How and where are virus particles assembled in an infected cell? To address this question, virologists are studying retroviruses and lentiviruses, enveloped viruses that contain RNA as their genetic material. The only viral protein that is needed to make a virus-like particle is the Gag protein (1, 2). In the lentivirus HIV-1, Gag is synthesized as a cytosolic 55-kDa precursor (Pr55gag) that contains four domains: p17 matrix (MA), p24 capsid (CA), p7 nucleocapsid (NC), and p6 (Fig. 1A). The newly synthesized Gag precursor targets to the plasma membrane in most cells, where it multimerizes by means of its CA and NC domains into large arrays that direct particle assembly and budding. How does HIV-1 Gag associate with cellular membranes? The membrane-binding (M) domain maps to the N-terminal region of MA. For the M domain to function, it must be covalently modified at its N terminus by myristate, a 14-carbon saturated fatty acid (3). Nearly all mammalian retroviral Gag proteins are myristoylated (4). However, myristoylation alone is not sufficient to promote stable membrane binding of proteins (5, 6). A second signal, which in the case of HIV-1 Gag is a cluster of basic residues within the M domain, synergizes with myristate to promote tight membrane binding (7). Structural studies of non-myristoylated forms of MA have revealed that part of the M domain forms an amphipathic β-pleated sheet, with the basic residues oriented on the top surface (8–10). Here they would be positioned to interact electrostatically with negatively charged headgroups of acidic phospholipids in the membrane. But where is the myristate? The article by Tang et al. in this issue of PNAS (11) now provides structural information for myrMA that reveals how the multimerization state of Gag regulates the orientation of the myristate moiety and Gag membrane binding during the virus life cycle.
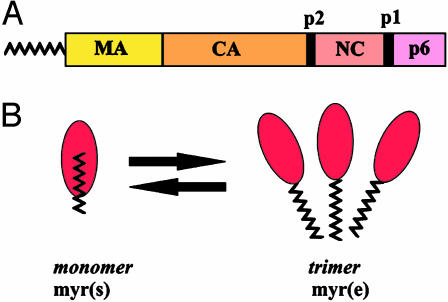
Fig. 1.
Structure of the HIV-1 Gag protein. (A) A schematic representation of Pr55gag and its sub-domains. (B) The entropic myristoyl switch for myrMA as proposed by Tang et al. (11). Note that ∼40% of the myristate moiety remains exposed in the myr(s) state.
As the virion matures, Gag is cleaved by the viral protease to generate mature cleavage products. Despite the fact that both Gag and MA are efficiently myristoylated, their membrane-binding properties are quite different: the Gag precursor binds well to membranes, whereas MA binds poorly (12). To explain the differential membrane binding of MA and Gag, it was proposed that accessibility of the myristate moiety is regulated by a “myristoyl switch” mechanism: myristate would be exposed in Gag but sequestered in the context of MA (12). Biochemical support for the myristoyl switch as a mechanism for regulation of Gag membrane binding has accumulated in the literature. For example, C-terminal truncations restore membrane binding to MA, presumably by flipping myristate “out” (12, 13). Conversely, cleavage of Gag by HIV-1 protease triggers the myristoyl switch “in,” thereby releasing MA from the membrane (14). Amino acid mutations near the N terminus of MA that block membrane binding can be suppressed by second site mutations downstream in MA (15, 16). Some of these mutants exhibit enhanced membrane binding and particle production compared with wild-type Gag (15, 17). All of these studies are consistent with the existence of a myristoyl switch for HIV-1 Gag, but until now confirmation from structural biology studies was lacking.
The new study by Tang et al. (11) provides direct support for a myristoyl switch model by demonstrating the existence of two states of myristoylated MA (myrMA): myr-exposed [myr(e)] and myr-sequestered [myr(s)]. Thus, MA joins a list of other myristoylated proteins where two state myristoylated structures have been solved: ADP ribosylation factor (ARF), recoverin, and c-Abl (18–20). Several findings make the Tang et al. (11) study particularly noteworthy and distinct from other reports describing myristoyl–protein structures. First, the tertiary structures of myr(e) and myr(s) MA are nearly identical. Unlike other myristoyl–protein structures, myrMA does not undergo a dramatic conformational change when switching between states. The myristate moiety inserts into a preexisting cavity and makes contact mostly with amino acids in the N-terminal half of MA. Chemical-shift data indicate that only a few residues, primarily Ser-9, Gly-10, and Gly-11, have altered conformations in the myr(e) vs. myr(s) states. In fact, mutation of these or adjacent residues has previously been shown to reduce Gag membrane binding, implying that this region is responsible for regulating the myristoyl switch.
