Abstract
The binding site of the dopamine D2 receptor (D2R), like those of homologous rhodopsin-like G protein-coupled receptors (GPCRs) that bind small molecules, is contained within a water-accessible crevice formed among its seven transmembrane segments (TMs). The high-resolution structure of bovine rhodopsin, however, revealed that the second extracellular loop (E2), which connects TM4 and TM5, folds down into the transmembrane domain and forms part of the ligand-binding surface for retinal. Whether E2 plays a related role in other rhodopsin-like GPCRs is unclear. To address this issue, we have now mutated to cysteine, one at a time, 10 consecutive residues in E2 of D2R. The reaction of five of these mutants with sulfhydryl reagents inhibited antagonist binding, and bound antagonist protected two, I184C and N186C, from reaction. The pattern of accessibility in E2 is consistent with a structure similar to that of bovine rhodopsin, in which the region C-terminal to the conserved disulfide bond is deeper in the binding-site crevice than is the N-terminal part of E2. Thus, E2 likely contributes to the binding site in the D2R and probably in other aminergic GPCRs as well. Knowledge of its detailed positioning and interactions with ligand would benefit GPCR molecular modeling and facilitate the design of novel drugs.
The extracellular loops are important in ligand binding in G protein-coupled receptors (GPCRs) with large molecular weight ligands, such as peptides (1). The role of these loops in aminergic GPCRs that bind small ligands has received much less attention, and it is widely believed that the transmembrane domain (TMD) is sufficient for ligand binding in these receptors (1). In the high-resolution bovine rhodopsin structure, however, the second extracellular loop (E2) folds down into the binding-site crevice to form a lid over retinal (2). It is unknown in aminergic GPCRs whether the E2 structure is similar to that of rhodopsin or whether E2 plays a role in ligand binding.
For nearly all rhodopsin-like GPCRs, the disulfide bond between Cys3.25 [Cys-107 in dopamine D2 receptor (D2R)] and the conserved Cys in E2 (Cys_e2, Cys-182 in D2R) connects E2 with the TMD§ (Fig. 1A), and this disulfide bond (SS-E2) is crucial to the structural integrity and function of many GPCRs. The removal of SS-E2 by mutagenesis severely disrupts ligand binding to muscarinic acetylcholine receptors (3, 4) and destabilizes the high-affinity state of the β2 adrenergic receptor (AR) (5). Moreover, antagonist protected the β2AR from the effects of reduction by DTT (6). Thus, SS-E2 is protected by a conformational change or steric block within the binding site.
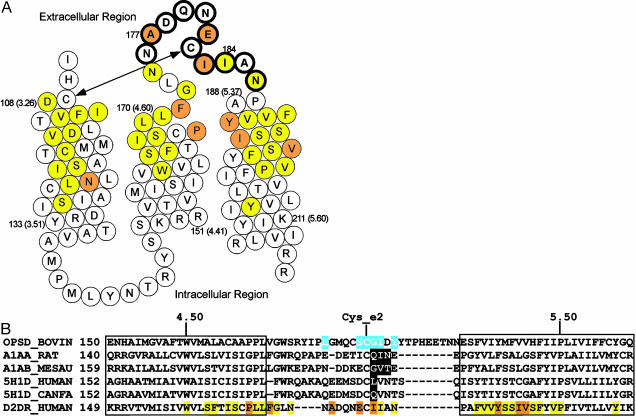
Fig. 1.
(A) Helical net representations of the residues in the TM3, TM4, E2, and TM5 of the D2R, summarizing the effects of all of the MTS reagents (see Fig. 2) on [3H]N-methylspiperone binding. Open circles indicated that these regents had no significant effect on binding; yellow or orange circles indicate that these reagents significantly inhibited binding; and yellow circles indicate sulpiride (TM3, TM4, and TM5) or YM-09151-2 (E2) protected residues. The conserved disulfide bond between TM3 and E2 (SS-E2) is indicated by an arrow. (B) Sequence alignment of D2R with receptors that have E2 residues found to be important in ligand binding (see Discussion). The retinal contact residues in the rhodopsin structure are shown highlighted in blue, and the residues found to be important in ligand binding in other receptors are shown highlighted in black. The color coding used for the D2R residues is the same as in A.
Although it had been argued that the presence of E2 within the TMD may be a feature unique to rhodopsin (7, 8), several reports implicate E2 in ligand specificity in aminergic and other small molecule-ligand GPCRs (reviewed in ref. 9 and Discussion). Our studies of transmembrane segment 4 (TM4) of the D2R also suggested a role for the extracellular end of TM4 and the N-terminal part of E2 in forming the binding-site crevice (10). Based on the length of E2 and the spatial constraints imposed by the conserved SS-E2, we proposed that E2 may enter into the binding-site crevice of aminergic and other rhodopsin-like GPCRs that have small molecule ligands (9).
