Abstract
To study the role of the transcription factor Myc-interacting protein 1 (MIZ-1) in activating various target genes after induction with the microtubule disrupting agent T113242, we have used small interfering RNA duplexes (siRNAs) to knockdown the expression of MIZ-1. As expected, depletion of MIZ-1 resulted in the inhibition of T113242-dependent activation of the low-density lipoprotein receptor (LDLR) gene in hepatocytes. Cells transfected with MIZ-1 siRNAs also exhibited growth arrest. In addition, inhibition of the extracellular signal-regulated kinase (ERK) pathway inhibited T113242-induced nuclear accumulation of MIZ-1 and activation of LDLR. Gene expression microarray analysis under various induction conditions identified other T113242-activated genes affected by a decrease in MIZ-1 and inhibition of the ERK pathway. We also found that the accumulation of MIZ-1 in the nucleus is influenced by cell–cell contact and/or growth. Taken together, our studies suggest that MIZ-1 regulates a specific set of genes that includes LDLR and that the ERK pathway plays a role in the activation of target promoters by MIZ-1.
The protooncogene c-myc has been extensively studied as a model for dissecting molecular mechanisms of neoplastic transformation. A transcription factor, Myc-interacting protein 1 (MIZ-1) was subsequently identified by virtue of its ability to bind Myc. A number of studies provided evidence to support a model in which Myc serves to repress the transcriptional activation function of MIZ-1 (1). An example of this type of regulation can be observed in the case of the cyclin-dependent kinase (CDK) inhibitor, p15ink4b (2, 3). Myc-mediated suppression of MIZ-1 transcriptional activation can be counteracted by transforming growth factor β signaling, which leads to decreasing levels of the inhibitory Myc protein. MIZ-1 transcriptional activity can also be modulated by UV exposure, which induces a decrease in topoisomerase II-binding protein (TopBP1). TopBP1, like Myc, associates with MIZ-1 and inhibits its ability to activate transcription of certain target genes such as the CDK inhibitor, p21cip1 (4). Thus, it appears that MIZ-1 is a tightly regulated transcription factor whose activity can be modulated by association with various regulatory partners.
Another level of MIZ-1 regulation involves the control of its subcellular localization. Cytoplasmic localization of MIZ-1 has been previously reported along with various factors, including the coactivator p300 and Myc, that alter MIZ-1 localization (1, 2). However, bona fide physiological signals that regulate the subcellular localization of endogenous MIZ-1 have been difficult to pinpoint. It is noteworthy that localization of Myc is affected by cell density. Specifically, in sparse cell densities, Myc is mainly localized to the nucleus, but accumulates in the cytoplasm upon growth at high density or serum withdrawal (5). Because Myc and MIZ-1 physically interact, this observation may be relevant to the control of MIZ-1 localization and activity.
In addition to these diverse mechanisms of MIZ-1 regulation, we have shown that MIZ-1 activity can also be regulated by its association with microtubules. According to this novel regulatory pathway, MIZ-1 is sequestered in the cytoplasm by association with microtubules. Upon drug-induced microtubule depolymerization, MIZ-1 is free to enter the nucleus where it binds to target sequences such as the low-density lipoprotein receptor (LDLR) gene promoter and activates transcription (6).
To further understand the microtubule-dependent activation of MIZ-1, we investigated several potential inhibitors of cellular signal transduction pathways, including one involving the extracellular signal-regulated kinase (ERK) pathway. This pathway was of particular interest because of reports showing activation of ERK after disruption of microtubules by using colchicine (7). Activation of Ras leading to activation of ERK also correlates with a decreased stability of microtubules (8). Because ERK has been shown to phosphorylate and regulate microtubule-associated proteins (9), we hypothesized that microtubule disruption by T113242 may activate ERK, which in turn leads to a cascade of events necessary for nuclear accumulation of MIZ-1.
Here, we report a series of experiments to better understand the regulation of MIZ-1 and to identify additional target genes of MIZ-1. First, we have used small interfering RNAs (siRNAs) to deplete endogenous MIZ-1 and investigate phenotypes associated with loss of this transcription factor. In combination with RNA interference (RNAi), we have also modulated MIZ-1 activity with a highly specific small molecule inhibitor (T113242) to analyze changes in gene expression across multiple conditions that activate and inhibit MIZ-1 function. We have tracked the movement of MIZ-1 within the cell and how its subcellular localization correlates with its transcriptional function. Our analysis confirms the connection between LDLR regulation and MIZ-1 activity and provides additional insights into MIZ-1 target genes as well as various mechanisms that modulate MIZ-1 activity.
Methods
Cell Culture. HepG2 cells were obtained from American Type Culture Collection and grown in DMEM supplemented with 10% heat-inactivated FBS, 2 mM l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin.