The second unique feature of the myrMA structure is the mode of regulation of the myristoyl switch. Sedimentation equilibrium studies revealed that myrMA exists in an equilibrium between monomeric and trimeric states. This is consistent with previous crystal structures of non-myrMA that have yielded both monomers and trimers. The striking finding reported here is that the monomer is in the myr(s) state, whereas the trimer is in the myr(e) state, implying that multimerization promotes exposure of the myristate (Fig. 1B). One would therefore predict that addition of the CA domain, which contains additional multimerization signals, should promote myristate exposure at lower protein concentrations than myrMA. This is exactly what Tang et al. (11) found. The monomer ↔ trimer equilibrium constant is 20 times higher for myrMA-CA than for myrMA. Thus, we can conclude that Gag–Gag interactions drive an entropic switch that shifts the myr(s) ↔ myr(e) equilibrium toward the myr(e) state.
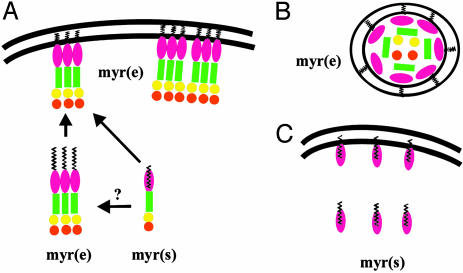
How is the myristoyl switch regulated during the HIV-1 life cycle? Consider the late stages of infection, where the nascent Gag polypeptide chain needs to have myristate exposed to bind to the plasma membrane and promote assembly (Fig. 2). At least two forces could provide the impetus for multimerization. Tethering Gag to the viral RNA template by means of the NC domain would serve to increase Gag–Gag interactions. In addition, association of Gag with lipid raft-like domains would increase the local protein concentration in the plane of the lipid bilayer and promote further multimerization (21, 22). Gag oligomers would then direct particle assembly at the membrane. During or shortly after virion release, the viral protease is activated, and Gag is cleaved into its mature domain substituents. The cramped quarters of the virus particle maintain MA in a high concentration (estimates range from 2 to 14 mM) and thereby likely keep it in a membrane-bound trimeric state (11, 14). However, once the virus infects a new cell, the contents of the particle are exposed to and diluted by the infected cell cytosol. As a result, myrMA shifts to the myr(s) state and is released from the membrane (Fig. 2).
Fig. 2.
Regulation of the myristoyl switch during the HIV-1 life cycle. A schematic representation of Gag is shown at the late stages of infection (A), in the virion (B), and in the newly infected cell (C).
The Tang et al. (11) study provides an elegant example of how a single viral protein can accomplish so much with so little. It explains how multimerization can enhance the efficiency of membrane binding and how maturation by proteolytic cleavage can cause release from the membrane by reversing the myristoyl switch. Yet a number of questions still remain: (i) When and where does multimerization of nascent Gag polypeptides begin, in the cytosol or at the membrane? Electron microscopy and biochemical studies suggest that newly synthesized Gag is present in small cytosolic complexes and that large-scale multimerization occurs at the membrane (23, 24). These data imply that newly synthesized Gag exists in the cytosol in a myr(s) state or perhaps as a small micelle of myr(e) Gag multimers. (ii) How does the orientation of myristate and the Gag domains change when cellular membranes are present? Ten of the 14 methylene groups of myristate typically penetrate into the lipid bilayer (5), so myr–myr interactions may be altered when Gag is membrane-bound. (iii) Will the structure of full-length myristoylated Pr55Gag, when it is ultimately solved, yield additional surprises? (iv) Are other retroviral and lentiviral Gag proteins also regulated by an entropic myristoyl switch? (v) Approximately 0.5% of all proteins in the human genome are myristoylated (25). Thus, are there cellular myristoylated proteins whose membrane binding may be regulated by an entropic switch? Further biochemical and structural studies should shed light on these issues.