In this study, we use the substituted-cysteine accessibility method (SCAM) (11) to identify systematically residues in E2 (Asn-176 to Asn-186) of the D2R that contribute to the binding-site crevice. Previously, we used SCAM to identify the residues in TM1–TM7 that form the surface of the binding-site crevice in the human D2R (reviewed in ref. 12). Consecutive residues are mutated to cysteine, one at a time, and the mutant receptors are expressed in heterologous cells. If ligand binding to a cysteine-substitution mutant is near normal, we assume that the structure of the mutant receptor is similar to that of WT and that the substituted cysteine lies in a similar orientation to that of the WT residue. The sulfhydryl of a cysteine facing into the binding-site crevice should react much faster with charged sulfhydryl-specific reagents than should sulfhydryls facing into the interior of the protein or into the lipid bilayer. For such reagents, we use derivatives of methanethiosulfonate (MTS): positively charged MTSethylammonium (MTSEA) and MTSethyltrimethylammonium (MTSET), negatively charged MTSethylsulfonate (MTSES) (13). These reagents form mixed disulfides with the cysteine sulfhydryl, covalently linking -SCH2CH2X. We infer that a WT residue forms part of the surface of the binding-site crevice if the reaction of its substituted cysteine with an MTS reagent alters binding irreversibly and if this reaction is retarded by the presence of ligand. Our SCAM analysis is presented here in the context of results from molecular modeling of E2, both of which support a role for E2 contributing to the binding site of the D2R.
Methods
Site-Directed Mutagenesis and Transfection. Cysteine mutations were generated as described (14). Mutations were confirmed by DNA sequencing. Mutants are named as (WT residue)(residue number)(mutant residue), where the residues are given in the single-letter code. The cDNA encoding the human dopamine D2short receptor or the appropriate cysteine mutant, epitope tagged at the amino terminus with the cleavable influenza-hemagglutinin signal sequence followed by the FLAG epitope (Sigma) (15) in the bicistronic expression vector pCIN4 (a gift from S. Rees, Glaxo) (16) was used for all transfections, which were performed as described (15). HEK293 cells were maintained and stably transfected pools of cells expressing the appropriate cysteine mutants were created as described (10). Cells were washed, dissociated, and prepared for subsequent assays as described (10).
[3H]N-methylspiperone Binding. Whole cells from a 35-mm plate were suspended in 400 μl of buffer A (25 mM Hepes/140 mM NaCl/5.4 mM KCl/1 mM EDTA/0.006% BSA, pH 7.4). Cells were then diluted with buffer A, typically 20-fold. [3H]N-methylspiperone (DuPont/NEN) binding was performed as described (10).
Reactions with MTS Reagents. Experiments were carried out on prewetted 96-well glass fiber (type B) filtration plates (Millipore). Aliquots (90 μl) of intact cell suspensions were incubated with freshly prepared MTS reagents (10 μl) (Biotium, Hayward, CA) at the stated concentrations at room temperature (RT) for 2 min. The cells were then washed with 200 μl of ice-cold buffer A twice. Cells were resuspended in 150 μl of buffer A without or with 2 μM (+)butaclamol (for nonspecific binding) by shaking the plate vigorously for 1 min. One hundred microliters of [3H]N-methylspiperone (final 100–150 pM) was added, and the plate was incubated at RT for 1 h. Finally, the cells were washed with 200 μl of ice-cold buffer A three times. After the plate was dried, the bottom Durapore membrane layer was removed, and 30 μl of SuperMix (Wallac) mixture was added to each well. After at least 5 h at RT, the plate was counted in a Wallac MicroBeta Liquid Scintillation Counter.
The fractional inhibition was calculated as 1 – [(specific binding after MTS reagent)/(specific binding without reagent)]. We used prism (GraphPad, San Diego) to analyze the effects of the MTS reagents by one-way ANOVA with Dunnett's post hoc test (P < 0.05). The second-order rate constant (k) for the reaction of MTSEA with each susceptible mutant was estimated as described (10).
For the protection assays, the cells were first incubated with 20 nM YM-09151-2 for 25–30 min at RT. After applying MTSEA (0.25 mM for I184C; 2.5 mM for N186C) for 2 min, the cells were washed three times with choline buffer (NaCl in buffer A replaced by cholineCl) for 15 min at RT and assayed for binding as described above.
Molecular Modeling. Because SS-E2 connects the extracellular end of TM3 to E2, which connects TM4 and TM5, the relative positions of these TMs are important determinants of the positioning of E2. We proposed previously that the extracellular portion of TM3 bends differently in rhodopsin and other aminergic receptors, depending on the Cys/Ser/Thr residue composition of TM3 (12). Based on computational simulations the extracellular region of TM3 in 5-hydroxytryptamine (5-HT)1A receptor has been inferred to bend more toward TM5 than in rhodopsin (17). Using similar methods (17) in molecular dynamics simulations, we observed a similar tendency for TM3 to bend toward TM5 in the D2R (data not shown). Therefore, we created, using modeller (18), a homology model of the TMD of the D2R based on both the rhodopsin structure and the individually modeled TM3 of D2R. The effect of the individually modeled TM3 was to reduce the distances between Cαs of Asp3.32 and Ser5.42/Ser5.43 by ∼2 Å, thereby better accommodating bound ligand. E2 of the D2R was then added to the TMD based on the structure of E2 of rhodopsin (using the alignment shown in Fig. 1B) and refined with the loop module of modeller (19). In contrast to our previous sequence alignment (12), TM4 has been shortened by one residue at its C-terminal end because in an updated sequence alignment we noted the presence of a gap after position 4.61 in other receptors, including the 5H1D receptors (see Fig. 1B). We also made TM5 one residue shorter at its N-terminal end to align the accessible and protected Ile-186 in D2R with the retinal contact residue Tyr-191 in rhodopsin. Given the recognized limitations of loop modeling in vacuo (20), our goal was not to predict the detailed structure of E2, but rather to explore our results in a structural context without significantly distorting the structure.