Antibodies. MIZ-1 antibodies used in immunoblots and immunofluorescence experiments were generated by rabbit injections with purified recombinant full-length MIZ-1 and affinity purified by using recombinant MIZ-1. GAPDH (Abcam) and BrdUrd (Becton Dickinson) antibodies were also used for immunoblots and immunofluorescence assays. Secondary antibodies labeled with CY3, FITC, and Texas red were obtained from Jackson ImmunoResearch.
RNase Protection Assays. Messenger RNA was extracted from cells by using TRI Reagent (Sigma) followed by mRNA purification (Oligotex, Qiagen, Valencia, CA) for the experiments with T113242 and PD98059. For RNA purification from siRNA-transfected cells, total RNA was purified from cells by using and RNeasy kit (Qiagen). Antisense probes for LDLR and GAPDH transcripts were labeled and gel purified by using a Maxiscript T7 kit (Ambion). The two probes were simultaneously hybridized with either equal amounts of RNA. RNase protected products were resolved by using the RPA III kit (Ambion, Austin, TX).
High-Density Oligonucleotide Microarray. Two micrograms of mRNA or 20 μg of total RNA per sample was prepared for hybridization following the manufacturer's protocols (Affymetrix). Twenty micrograms of fragmented cRNA was hybridized to HGU133A arrays and scanned by using a Hewlett Packard GeneArray Scanner. Expression data were analyzed by using genechip analysis suite v5 and data mining tool v4 (Affymetrix).
RNAi. HepG2 cells (2 × 104 cells per six wells) were transfected with 6 μl of 20 μM siRNA (Dharmacon, Lafayette, CO) and 10 μl of Oligofectamine (Invitrogen) in 248 μl of Opti-MEM media (GIBCO/BRL) in 3 ml of media. MIZ106 and MIZ2078 refer to the starting base number of the MIZ mRNA target region. For Western assays, cells were lysed in 1% Nonidet P-40/1% deoxycholate/0.1% SDS/0.15 M NaCl/1× PBS/2 mM EDTA/0.25 mM PMSF/0.5 mM benzamidine. Equal amounts of total protein from cleared lysates were loaded onto an 8% acrylamide gel.
Flow Cytometry. HepG2 cells transfected with siRNAs were fixed with 70% EtOH, washed in PBS, and stained in 0.1% Triton X-100/0.2 mg/ml RNase A/0.02 mg/ml propidium iodide. Over 15,000 cells were analyzed by using an EPICS XL-MCL flow cytometer (Beckman Coulter). Data were analyzed by using f lowjo (TreeStar) and cell cycle calculations were made by using a Dean–Jett–Fox algorithm. For BrdUrd experiments, cells were labeled with BrdUrd 24 h before processing cells by using a BrdUrd Flow kit (BD Bioscience).
Indirect Immunofluorescence. HepG2 cells were grown on eight-chamber slides coated with poly(lysine) (Lab-Tek), and the growth surface was scraped with a pipette tip and treated with BrdUrd 1.5 h before harvest. BrdUrd was detected by using reagents from a BrdUrd flow kit (BD Bioscience) and other immunofluorescence procedures were as described (6).
Footprint Assays. Up to 40 ng of recombinant MIZ-1 was incubated with ≈0.03 pmol of labeled probe corresponding to the spectrin promoter. Protocol was as described (6).
Transient Transfections. Appropriate promoters were PCR-amplified and inserted in the pGL3-Basic vector (Promega). The dual luciferase protocol was as described (6). Luciferase values were normalized by using an internal Rous sarcoma virus promoter–reporter construct.
Results
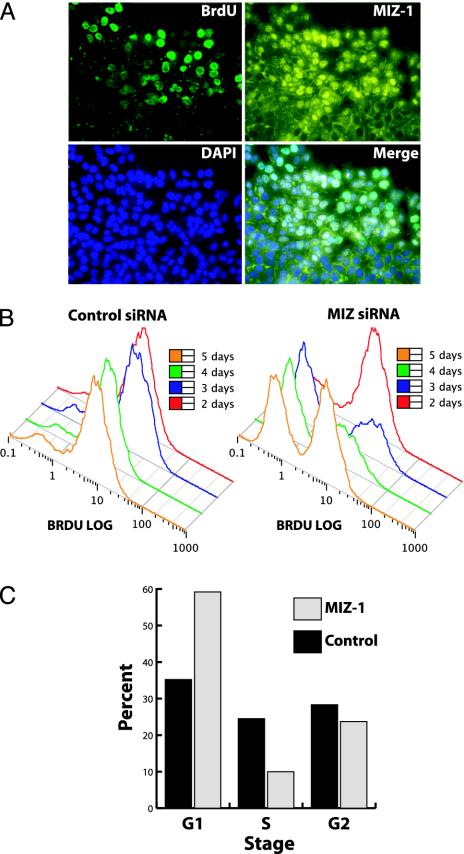
Multiple Signals Govern MIZ-1 Localization. Having previously demonstrated the effect of drug (T113242)-induced microtubule disruption on MIZ-1 subcellular localization (6), we sought to probe physiological conditions that might influence MIZ-1 nuclear translocation. During the course of our immunofluorescence studies, we consistently observed a differential pattern of nuclear MIZ-1 in cells adjacent to the perimeter of confluent colonies. These perimeter cells frequently displayed nuclear localization of MIZ-1, whereas cells in the center of the colony (i.e., contact inhibited) showed largely cytoplasmic localization of MIZ-1. In addition, most isolated and dispersed cells showed predominately nuclear localization (data not shown). To further examine