See companion article on page 517.
References
- 1.Freed, E. O. (1998) Virology 251, 1–15. [DOI] [PubMed] [Google Scholar]
- 2.Garnier, L., Bowzard, J. B. & Wills, J. W. (1998) AIDS 12, S5–S16. [PubMed] [Google Scholar]
- 3.Gottlinger, H. G., Sodroski, J. G. & Haseltine, W. A. (1989) Proc. Natl. Acad. Sci. USA 86, 5781–5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Resh, M. D. (1999) Biochim. Biophys. Acta 1451, 1–16. [DOI] [PubMed] [Google Scholar]
- 5.Peitzsch, R. M. & McLaughlin, S. (1993) Biochemistry 32, 10436–10443. [DOI] [PubMed] [Google Scholar]
- 6.Resh, M. D. (1994) Cell 76, 411–413. [DOI] [PubMed] [Google Scholar]
- 7.Zhou, W., Parent, L. J., Wills, J. W. & Resh, M. D. (1994) J. Virol. 68, 2556–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Massiah, M. A., Starich, M. R., Paschall, C., Summers, M. F., Christensen, A. M. & Sundquist, W. I. (1994) J. Mol. Biol. 244, 198–223. [DOI] [PubMed] [Google Scholar]
- 9.Matthews, S., Barlow, P., Boyd, J., Barton, G., Russell, R., Mills, H., Cunningham, M., Meyers, N., Burns, N., et al. (1994) Nature 370, 666–668. [DOI] [PubMed] [Google Scholar]
- 10.Hill, C. P., Worthylake, D., Bancroft, D. P., Christensen, A. M. & Sundquist, W. I. (1996) Proc. Natl. Acad. Sci. USA 93, 3099–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang, C., Loeliger, E., Luncsford, P., Kinde, I., Beckett, D. & Summers, M. F. (2004) Proc. Natl. Acad. Sci. USA 101, 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou, W. & Resh, M. D. (1996) J. Virol. 70, 8540–8548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spearman, P., Horton, R., Ratner, L. & Kuli-Zade, I. (1997) J. Virol. 71, 6582–6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hermida-Matsumoto, L. & Resh, M. D. (1999) J. Virol. 73, 1902–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reil, H., Bukovsky, A. A., Gelderblom, H. R. & Gottlinger, H. G. (1998) EMBO J. 17, 2699–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ono, A. & Freed, E. O. (1999) J. Virol. 73, 4136–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paillart, J.-C. & Gottlinger, H. G. (1999) J. Virol. 73, 2604–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldberg, J. (1998) Cell 95, 237–248. [DOI] [PubMed] [Google Scholar]
- 19.Ames, J. B., Ishima, R., Tanaka, T., Gordon, J. I., Stryer, L. & Ikura, M. (1997) Nature 389, 198–202. [DOI] [PubMed] [Google Scholar]
- 20.Hantschel, O., Nagar, B., Guettler, S., Kretzschmar, J., Dorey, K., Kuriyan, J. & Superti-Furga, G. (2003) Cell 112, 845–857. [DOI] [PubMed] [Google Scholar]
- 21.Sandefur, S., Varthakavi, V. & Spearman, P. (1998) J. Virol. 72, 2723–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindwasser, O. W. & Resh, M. D. (2001) J. Virol. 75, 7913–7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tritel, M. & Resh, M. D. (2000) J. Virol. 74, 5845–5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nermut, M. V., Zhang, W. H., Francis, G., Ciampor, F., Morikawa, Y. & Jones, I. M. (2003) Virology 305, 219–227. [DOI] [PubMed] [Google Scholar]
- 25.Maurer-Stroh, S., Eisenhaber, B. & Eisenhaber, F. (2002) J. Mol. Biol. 317, 541–557. [DOI] [PubMed] [Google Scholar]