Results
Effects of Cysteine Substitution on Antagonist Binding. In a background of the mutant C1183.36S, which is relatively insensitive to the MTS reagents (14), we mutated to cysteine, one at a time, 10 residues, Asn-176 to Asn-186 in E2. (Note that all of these constructs contain Cys-182_e2, an endogenous cysteine that is disulfide-bonded with Cys-1073.25.) Each mutant receptor was stably expressed in HEK293 cells, and the KD and BMAX characterizing the equilibrium binding of the radiolabeled antagonist, [3H]N-methylspiperone, were determined. At eight positions, the KD of the cysteine-substitution mutant was between 0.8 and 1.5 times the KD of C118S (statistically insignificant differences). For I183C and I184C, the KD was 1.7 (P < 0.05) and 3.6 (P < 0.01) times that of C118S, respectively. At all positions, the BMAX ranged from 81% to 254% of that obtained with C118S (Table 1). The KI of the antagonist YM-09151-2 in competition with [3H]N-methylspiperone was also determined (Table 2). For A177C, the KI was 4.5-fold (P < 0.05) that of C118S; for I183C and I184C, the KI values were 11- and 12-fold (P < 0.01) that of C118S (Table 2). At the other seven positions the effects on YM-09151-2 affinity were not significantly different from that of C118S.
Table 1. Characteristics of [3H]N-methylspiperone binding.
| Mutant | KD, pM | BMAX, pmol/mg protein | KMUT/KC118S | n |
|---|---|---|---|---|
| C118S* | 93 ± 7 | 5.2 ± 0.3 | 1.0 | 7 |
| N176C | 93 ± 19 | 7.6 ± 1.1 | 1.0 | 3 |
| A177C | 125 ± 16 | 13.2 ± 1.4 | 1.3 | 3 |
| D178C | 79 ± 11 | 5.7 ± 0.6 | 0.8 | 3 |
| Q179C | 72 ± 7 | 6.9 ± 0.2 | 0.8 | 3 |
| N180C | 92 ± 12 | 7.4 ± 1.1 | 1.0 | 3 |
| E181C | 129 ± 30 | 4.2 ± 0.2 | 1.4 | 3 |
| I183C† | 163 ± 13 | 6.5 ± 0.6 | 1.7 | 3 |
| I184C‡ | 345 ± 14 | 6.7 ± 0.5 | 3.6 | 4 |
| A185C | 147 ± 27 | 6.4 ± 0.7 | 1.5 | 3 |
| N186C | 96 ± 12 | 6.6 ± 0.4 | 1.0 | 3 |
[3H]N-methylspiperone binding to HEK293 cells stably expressing the appropriate D2R mutant was determined as described in Methods. Data were fit to the binding isotherm by nonlinear regression. The means and SEM are shown for three to seven independent experiments, each with duplicate determination.
Note that Cys-182, which is disulfide-bonded to Cys3.25 (see text) is present in the background construct, C118S, and thus in all of the cysteine mutants in E2.
KD was significantly different (P < 0.05) from C118S by one-way ANOVA and Dunnett's post hoc test.
KD was significantly different (P < 0.01) from C118S by one-way ANOVA and Dunnett's post hoc test.
Table 2. Inhibitory potency of YM-09151-2 on [3H]N-methylspiperone binding.
| Mutant | KI, nM | KI(MUT)/KI(C118S) |
|---|---|---|
| C118S* | 0.096 ± 0.021 | 1.0 |
| N176C | 0.42 ± 0.10 | 4.3 |
| A177C† | 0.43 ± 0.095 | 4.5 |
| D178C | 0.28 ± 0.079 | 2.9 |
| Q179C | 0.23 ± 0.044 | 2.4 |
| N180C | 0.19 ± 0.055 | 2.0 |
| E181C | 0.33 ± 0.031 | 3.4 |
| I183C‡ | 1.1 ± 0.13 | 11 |
| I184C‡ | 1.1 ± 0.12 | 12 |
| A185C | 0.16 ± 0.034 | 1.6 |
| N186C | 0.22 ± 0.071 | 2.2 |
Cells transfected with the appropriate receptor were assayed with [3H]N-methylspiperone (150 pM) in the presence of nine concentrations of YM-09151-2. The apparent KI was determined by the method of Cheng and Prusoff (40) using the IC50 value obtained by fitting the data to a variable slope competition model by nonlinear regression. The mean and SEM are shown for three independent experiments, each with duplicate determinations.
Note that Cys-182, which is disulfide-bonded to Cys3.25 (see text) is present in the background construct, C118S, and thus in all of the cysteine mutants in E2.
KI was significantly different (P < 0.05) from C118S by one-way ANOVA and Dunnett's post hoc test.
KI was significantly different (P < 0.01) from C118S by one-way ANOVA and Dunnett's post hoc test.