this phenomenon, we performed in vitro scrape wound assays (10). These “wound-induced” cultures were then treated with BrdUrd to label replicating cells and discriminate them from stationary cells. Using indirect immunofluorescence, we stained for BrdUrd and endogenous MIZ-1 as well as 4′,6-diamidino-2-phenylindole (DAPI) to visualize nuclei. As expected, most cells along the edge of the scrape were positive for BrdUrd staining. Importantly, as expected, the majority of these cells also showed nuclear localization of MIZ-1 (Fig. 1A). By contrast, interior confluent cells were quiescent and largely retained a cytoplasmic localization of MIZ-1. These results suggest that MIZ-1 localization may be correlated with cell–cell contact and/or growth state. Thus, these physiological signals may in part influence MIZ-1 localization by a mechanism that is likely to be distinct from the drug-induced nuclear localization of MIZ-1 we observed previously, although both may involve microtubule integrity. At present, the molecular basis for this putative cell contact of growth state mediated localization of MIZ-1 remains unknown.
Fig. 1.
MIZ-1 localization and RNAi-induced growth arrest. (A) Cells were grown to confluency, and the growth surface was scraped (Upper Right), exposed to BrdUrd, and then fixed. BrdUrd and MIZ-1 were visualized by using indirect immunofluorescence. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). (B) HepG2 cells were transfected with control (scramble) or MIZ-1 siRNAs, labeled with BrdUrd, and analyzed for BrdUrd incorporation by using flow cytometry after 2, 3, 4, and 5 days of transfection with siRNAs. There are two distinct populations of cells. (Left) One population with the weaker BrdUrd signal. (Right) Cells that have incorporated BrdUrd S-phase and, thus, represent actively growing cells. (C) Cells were transfected with siRNAs for 4 days, stained with propidium iodide, and analyzed by flow cytometry. Cell cycle statistics were generated by using a Dean–Jett–Fox algorithm.
siRNA-Mediated Knock-Down of MIZ-1 Inhibits Cell Proliferation. Because cell–cell contact and/or growth state appear to be correlated with MIZ-1 localization, we next asked whether MIZ-1 can influence cell growth. To complement previous cell growth studies that relied on overproduction of MIZ-1, we decided to look for cell growth phenotypes associated with a loss of MIZ-1. We used siRNAs to knock down expression of endogenous MIZ-1 (see Fig. 3A) in HepG2 cells. Cells were treated with BrdUrd to discriminate dividing from nondividing cells with flow cytometry. Measurements were taken over a period of 5 days after transfections with specific siRNAs (Fig. 1B). Compared to control cells (transfected with nonspecific siRNAs), the cells transfected with MIZ-1 siRNAs consistently showed an increase in nondividing (BrdUrd–) cells and a decrease in dividing (BrdUrd+) cells. This growth arrest trend peaked between 3 and 4 days of MIZ-1 depletion. After 5 days, some of the MIZ siRNA-treated cells began to recover and divide. Resumption of growth corresponded to an increase in MIZ-1 levels observed by using Western blots (data not shown) after the transient knock-down of MIZ-1.
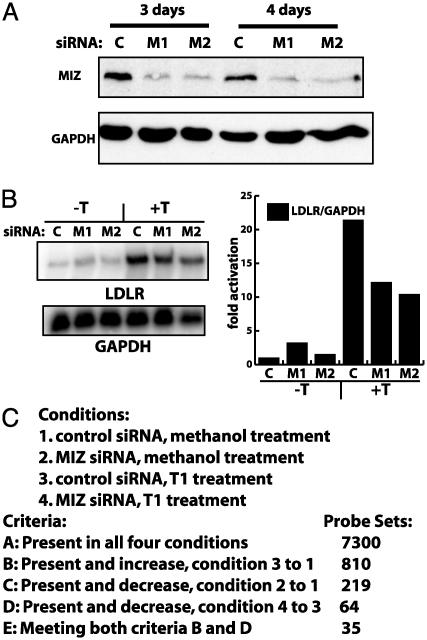
Fig. 3.
RNAi of MIZ-1 inhibits T113242 activation of LDLR. (A) Western blot analysis of MIZ-1 from HepG2 cells transfected for 3 or 4 days with either control or MIZ-1-directed siRNAs (M1 and M2). M1 and M2 are two different siRNAs directed against two different regions of the MIZ-1 transcript. Equal amounts of soluble protein were loaded in each lane and blotted for GAPDH as a loading standard. (B) HepG2 were transfected with siRNAs as in A, and RNA was analyzed by RNase protection assays. Total RNA was simultaneously hybridized with probes for LDLR and GAPDH transcripts. LDLR transcript levels were normalized to GAPDH levels, and quantitation is shown in the graph. (C) Summary of microarray data from RNAi and T113242 experiments. Methanol was the drug vehicle control. Probe sets indicate the number of genes meeting each criteria.
To confirm these findings, we also investigated the MIZ-1 RNAi-induced growth phenotype by using propidium iodide staining. HepG2 cells were treated in a manner similar to the BrdUrd labeling experiments and DNA content was measured with propidium iodide staining to assess changes in cell cycle profiles. Compared to control cells (Fig. 1C), transfection of MIZ-1 siRNAs caused cells to accumulate in G1, with less cells in S phase and G2 after 4 days. The RNAi-mediated loss of MIZ-1 expression appears to induce cell cycle arrest before S phase. Thus, we observed a good correlation between MIZ-1 levels and cell proliferation. Thus, at least for HepG2 cells, our results using multiple assays indicate that loss of MIZ-1 can lead to inhibition of cell proliferation. Paradoxically, our findings contrast previous reports that overexpression of MIZ-1 can also lead to cell arrest (1, 2). Perhaps the intracellular levels of MIZ-1 or its binding partners impact gene expression programs that control cell growth. Alternatively, the growth arrest phenotype associated with MIZ-1 may be cell type dependent.
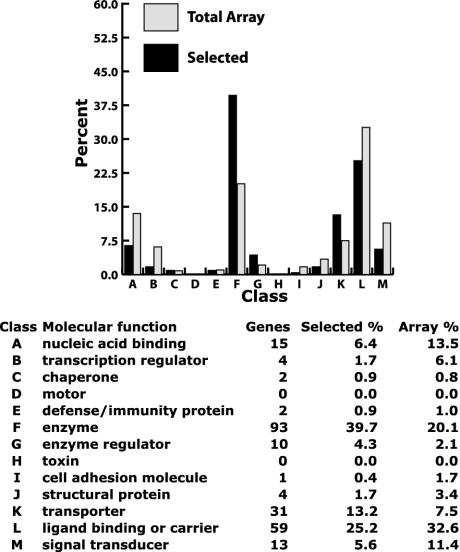
Analysis of Genes Down-Regulated by Depletion of MIZ-1. We next took a different approach to investigate the role of MIZ-1 in the regulation of HepG2 gene expression. Because loss of MIZ-1 in these cells can modulate growth, we wanted to identify specific genes affected by the loss of MIZ-1. To address this question, we purified RNA from cells transfected with MIZ-1 siRNAs in two independent sets of experiments. These RNAs were subsequently analyzed by using microarrays containing 15,000 human genes. Fig. 2 summarizes the molecular function associated with the genes that are down-regulated due to depletion of MIZ-1 in multiple independent experiments. Some of these genes likely represent direct target genes of MIZ-1. However, because this assay does not discriminate between direct effects versus secondary effects induced by MIZ-1 loss, the affected genes should mainly be viewed as a phenotypic fingerprint corresponding to loss of MIZ-1 expression.
Fig. 2.
Microarray analysis of down-regulated genes in MIZ-1-depleted cells. Gene ontology classifications for down-regulated genes in MIZ-1 depletion experiments are shown. Solid bars show classification distribution for transcripts meeting certain selected criteria. Light bars show classification distribution of all genes represented on the H133A microarray. Selected genes were down-regulated in two independent assays when MIZ-1 was depleted by using siRNAs. Of the 219 selected genes, 146 had molecular function descriptions.
Compared to the distribution of all classes of genes represented on the microarray, we observed that the most common genes that met our criteria were enzymes, transporters, and metabolic genes. Within this list of down-regulated genes are several that have been implicated in cytoskeletal structure and growth regulation, including anaphase promoting complex subunit 5, splicing factor arginine/serine-rich (SFRS) protein kinase 1, cytoskeleton-associated protein 4 (p63, a p53 homolog), katanin p80, tubulin (γ complex associated protein 3), testis thymus kinase (TTK) protein kinase, serine/threonine kinase 6, survivin, fibroblast growth factor receptor 3, macrophage stimulating 1 (hepatocyte growth factor-like), activin β E, density-regulated protein, and BCL2. Several of these genes could represent direct MIZ-1 target genes and/or genes responsible for the growth arrest observed after MIZ-1 depletion.
RNAi of MIZ-1 and T113242 Activation of LDLR. To follow-up on our preliminary microarray analysis, we decided to develop more stringent methods for filtering the microarray data to identify putative direct MIZ-1 target genes. We also wanted to confirm the role that MIZ-1 has been reported to play in the T113242 activation of LDLR transcription (6). Specifically, we asked whether knocking down MIZ-1 with RNA interference also disrupts T113242 activation of LDLR transcription. Western blot analysis confirmed that (Fig. 3A), siRNAs directed against MIZ-1 specifically knocked down MIZ-1 expression in HepG2 cells compared to a nonspecific control siRNA. As expected, control siRNAs do not affect any specific set of sequences in the blast database. We transfected cells with siRNAs directed against two different regions of the MIZ-1 transcript and then treated these cells with T113242. RNase protection assays revealed a modest (2- to 3-fold) but reproducible decrease in T113242 activation of LDLR in cells transfected with MIZ siRNAs (Fig. 3B). Throughout these experiments, cells were treated with 25-hydroxycholesterol, which inhibits the sterol regulatory element-binding protein (SREBP)-mediated activation of LDLR transcription (11), a mechanism that is independent of MIZ-1. These results indicate that depleting the amount of MIZ-1 in HepG2 cells indeed inhibits T113242 activation of LDLR and support our earlier conclusion that MIZ-1 plays a role in T113242 activation of LDLR independent of SREBP activation and cholesterol.
Next, we used microarray analysis to identify other genes exhibiting this type of dual sensitivity to T113242 and MIZ-1. As with the RNase protection assays, cells were transfected with siRNAs and either mock treated or treated with T113242 for 8 h. We compiled one list of genes that were up-regulated when comparing the scrambled siRNA T113242-treated cells to the scrambled siRNA mock treated cells (Fig. 3C, list B). A second list was generated for genes that were down-regulated when comparing the MIZ siRNA T113242 treated cells to the scrambled siRNA T113242 treated cells (Fig. 3C, list D). A third list was generated that only contain genes found in both of the first two lists. This class (Fig. 3C, list E) represents genes whose activation by T113242 is inhibited when MIZ-1 expression is decreased. Among this list of genes is the LDLR (P value 0.026), which further confirms the role of MIZ-1 in mediating T113242 activation of LDLR. Other gene products identified in this list include transcription factors (BHLHB2 and EGR1), cytoskeleton-associated proteins (ZFP36, SPTBN1), and proteins associated with cell proliferation (BTG3, OSMR, MCP, IER3), which are of particular interest given the growth arrest phenotype associated with MIZ-1 depletion and T113242-induced changes in the cytoskeleton.
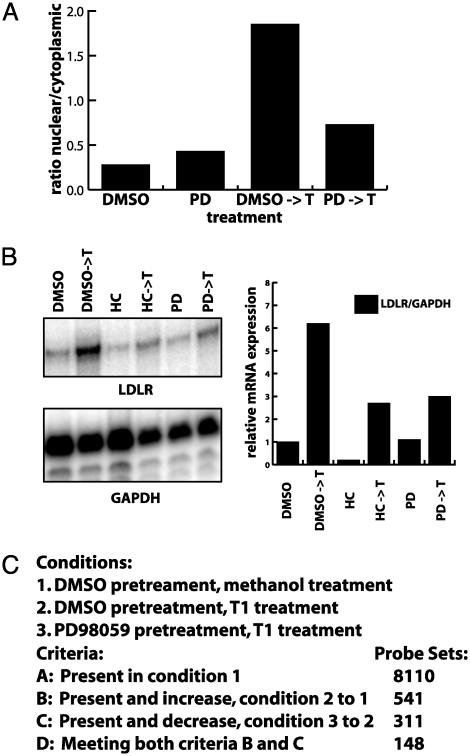
Inhibition of ERK Abrogates MIZ-1 Activation. In addition to identifying MIZ-1-regulated genes, we have also been interested in identifying other signaling pathways that might influence the activation of MIZ-1. After testing several cell growth and drug treatment conditions, we found one specific inhibitor (PD98059) of a signal transduction pathway that had a significant affect on MIZ-1 activity. Our initial rationale for using the ERK pathway inhibitor, PD98059, derived from the finding that microtubule disrupting drugs like nocodazole and colchicine not only activate MIZ-1, but also induce ERK (7). In addition, LDLR gene expression has been reported to respond to stimulation of the ERK cascade (12). We therefore sought to determine whether ERK activation in some way modulates MIZ-1 subcellular localization in response to microtubule depolymerization by T113242. Cells were pretreated with DMSO alone or with the ERK pathway inhibitor, PD98059 dissolved in DMSO, which inhibits activation of MEK1/2 and prevents further activation of ERK. After PD98059 pretreatment, cells were mock treated or treated with the microtubule disrupting agent T113242. Subcellular localization of MIZ-1 in transfected cells was subsequently determined by using immunofluorescence (Fig. 4A). Whereas PD98059 alone did not significantly alter MIZ-1 localization, pretreatment with PD98059 followed by T113242 treatment inhibited nuclear accumulation of MIZ-1, suggesting that the ERK pathway may indeed play a role in nuclear accumulation of MIZ-1.
Fig. 4.
PD98059 inhibits T113242-induced MIZ-1 nuclear accumulation and activation of LDLR transcription. (A) Indirect immunofluorescence assays were used to score the subcellular localization of MIZ-1. The ratio of HepG2 cells showing nuclear MIZ-1 versus cytoplasmic was plotted for cells treated with various drugs. Cells were pretreated with DMSO or PD98059 (PD) and then treated with either methanol (vehicle) or T113242. (B) Cells were treated as in A. RNA was purified and used in RNase protection assays with probes for LDLR and GAPDH. Products were quantitated, and the relative amounts of RNA were adjusted for the internal GAPDH probe and plotted for the different conditions. Some conditions included treatment with 25-hydroxycholesterol (HC), an inhibitor of SREBP activation of LDLR expression. (C) Summary of four independent sets of microarray analysis. RNA was purified from cells after treatments described in A and analyzed by using the Affymetrix system. A probe set represents a single gene, but a gene may be represented by multiple probe sets resulting in less distinct genes than the probe set number indicates. The probe sets shown indicate the number of probe sets meeting the appropriate criteria in three of four experiments.
Because MIZ-1 nuclear accumulation induced by T113242 was inhibited by pretreatment with PD98059, we next tested its effects on T113242 activation of LDLR transcription. Relative transcript levels were normalized by using an internal probe for GAPDH. There was a marked decrease in T113242 activation of LDLR transcription when cells were pretreated with PD98059, but not in the presence of 25-hydroxycholesterol, a known inhibitor of SREBP activation of LDLR (11). These results suggest that ERK activation which affects MIZ-1 nuclear localization also mediates T113242 activation of LDLR transcription. Thus, there appears to be a strong correlation between inhibition of T113242-induced MIZ-1 nuclear accumulation and inhibition of T113242-stimulated LDLR transcription by disrupting the ERK signaling pathway.
RNA from these experiments was next used to identify other genes that are activated by T113242 and inhibited by PD98059. For these experiments, we used RNA from cells treated in two ways. Cells were pretreated with PD98059 or vehicle for 2 or 5 h (pretreatment), and then treated with T113242 or vehicle for 6 or 9.5 h (treatment), respectively. Duplicate independent samples were used for each condition in each set. As before, we used microarray analysis to detect changes in transcript levels. Specifically, we identified genes that were up-regulated when comparing T113242 treated to mock-treated cells (both pretreated with DMSO) and down-regulated when comparing PD98059-pretreated cells to DMSO-pretreated cells. Transcripts meeting these different criteria obtained from multiple experiments are summarized in Fig. 4C, list D. As expected from our previous RNase protection assays, the LDLR transcript was activated by T113242 and inhibited by pretreatment with PD98059. These results indicate that there are a number of genes that display the same pattern of sensitivity to T113242 and PD98059 as LDLR, further supporting the notion that ERK activation plays a role in T113242-mediated activation of a subset of genes that includes LDLR. Other gene products in this list include apoptosis regulators (LAGLS1, MCL1, IER3, BCL2L1, GG2-1), signal transducers (INPP1, EPHA2, LIF, IGFBP1, ACTN1, GPR126, GPRC5C, SFN), transcription factors (SOX4, ZFP36L1, MAFF), cytoskeleton-associated proteins (SPTBN1, KRT18, VASP, ARPC2, TUBA3), cell cycle regulators (TIMP1, CKS2, AREG, CDKN1B, BTG3), cell adhesion proteins (LAMB1, ITGAN, ICAM1, IL-8, MCAM, FN1), and metabolic proteins (OSBPL10, FACL5, KYNU, GK, GOT1, ARG2). These classes are noteworthy given that extended exposure to T113242 induces apoptosis and disrupts microtubules, both of which are associated with MIZ-1 and cell proliferation.
Combining Microarray Data from RNAi, T113242, and PD98059 Experiments. One goal of performing these different microarray experiments was to identify MIZ-1 target genes. The most conservative approach to interpreting the microarray data were to identify those genes that would meet multiple criteria from independent experiments carried out in duplicate/triplicate. The criteria of interest were: (i) transcripts activated by T113242 alone, (ii) transcripts down regulated by the loss of MIZ-1 via siRNAs depletion after treatment with T113242, (iii) transcripts activated by T113242 when pretreated with DMSO, and (iv) transcripts inhibited by PD98059 pretreatment before T113242 treatment. Only a small subset of seven genes satisfied all of these criteria in multiple independent experiments after interrogating ≈8,500 genes. As expected, the LDLR gene was present in these MIZ-1 target genes: SPTBN1, IER3, LDLR, SERPINE1, JAG1, GK, and SLC38A1 (Locuslink ID). Other gene products found in this list include a cytoskeletal protein (spectrin), an apoptosis inhibitor (immediate early response 3), and a protein in the transforming growth factor β signaling pathway (serine plasminogen activator inhibitor). Note that some potential MIZ-1 target genes identified via previous studies (2–4, 13), p15ink4b and p21cip1, failed to meet this stringent criteria. One explanation is that certain transcripts (such as p15ink4b) were below the detection limit of the microarray analysis. On the other hand, the presence of LDLR on this list generated by using the most stringent criteria adds validity to the screen and helps confirm the role of MIZ-1 in regulating specific genes.
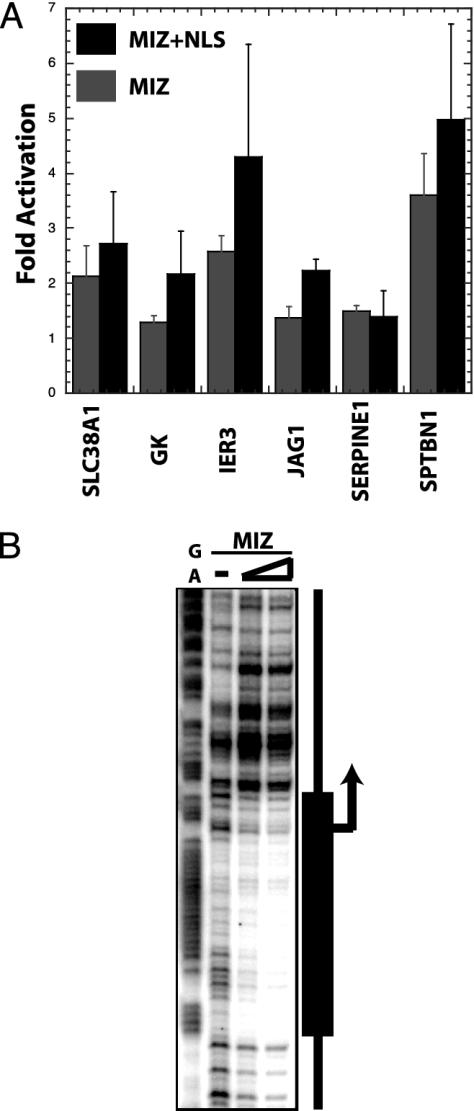
Identifying Potential Direct MIZ-1 Target Genes. Because the potential MIZ-1 target gene list was generated by using multiple independent strategies for activating and inhibiting MIZ-1 function, we felt confident that some of the identified genes would be direct MIZ-1 target genes. To validate these microarrays hits, we cloned the promoters of several of these putative MIZ-1 target genes into luciferase reporters. Cotransfection with control plasmids and MIZ expression plasmids were carried out with either wild-type MIZ-1 or a MIZ-1 construct containing a simian virus 40 nuclear localization signal. The immediate early response 3 and spectrin genes were significantly activated when cotransfected with MIZ-1 (Fig. 5A). As expected, the MIZ–NLS (nuclear localization signal) construct also directed a high level of activation with these two putative MIZ-1 target genes, supporting the notion that subcellular localization is an important component of the mechanism that regulates MIZ-1 transcriptional activity.
Fig. 5.
Validation of microarray hits using reporter and footprint assays. (A) Luciferase-reporter assays were performed by using promoters of selected genes. The promoter reporter plasmids, an internal control reporter vector, and MIZ-1 expression plasmids were cotransfected into HepG2 cells. Luciferase values were normalized compared to the internal control reporter, and the fold activation is shown is relative to the negative control plasmid lacking the MIZ-1 gene. The following promoter reporter constructs were used: amino acid transporter (SLC38A1), glycerol kinase (GK), immediate early response 3 (IER3), jagged (JAG1), serine proteinase inhibitor (SERPINE1), and spectrin (SPTBN1). (B) Purified recombinant MIZ-1 was incubated with a labeled spectrin promoter fragment and subjected to DNaseI digestion. Protected fragments are shown by the promoter diagram on the right.
The spectrin reporter was highly responsive to MIZ-1 transactivation, so we assayed this promoter for direct MIZ-1 binding. We used DNase I footprinting assays with the spectrin promoter and purified recombinant MIZ-1 to identify at least one MIZ-1 binding site in the spectrin promoter (Fig. 5B). The location of the binding site overlaps the transcriptional start site, which is very reminiscent of the arrangement seen with other MIZ-1 target promoters (i.e., LDLR, p21 and p15; refs. 6, 13).
Discussion
A combination of RNAi depletion assays together with various methods of MIZ-1 activation and inactivation coupled with microarray analysis has allowed us not only to validate LDLR as a bona fide MIZ-1 target gene, but also identified several new target genes. It was surprising to find little overlap between the list of genes that were down-regulated by RNAi depletion of MIZ-1 alone (Fig. 2) and the short list of seven genes (SPTBN1, IER3, LDLR, SERPINE1, JAG1, GK, and SLC38A1) meeting the multiple criteria of drug-dependent induction, ERK sensitivity, and sensitivity to MIZ-1 levels, because both classes of genes should include potential MIZ-1 target genes. However, the activation of the seven genes listed above are dependent on activation by T113242. The genes that were down-regulated by MIZ-1 depletion alone (Fig. 2) could be activated by MIZ-1 through a T113242-independent mechanism. Therefore, our analysis revealed at least two distinct sets of genes. One group meets the criteria of activation by T113242 and MIZ-1 and the other set of genes induced by a different mechanism also involving MIZ-1. In this latter case, MIZ-1 may need to cooperate with additional activators and cofactors other than those involved with activation by T113242.
The connection between ERK, MIZ-1 and LDLR correlates well with several other observations. For example, activation of ERK using an oncogenic form of Raf-1 resulted in growth arrest and increased p21cip1 expression (12)reminiscent of the response observed with MIZ-1 activation (4). As reported previously, UV damage also triggers stress activated kinases that feed into the ERK pathway. This agrees with the finding that UV damage derepresses MIZ-1 (4). Interestingly, one gene from the list of potential MIZ-1 targets (SPTBN1, IER3, LDLR, SER-PINE1, JAG1, GK, and SLC38A1) is the immediate early response 3 (IER3) gene, which is expressed after UV exposure (14). Therefore, the cell cycle arrest observed upon ERK activation may be due in part to stimulation of MIZ-1 activity. Although MIZ-1 depletion also leads to growth suppression, the possibility exists that this growth suppression is caused by other functions of MIZ-1 unrelated to transcription of the CDK inhibitor. Taken together, these data present the possibility that ERK activation contributes to the activation of MIZ-1 through phosphorylation, leading to the nuclear accumulation of MIZ-1, thus increasing its ability to activate transcription of target genes.
We previously reported that MIZ-1 can regulate LDLR transcription via a mechanism involving microtubule association, but the physiological rationale for this link was not obvious. Our new findings uncovering a connection between MIZ-1 and the spectrin gene may provide a possible rationale. Genetic and cell biological experiments have revealed various roles of spectrin, including cell polarity, cell adhesion, association of the actin cytoskeleton with the plasma membrane, stabilization or activation of signaling proteins, and fortification of the lipid bilayer (15). Its function in the lipid bilayer may be related to the function of LDLR regulating membrane fluidity through the control of cholesterol import. Given the cytoskeletal changes induced by T113242, it is reasonable to speculate that MIZ-1 may play a role in activating the spectrin promoter to minimize drug-induced morphological and cytoskeletal changes. Additional experiments will need to be carried out before we can fully appreciate the role MIZ-1 plays in regulating cell growth and signaling.
Acknowledgments
We thank Mallory Haggart for oligonucleotide synthesis and DNA sequencing, and K. Geles, B. Glover, Y. Isogai, W. Liu, and O. Puig for advice and comments on the manuscript. This work was supported in part by National Institutes of Health Grant CA25417 (to R.T.).
Abbreviations: MIZ-1, myc-interacting protein 1; CDK, cyclin-dependent kinase; LDLR, low-density lipoprotein receptor; ERK, extracellular signal-regulated kinase; siRNA, small interfering RNA; RNAi, RNA interference; SREBP, sterol regulatory element-binding protein.
References
- 1.Peukert, K., Staller, P., Schneider, A., Carmichael, G., Hänel, F. & Eilers, M. (1997) EMBO J. 16, 5672–5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Staller, P., Peukert, K., Kiermaier, A., Seoane, J., Lukas, J., Karsunky, H., Moroy, T., Bartek, J., Massague, J., Hanel, F. & Eilers, M. (2001) Nat. Cell Biol. 3, 392–399. [DOI] [PubMed] [Google Scholar]
- 3.Seoane, J., Pouponnot, C., Staller, P., Schader, M., Eilers, M. & Massague, J. (2001) Nat. Cell Biol. 3, 400–408. [DOI] [PubMed] [Google Scholar]
- 4.Herold, S., Wanzel, M., Beuger, V., Frohme, C., Beul, D., Hillukkala, T., Syvaoja, J., Saluz, H. P., Haenel, F. & Eilers, M. (2002) Mol. Cell 10, 509–521. [DOI] [PubMed] [Google Scholar]
- 5.Vriz, S., Lemaitre, J. M., Leibovici, M., Thierry, N. & Mechali, M. (1992) Mol. Cell. Biol. 12, 3548–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ziegelbauer, J., Shan, B., Yager, D., Larabell, C., Hoffmann, B. & Tjian, R. (2001) Mol. Cell 8, 339–349. [DOI] [PubMed] [Google Scholar]
- 7.Schmid-Alliana, A., Menou, L., Manie, S., Schmid-Antomarchi, H., Millet, M. A., Giuriato, S., Ferrua, B. & Rossi, B. (1998) J. Biol. Chem. 273, 3394–3400. [DOI] [PubMed] [Google Scholar]
- 8.Harrison, R. E. & Turley, E. A. (2001) Neoplasia 3, 385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reszka, A. A., Seger, R., Diltz, C. D., Krebs, E. G. & Fischer, E. H. (1995) Proc. Natl. Acad. Sci. USA 92, 8881–8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Funaki, T., