this replacement involved an adaptative advantage that PϕB imparted to phytochromes found in land plant species, we must leave this question to be answered by plant ecologists.
Our studies show that the phenotypic consequence of the blue-shifted spectra of the PCB-apophytochrome adducts was most pronounced for the phyA-mediated FR-HIR. More detailed VLFR, LFR, and HIR action spectra over a wider range of wavelengths will be needed to fully determine whether there are quantitative differences between phyA- and phyB-mediated responses in Ler WT with corresponding responses of their unnatural PCB adducts in PCYA1. Because PCB adducts of phyB will be blue-shifted, we expect that the action spectra for phyB-mediated responses in PcyA-expressing plants will also reflect this spectral shift. Such action spectra could be performed by using PcyA-expressing hy2 transgenic plants in which one or more phytochromes are missing. By comparing the phenotypes of these transgenic plants with phytochrome-deficient mutant plants in HY2 backgrounds, the photobiological activities of individual native phytochromes can be quantitatively compared with those of their PCB-adduct counterparts. Although the present studies did not reveal gain-of-function phenotypes due to constitutive and/or ectopic expression of PcyA, these investigations may uncover such effects. Such analyses will be particularly interesting vis à vis the possible alteration of plastid-nuclear signaling, in view of the well established regulatory role of tetrapyrroles in this response (51, 52).
Acknowledgments
We thank Hiroko Hanzawa and Masaki Furuya for valuable advice for the physiological studies; Akira Nagatani for providing the monoclonal phyA antibody; Norio Murata and Masakatsu Watanabe for hosting our studies at the National Institute of Basic Biology; Shoichi Higashi for help with operation of the Okazaki Large Spectrograph; and Naoko Iwata, Toshikazu Kobayashi, Ayumi Kagawa, and Junko Kamikubo for technical assistance. This work was supported by Research for the Future Program 00L01605 from the Japan Society for the Promotion of Science (to T.K.), National Institute of Basic Biology Cooperative Program for the Okazaki Large Spectrograph Grant 03-512 (to T.K.), Ministry of Agriculture, Forestry, and Fisheries of Japan Rice Genome Project Grants SY1108 and IP1006 (to T.S.), and Grant AMD-0103397 from the U.S. Department of Agriculture Competitive Research Grant Initiative (to J.C.L.).
Abbreviations: BV, biliverdin; FR, far-red; FRc, continuous FR irradiation; HIR, highirradiance response; LD, long-day; LED, light-emitting diode; LF, low-fluence; LFR, LF response; PCB, phycocyanobilin; PϕB, phytochromobilin; R, red; Rc, continuous R irradiation; VLF, very-low-fluence; VLFR, VLF response.
References
- 1.Smith, H. (2000) Nature 407, 585–591. [DOI] [PubMed] [Google Scholar]
- 2.Fankhauser, C. (2001) J. Biol. Chem. 276, 11453–11456. [DOI] [PubMed] [Google Scholar]
- 3.Quail, P. H. (2002) Nat. Rev. Mol. Cell Biol. 3, 85–93. [DOI] [PubMed] [Google Scholar]
- 4.Casal, J. J., Luccioni, L. G., Oliverio, K. A. & Boccalandro, H. E. (2003) Photochem. Photobiol. Sci. 2, 625–636. [DOI] [PubMed] [Google Scholar]
- 5.Clack, T., Mathews, S. & Sharrock, R. A. (1994) Plant Mol. Biol. 25, 413–427. [DOI] [PubMed] [Google Scholar]
- 6.Mathews, S. & Sharrock, R. A. (1997) Plant Cell Environ. 20, 666–671. [Google Scholar]
- 7.Lagarias, J. C. & Rapoport, H. (1980) J. Am. Chem. Soc. 102, 4821–4828. [Google Scholar]
- 8.Terry, M. J. (1997) Plant Cell Environ. 20, 740–745. [Google Scholar]
- 9.Terry, M. J., Wahleithner, J. A. & Lagarias, J. C. (1993) Arch. Biochem. Biophys. 306, 1–15. [DOI] [PubMed] [Google Scholar]
- 10.Frankenberg, N. F. & Lagarias, J. C. (2003) in The Porphyrin Handbook, eds. Kadish, K. M., Smith, K. M. & Guilard, R. (Academic, New York), Vol. 13, pp. 211–235. [Google Scholar]
- 11.Muramoto, T., Kohchi, T., Yokota, A., Hwang, I. H. & Goodman, H. M. (1999) Plant Cell 11, 335–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis, S. J., Kurepa, J. & Vierstra, R. D. (1999) Proc. Natl. Acad. Sci. USA 96, 6541–6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muramoto, T., Tsurui, N., Terry, M. J., Yokota, A. & Kohchi, T. (2002) Plant Physiol. 130, 1958–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kohchi, T., Mukougawa, K., Frankenberg, N., Masuda, M., Yokota, A. & Lagarias, J. C. (2001) Plant Cell 13, 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koornneef, M., Rolff, E. & Spruit, C. J. P. (1980) Z. Pflanzenphysiol. 100, 147–160. [Google Scholar]
- 16.Frankenberg, N., Mukougawa, K., Kohchi, T. & Lagarias, J. C. (2001) Plant Cell 13, 965–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beale, S. I. (1993) Chem. Rev. 93, 785–802. [Google Scholar]
- 18.Frankenberg, N. & Lagarias, J. C. (2003) J. Biol. Chem. 278, 9219–9226. [DOI] [PubMed] [Google Scholar]
- 19.Hughes, J., Lamparter, T., Mittmann, F., Hartmann, E., Gartner, W., Wilde, A. & Borner, T. (1997) Nature 386, 663. [DOI] [PubMed] [Google Scholar]
- 20.Yeh, K.-C., Wu, S.-H., Murphy, J. T. & Lagarias, J. C. (1997) Science 277, 1505–1508. [DOI] [PubMed] [Google Scholar]
- 21.Hubschmann, T., Borner, T., Hartmann, E. & Lamparter, T. (2001) Eur. J. Biochem. 268, 2055–2063. [DOI] [PubMed] [Google Scholar]
- 22.Montgomery, B. L. & Lagarias, J. C. (2002) Trends Plant Sci. 7, 357–366. [DOI] [PubMed] [Google Scholar]
- 23.Elich, T. D. & Lagarias, J. C. (1989) J. Biol. Chem. 264, 12902–12908. [PubMed] [Google Scholar]
- 24.Wahleithner, J. A., Li, L. & Lagarias, J. C. (1991) Proc. Natl. Acad. Sci. USA 88, 10387–10391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lagarias, J. C. & Lagarias, D. M. (1989) Proc. Natl. Acad. Sci. USA 86, 5778–5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, L. & Lagarias, J. C. (1992) J. Biol. Chem. 267, 19204–19210. [PubMed] [Google Scholar]
- 27.Li, L., Murphy, J. T. & Lagarias, J. C. (1995) Biochemistry 34, 7923–7930. [DOI] [PubMed] [Google Scholar]
- 28.Wu, S.-H., McDowell, M. T. & Lagarias, J. C. (1997) J. Biol. Chem. 272, 25700–25705. [DOI] [PubMed] [Google Scholar]
- 29.Hanzawa, H., Shinomura, T., Inomata, K., Kakiuchi, T., Kinoshita, H., Wada, K. & Furuya, M. (2002) Proc. Natl. Acad. Sci. USA 99, 4725–4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carrington, J. C., Freed, D. D. & Oh, C.-C. (1990) EMBO J. 9, 1347–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagarias, D. M., Crepeau, M. W., Maines, M. D. & Lagarias, J. C. (1997) Plant Cell 9, 675–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Becker, D. (1990) Nucleic Acids Res. 18, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bechtold, N., Ellis, J. & Pelletier, G. (1993) C. R. Acad. Sci. Ser. III 316, 1194–1199. [Google Scholar]
- 34.Nagatani, A., Reed, J. W. & Chory, J. (1993) Plant Physiol. 102, 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bradford, M. M. (1976) Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 36.Berkelman, T. R. & Lagarias, J. C. (1986) Anal. Biochem. 156, 194–201. [DOI] [PubMed] [Google Scholar]
- 37.Shinomura, T., Nagatani, A., Chory, J. & Furuya, M. (1994) Plant Physiol. 104, 363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watanabe, M., Furuya, M., Muiyoshi, Y., Inoue, Y., Iwahashi, I. & Matsumoto, K. (1982) Photochem. Photobiol. 36, 491–498. [Google Scholar]
- 39.Pratt, L. H. (1995) Photochem. Photobiol. 61, 10–21. [Google Scholar]
- 40.Hanzawa, H., Inomata, K., Kinoshita, H., Kakiuchi, T., Jayasundera, K. P., Sawamoto, D., Ohta, A., Uchida, K., Wada, K. & Furuya, M. (2001) Proc. Natl. Acad. Sci. USA 98, 3612–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Furuya, M. (1989) Adv. Biophys. 25, 133–167. [DOI] [PubMed] [Google Scholar]
- 42.Sharrock, R. A. & Clack, T. (2002) Plant Physiol. 130, 442–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Furuya, M. & Schafer, E. (1996) Trends Plant Sci. 1, 301–307. [Google Scholar]
- 44.Shinomura, T., Nagatani, A., Hanzawa, H., Kubota, M., Watanabe, M. & Fururya, M. (1996) Proc. Natl. Acad. Sci. USA 93, 8129–8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Botto, J. F., Sanchez, R. A., Whitelam, G. C. & Casal, J. J. (1996) Plant Physiol. 110, 439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quail, P. H., Boylan, M. T., Parks, B. M., Short, T. W., Xu, Y. & Wagner, D. (1995) Science 268, 675–680. [DOI] [PubMed] [Google Scholar]
- 47.Shinomura, T., Uchida, K. & Furuya, M. (2000) Plant Physiol. 122, 147–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maloof, J. N., Borevitz, J. O., Dabi, T., Lutes, J., Nehring, R. B., Redfern, J. L., Trainer, G. T., Wilson, J. M., Asami, T., Berry, C. C., et al. (2001) Nat. Genet. 29, 441–446. [DOI] [PubMed] [Google Scholar]
- 49.Ballare, C. L. (1999) Trends Plant Sci. 4, 97–102. [DOI] [PubMed] [Google Scholar]
- 50.Yanovsky, M. J., Luppi, J. P., Kirchbauer, D., Ogorodnikova, O. B., Sineshchekov, V. A., Adam, E., Kircher, S., Staneloni, R. J., Schafer, E., Nagy, F. & Casal, J. J. (2002) Plant Cell 14, 1591–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mochizuki, N., Brusslan, J. A., Larkin, R., Nagatani, A. & Chory, J. (2001) Proc. Natl. Acad. Sci. USA 98, 2053–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strand, A., Asami, T., Alonso, J., Ecker, J. R. & Chory, J. (2003) Nature 421, 79–83. [DOI] [PubMed] [Google Scholar]
- 53.An, Y.-Q., McDowell, J. M., Huang, S., McKinney, E. C., Chambliss, S. & Meagher, R. B. (1996) Plant J. 10, 107–121. [DOI] [PubMed] [Google Scholar]
- 54.Finney, D. J. (1952) Probit Analysis (Cambridge Univ. Press, Cambridge, U.K.), 2nd Ed.