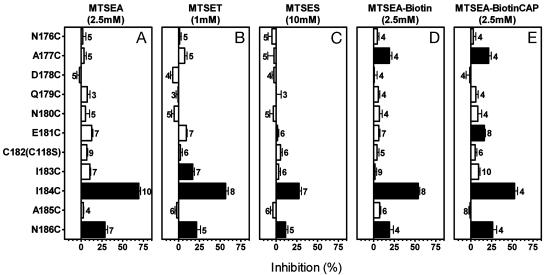
Reactions of MTS Reagents with the Mutants. In two of 10 cysteine-substitution mutants (Fig. 2A), I184C and N186C, a 2-min application of 2.5 mM MTSEA significantly inhibited [3H]N-methylspiperone binding. The rates of reaction of these two cysteines were relatively fast, compared to most accessible TMD residues in the D2R, with rate constants of 160 ± 25 M–1·s–1 (n = 4) and 140 ± 30 M–1·s–1 (n = 3) for I184C and N186C, respectively. The antagonist YM-09151-2 at 20 nM significantly protected against the reaction with MTSEA by 62 ± 3% (n = 4, P < 0.01) and 79 ± 10% (n = 4, P < 0.01) for I184C and N186C, respectively.
Fig. 2.
Inhibition of specific[3H]N-methylspiperone (150 pM) binding to intact cells transfected with the background construct, C118S, or with the substituted-cysteine mutant D2Rs resulting from 2-min applications of 2.5 mM MTSEA (A), 1.0 mM MTSET (B), 10 mM MTSES (C), 2.5 mM MTSEA-Biotin (D), and 2.5 mM 2-((6-((biotinyl)amino)hexanoyl)amino)ethyl MTS (MTSEA-BiotinCAP) (E). The mean and SEM are shown. The number of independent experiments for each mutant is shown next to the bars. Solid bars indicate mutants for which inhibition was significantly different (P < 0.05) from C118S by one-way ANOVA and Dunnett's post hoc test.
At 10 mM, MTSES, a negatively charged MTS derivative, significantly inhibited (Fig. 2C) binding to the same two mutants that reacted with MTSEA (Fig. 2A). In addition to I184C and N186C, I183C was significantly inhibited by treatment with 1 mM MTSET (Fig. 2B). MTSET is positively charged like MTSEA but is bulkier (11).
Because we use the effects on ligand binding as an indirect readout of reaction, there is a chance of false-negative determinations of accessibility with MTSEA, MTSES, and MTSET, especially in a loop (see below). We reasoned that reaction of a substituted Cys with a MTS reagent that adds a longer and/or bulkier group to the sulfhydryl might impact more on subsequent ligand binding and therefore be detectable. Indeed, a 2-min application of 2.5 mM 2-((biotinyl)amino)ethyl MTS (MTSEA-Biotin) (21) inhibited [3H]N-methylspiperone binding to A177C and I184C and N186C (Fig. 2D), and the even longer 2-((6-((biotinyl)amino)hexanoyl)amino)ethyl MTS (MTS-Biotin CAP) also significantly inhibited [3H]N-methylspiperone binding to E181C, in addition to A177C, I184C, and N186C (Fig. 2E).
I184C Does Not Form a Mismatched Disulfide Bond with Either Cys-1073.25 or Cys-182_e2. The substituted cysteine in the most reactive mutant, I184C, is very close to Cys-182_e2, the Cys that forms the conserved SS-E2 with Cys-1073.25. It is conceivable that I184C disrupts SS-E2 by forming a mismatched disulfide bond with either Cys-182_e2 or Cys-1073.25, which would then expose either Cys-1073.25 or Cys-182_e2 instead of I184C itself. To exclude this possibility, we mutated Cys-182_e2 to Ala in both the background construct (C118S) and I184C. Mutation of Cys-182_e2 to Ala did not affect binding affinity for [3H]N-methylspiperone, and the Bmax was near normal as well (55% of the background level). The affinity for YM-09151-2, however, was significantly decreased (21-fold) (Table 3). Consistent with the reduction of YM-09151-2 affinity in I184C (12-fold) (Table 2), the combined mutation I184C/Cys-182_e2A decreased the affinity for YM-09151-2 (200-fold) (Table 3).
Table 3. Pharmacological characterization of the C182A mutants.
| Mutant | KD, pM N-methylspiperone | BMAX, pmol/mg protein | KMUT/KC118S | KI, nM YM-09151-2 | KI(MUT)/KI(C118S) |
|---|---|---|---|---|---|
| C118S* | 93 ± 7 | 5.2 ± 0.3 | 1.0 | 0.096 ± 0.021 | 1.0 |
| C182A | 163 ± 15 | 2.9 ± 0.5 | 1.8 | 2.1 ± 0.3 | 21 |
| I184C/C182A | 198 ± 21 | 6.0 ± 0.8 | 2.1 | 19 ± 3.1 | 200 |
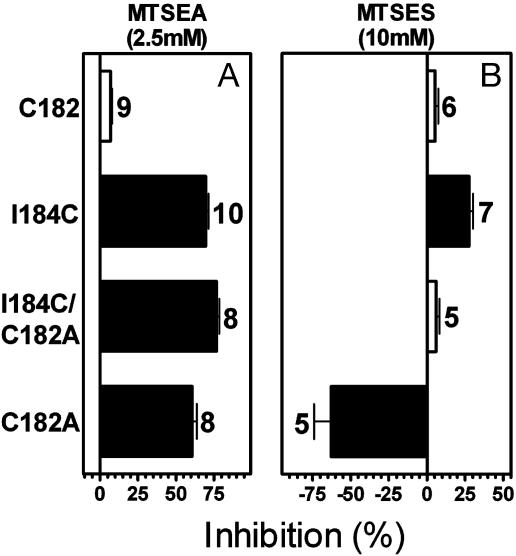
The binding of N-methylspiperone to Cys-182A was significantly inhibited by 2.5 mM MTSEA (Fig. 3A), likely as a result of the reaction of Cys-1073.25, the normal disulfide bonding partner of Cys-182_e2. N-methylspiperone binding to I184C/C182A was also strongly inhibited by 2.5 mM MTSEA (Fig. 3A). If 184C was disulfide-bonded to Cys-1073.25, then in the I184C/C182A background no cysteine would be available for reaction with MTSEA. Therefore, the inhibition of binding to I184C/C182A by MTSEA strongly argues against the existence of a mismatched disulfide between I184C and Cys-1073.25. Because these residues do not form a disulfide bond in the absence of Cys-182_e2, they are unlikely to form a disulfide bond in the presence of Cys-182_e2 in the I184C construct. Therefore, I184C (or conceivably Cys-1073.25, but see below) reacts in the I184C construct.
Fig. 3.
Inhibition of specific[3H]N-methylspiperone (150 pM) binding to intact cells transfected with the background construct, C118S, or with I184C, C182A/I184C, and C182A D2Rs resulting from 2-min applications of 2.5 mM MTSEA (A)or 10 mM MTSES (B). The mean and SEM are shown. The number of independent experiments for each mutant is shown next to the bars. Solid bars indicate mutants for which inhibition was significantly different (P < 0.05) from C118S by one-way ANOVA and Dunnett's post hoc test.
C182A was significantly potentiated by MTSES (Fig. 3B), an effect similar to that seen with D1083.26C (14), the position immediately adjacent to Cys-1073.25. This unusual potentiating effect of MTSES, which we hypothesized results from the addition of a negative charge, supports the accessibility of Cys-1073.25 in C182A. In contrast, in the I184C construct, MTSES had a strong inhibitory effect on N-methylspiperone binding (Fig. 2C), which is not consistent with the reaction of Cys-1073.25 but rather with that of 184C. Therefore, Cys-1073.25 in I184C is not accessible and likely forms a disulfide bond with Cys-182_e2 (or conceivably with I184C). Because, as we argued above, Cys-1073.25 is unlikely to form a disulfide bond with I184C, the most likely scenario is that Cys-1073.25 and Cys-182_e2 form a normal SS-E2 in I184C, and that the reactivity of this mutant results from the substituted cysteine at position 184.
Discussion
Our previous studies identified the residues within the TMD of the D2R that form the surface of the water-accessible binding-site crevice (reviewed in ref. 12). The high-resolution structure of bovine rhodopsin, however, indicated that E2 also forms part of the binding site for retinal (2). Several reports have implicated E2 in ligand specificity in aminergic and other small molecule-ligand GPCRs. Zhao et al. (22) found that substitution of three consecutive residues in E2 interconverted the ligand specificity for particular antagonists between that of α1BAR and α1AAR. Substitution of a single residue in E2 was sufficient to interconvert the pharmacological specificities of canine 5-HT1D and human 5-HT1D receptor (23) (Fig. 1B). Similarly, substitution of E2 and TM5 changed the subtype specificity of the 5-HT1D receptor to that of the 5-HT1B receptor and vice versa (24). In adenosine receptor, in which the binding site is also formed in the TMD, several glutamate residues in E2 are critical for ligand recognition (25, 26). Thus, although it has been argued that the presence of E2 within the TMD may be a feature unique to rhodopsin (7, 8), we proposed that E2 may contribute directly to forming the binding site of aminergic and certain other small molecule-ligand GPCRs (9).
Our SCAM results on E2 of the D2R supported this hypothesis. Ligand binding to 5 of the 10 cysteine-substitution mutants in E2 was inhibited by one or more of the sulfhydryl reagents tested. These include A177C, E181C, I183C, I184C, and N186C. The reaction of I184C and N186C with MTSEA strongly inhibited [3H]N-methylspiperone binding. MTSET, which is bulkier than MTSEA, identified an additional residue, I183C, the reaction of which inhibited binding. The much longer and bulkier MTSEA-Biotin and 2-((6-((biotinyl)amino)hexanoyl)amino)ethyl MTS (MTS-BiotinCAP) inhibited binding to A177C and/or E181C, whereas the reactions of smaller MTS reagents at these positions were not detected.
The rate constants for the reaction of MTSEA with I184C and N186C, 160 and 140 M–1·s–1, respectively, were comparable to the fastest rate constants we have determined in the TMD of the D2R, for which the range was 0.9 to 290 M–1·s–1 (10, 14, 15, 27–31). Thus, these two residues are as exposed as any in the binding-site crevice but not more so. These rate constants are in the range of rate constants for MTSEA reaction with substituted cysteines in the open channel of the nicotinic acetylcholine receptor (32), but they are much less than the rate constant for the reaction of MTSEA with 2-mercaptoethanol in solution (∼76,000 M–1·s–1) (32). This finding suggests that the residues in the E2 loop that contribute to ligand binding, like the residues in the D2R binding-site crevice and the residues in the acetylcholine receptor channel, are not freely accessible. Thus, E2 is likely to be folded into the binding-site crevice a significant fraction of the time.
This pattern is quite similar to that of bovine rhodopsin (Fig. 1B), in which the residues N-terminal to Cys_e2 are more extracellularly located relative to the stretch C-terminal to Cys_e2. Thus, the residues after Cys-E2, Ile-184 and Asn-186, are likely close to the core of the ligand-binding pocket, consistent with the strong impact on ligand binding of the attachment of –SCH2CH2NH3+. The residues Gly-173 and Asn-175, at the extracellular end of TM4, are also likely located close to the core of the ligand-binding pocket, as the corresponding cysteine mutants have similar reaction profiles with the MTS reagents as do those for Ile-184 and Asn-186 (Fig. 1A) (10). In contrast, Ala-177 and Glu-181 are likely more distant from the binding site, consistent with the need for attachment of the bulkier biotin moieties to perturb ligand binding (Fig. 4).
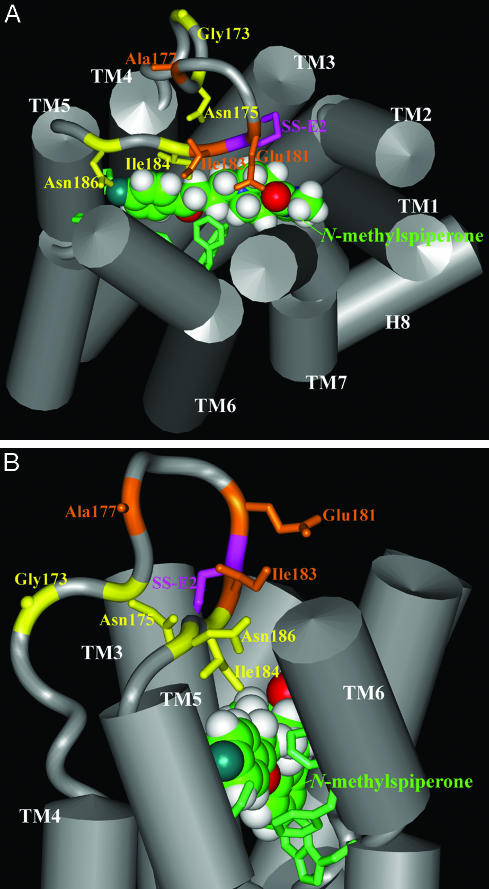
Fig. 4.
Extracellular view (A) and side view (B) of 3D molecular representation of a D2R model showing the TMD and E2. The helical regions of TMD are represented by gray cylinders. E2 is shown as a gray ribbon, with the Cα shown in the same color as the side chains, as described below. The two residues forming the conserved SS-E2 (Cys-182_e2–Cys-1073.25) are connected with purple sticks. The side chains of protected residues (Gly-173, Asn-175, Ile-184, and Asn-186) are shown as yellow sticks, and the other accessible residues in E2 (Ala-177, Glu-191, and Ile-183) are shown as orange sticks. The antagonist ligand N-methylspiperone is shown in space-filling representation, manually docked within the TMD, and oriented by the key residues, shown as green sticks, that have been found to be essential for ligand binding (see ref. 31).
Consistent with this interpretation, our modeling results in the D2R show Ile-184 within contact distance of bound N-methylspiperone. The other reactive and protected residues, Asn-175 and Asn-186, are located in close proximity to bound ligand. The reactive residues Ala-177, Glu-181, and Ile-183 also face the ligand-binding pocket (Fig. 4), and the chemically modified side chains likely extend sufficiently to disrupt ligand binding. The positioning in the model of the N-terminal portion of E2 containing Gly-173 and Asn-175 is likely affected by the recognized limitations of loop modeling in vacuum (20), and this portion of E2 may be packed closer to bound ligand in the actual structure. In contrast, the spatial constraints imposed by SS-E2, the distance between TM3 and TM5, and the small number of residues between Cys_e2 and TM5 give little freedom to this C-terminal portion of E2, consistent with the convergence between our data and the model.
Because the MTS reagents are a billion times more reactive with the thiolate than with thiol (33) and only water-accessible cysteines are likely to ionize to a significant extent, and because the reagents we generally use are charged, we assume that these MTS reagents react much faster with water-accessible cysteine residues than with cysteines facing the protein interior or lipid. This method has been successfully applied to the TMD for a number of GPCRs (12, 34) and other membrane proteins (11). In extracellular loops, however, the possibility of a false-negative determination of accessibility seems much greater than in the TMD, especially if the loops are unstructured and flexible. Thus, although we cannot infer that the residues for which reaction gave no inhibition of binding are not water accessible, we infer that the detectably reactive residues are not only water accessible but also line the binding-site crevice, based on the impact of reaction on binding and the ability of ligand to protect against reaction. Our E2 SCAM results, therefore, are consistent with the existence of the structural constraint posed by the SS-E2 on the positioning of the C-terminal portion of E2 as a lid over bound ligand and show that SCAM is applicable to an extracellular loop of a GPCR.
We cannot rule out the possibility that the protection at positions 184 and 186 results from a ligand-induced conformational rearrangement of E2 or another part of the receptor, such as the extracellular ends of the TMs and/or other extracellular loop structures, that limits access of MTSEA to the substituted cysteines at these positions. However, for three of the mutants, A177C, I183C, and I184C, the binding affinity for the antagonist YM-09151-2 was significantly lower than that of the background receptor (Table 2), consistent with a possible role of these WT side chains in the direct binding of this antagonist. These mutants are a subset of the five reactive substituted cysteines n E2. Although an effect of mutation on binding does not guarantee that the mutated residues contact ligand directly, Ile-183 and Ile-184, the two residues immediately after Cys-182_e2 are aligned with positions in E2 previously identified as determinants of ligand selectivity and direct retinal contacts (Fig. 1B). Consistent with our findings with the antagonist YM-09151-2, we have shown that the affinity of I184C for another antagonist, sulpiride, is reduced ∼21-fold compared to C118S and that sulpiride at 100 μM also protected I184C by ∼40% (data not shown).
An understanding of the structural and functional roles of extracellular loops may help us to explain certain aspects of ligand selectivity that cannot be fully explained by interactions with the TMs. Unlike the TMs, the extracellular loops, including E2, are highly divergent among rhodopsin-like GPCRs, despite the presence of the highly conserved SS-E2. Thus, the loops may provide more ligand contact variety among receptor subtypes than do the TMs. Taken together, our experimental data, the positioning of retinal contact residues near SS-E2 in rhodopsin, and the positioning of potential ligand contact residues near SS-E2 in various aminergic receptors (Fig. 1B) raise the possibility that E2, at least in this region, has a related structure in rhodopsin and aminergic receptors. In addition, three residues in E2 of the β2AR that form a metal binding site at which zinc slows the dissociation of bound antagonist were recently identified, providing further evidence that E2 forms a lid over bound ligand in aminergic receptors (G. Swaminath, T. W. Lee, L.S., J.A.J., and B. Kobilka, unpublished work).
The entrance route of ligands into the binding-site crevice and the conformational rearrangements of E2 that must accompany binding and/or activation are currently unknown and require further study. Interestingly, the extracellular end of TM4 contributes to the dimer interface of the D2R (35), and the dimer interface in mouse rhodopsin, as determined by atomic force microscopy of native retinal membranes (36), was inferred to be TM4 and TM5. Therefore, E2 must be positioned quite near the dimer interface. This finding is particularly intriguing given results in the family C heterodimeric γ-aminobutyric acid (GABA) type B receptor showing that binding of GABA to the GB1 subunit transactivates G protein via an interaction with the GB2 subunit (37) Thus E2, which lines the binding-site crevice and likely interacts directly with bound ligand, may be involved in information transfer between GPCRs in a dimeric or oligomeric complex. Also of note, an antibody directed against an epitope in E2 of the β2AR acted as an agonist, whereas Fab fragments of this antibody acted as an inverse agonist (38), further suggesting that conformational perturbation of E2 can affect the activation state of GPCRs.
Acknowledgments
We thank Marta Filizola, Ulrik Gether, Arthur Karlin, and Harel Weinstein for valuable discussion and comments on the manuscript, Hui Xie for technical support, and Xavier Deupi and Leonardo Pardo for advice on our simulations of D2R TM3. This work was supported by National Institutes of Health Grants MH57324 and MH54137, the Lebovitz Fund, and the Lieber Center.
Abbreviations: GPCR, G protein-coupled receptor; D2R, dopamine D2 receptor; TMD, transmembrane domain; TM, transmembrane segment; SCAM, substituted-cysteine accessibility method; MTS, methanethiosulfonate; MTSEA, MTSethylammonium; MTSET, MTS-ethyltrimethylammonium; MTSES, MTSethylsulfonate; MTSEA-Biotin, 2-((biotinyl)amino)-ethyl MTS; RT, room temperature; 5HT, 5-hydroxytryptamine; AR, adrenergic receptor.
Footnotes
For the residues in the TMD, we use the Ballesteros and Weinstein indexing system (39), which is shown in superscript.
References
- 1.Gether, U. (2000) Endocr. Rev. 21, 90–113. [DOI] [PubMed] [Google Scholar]
- 2.Palczewski, K., Kumasaka, T., Hori, T., Behnke, C. A., Motoshima, H., Fox, B. A., Le Trong, I., Teller, D. C., Okada, T., Stenkamp, R. E., et al. (2000) Science 289, 739–745. [DOI] [PubMed] [Google Scholar]
- 3.Savarese, T. M., Wang, C. D. & Fraser, C. M. (1992) J. Biol. Chem. 267, 11439–11448. [PubMed] [Google Scholar]
- 4.Zeng, F. Y., Soldner, A., Schoneberg, T. & Wess, J. (1999) J. Neurochem. 72, 2404–2414. [DOI] [PubMed] [Google Scholar]
- 5.Noda, K., Saad, Y., Graham, R. M. & Karnik, S. S. (1994) J. Biol. Chem. 269, 6743–6752. [PubMed] [Google Scholar]
- 6.Lin, S., Gether, U. & Kobilka, B. K. (1996) Biochemistry 35, 14445–14451. [DOI] [PubMed] [Google Scholar]
- 7.Menon, S. T., Han, M. & Sakmar, T. P. (2001) Physiol. Rev. 81, 1659–1688. [DOI] [PubMed] [Google Scholar]
- 8.Bourne, H. R. & Meng, E. C. (2000) Science 289, 733–734. [DOI] [PubMed] [Google Scholar]
- 9.Shi, L. & Javitch, J. A. (2002) Annu. Rev. Pharmacol. Toxicol. 42, 437–467. [DOI] [PubMed] [Google Scholar]
- 10.Javitch, J. A., Shi, L., Simpson, M. M., Chen, J., Chiappa, V., Visiers, I., Weinstein, H. & Ballesteros, J. A. (2000) Biochemistry 39, 12190–12199. [DOI] [PubMed] [Google Scholar]
- 11.Karlin, A. & Akabas, M. H. (1998) Methods Enzymol. 293, 123–145. [DOI] [PubMed] [Google Scholar]
- 12.Ballesteros, J. A., Shi, L. & Javitch, J. A. (2001) Mol. Pharmacol. 60, 1–19. [PubMed] [Google Scholar]
- 13.Stauffer, D. A. & Karlin, A. (1994) Biochemistry 33, 6840–6849. [DOI] [PubMed] [Google Scholar]
- 14.Javitch, J. A., Fu, D., Chen, J. & Karlin, A. (1995) Neuron 14, 825–831. [DOI] [PubMed] [Google Scholar]
- 15.Javitch, J. A., Ballesteros, J. A., Weinstein, H. & Chen, J. (1998) Biochemistry 37, 998–1006. [DOI] [PubMed] [Google Scholar]
- 16.Rees, S., Coote, J., Stables, J., Goodson, S., Harris, S. & Lee, M. G. (1996) BioTechniques 20, 102–110. [DOI] [PubMed] [Google Scholar]
- 17.Lopez-Rodriguez, M. L., Vicente, B., Deupi, X., Barrondo, S., Olivella, M., Morcillo, M. J., Behamu, B., Ballesteros, J. A., Salles, J. & Pardo, L. (2002) Mol. Pharmacol. 62, 15–21. [DOI] [PubMed] [Google Scholar]
- 18.Sali, A. & Blundell, T. L. (1993) J. Mol. Biol. 234, 779–815. [DOI] [PubMed] [Google Scholar]
- 19.Fiser, A., Do, R. K. & Sali, A. (2000) Protein Sci. 9, 1753–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mehler, E. L., Periole, X., Hassan, S. A. & Weinstein, H. (2002) J. Comput. Aided Mol. Des. 16, 841–853. [DOI] [PubMed] [Google Scholar]
- 21.Boileau, A. J., Evers, A. R., Davis, A. F. & Czajkowski, C. (1999) J. Neurosci. 19, 4847–4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao, M. M., Hwa, J. & Perez, D. M. (1996) Mol. Pharmacol. 50, 1118–1126. [PubMed] [Google Scholar]
- 23.Wurch, T. & Pauwels, P. J. (2000) J. Neurochem. 75, 1180–1189. [DOI] [PubMed] [Google Scholar]
- 24.Wurch, T., Colpaert, F. C. & Pauwels, P. J. (1998) Mol. Pharmacol. 54, 1088–1096. [DOI] [PubMed] [Google Scholar]
- 25.Olah, M. E., Jacobson, K. A. & Stiles, G. L. (1994) J. Biol. Chem. 269, 24692–24698. [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, J., Jiang, Q., Glashofer, M., Yehle, S., Wess, J. & Jacobson, K. A. (1996) Mol. Pharmacol. 49, 683–691. [PMC free article] [PubMed] [Google Scholar]
- 27.Javitch, J. A., Ballesteros, J. A., Chen, J., Chiappa, V. & Simpson, M. M. (1999) Biochemistry 38, 7961–7968. [DOI] [PubMed] [Google Scholar]
- 28.Fu, D., Ballesteros, J. A., Weinstein, H., Chen, J. & Javitch, J. A. (1996) Biochemistry 35, 11278–11285. [DOI] [PubMed] [Google Scholar]
- 29.Javitch, J. A., Fu, D. & Chen, J. (1995) Biochemistry 34, 16433–16439. [DOI] [PubMed] [Google Scholar]
- 30.Javitch, J. A., Li, X., Kaback, J. & Karlin, A. (1994) Proc. Natl. Acad. Sci. USA 91, 10355–10359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi, L., Simpson, M. M., Ballesteros, J. A. & Javitch, J. A. (2001) Biochemistry 40, 12339–12348. [DOI] [PubMed] [Google Scholar]
- 32.Pascual, J. M. & Karlin, A. (1998) J. Gen. Physiol. 111, 717–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberts, D. D., Lewis, S. D., Ballou, D. P., Olson, S. T. & Shafer, J. A. (1986) Biochemistry 25, 5595–5601. [DOI] [PubMed] [Google Scholar]
- 34.Javitch, J. A., Shi, L. & Liapakis, G. (2002) Methods Enzymol. 343, 137–156. [DOI] [PubMed] [Google Scholar]
- 35.Guo, W., Shi, L. & Javitch, J. A. (2003) J. Biol. Chem. 278, 4385–4388. [DOI] [PubMed] [Google Scholar]
- 36.Liang, Y., Fotiadis, D., Filipek, S., Saperstein, D. A., Palczewski, K. & Engel, A. (2003) J. Biol. Chem. 278, 21655–21662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duthey, B., Caudron, S., Perroy, J., Bettler, B., Fagni, L., Pin, J. P. & Prezeau, L. (2002) J. Biol. Chem. 277, 3236–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mijares, A., Lebesgue, D., Wallukat, G. & Hoebeke, J. (2000) Mol. Pharmacol. 58, 373–379. [DOI] [PubMed] [Google Scholar]
- 39.Ballesteros, J. & Weinstein, H. (1995) Methods Neurosci. 25, 366–428. [Google Scholar]
- 40.Cheng, Y. & Prusoff, W. H. (1973) Biochem. Pharmacol. 22, 3099–3108. [DOI] [PubMed] [Google Scholar]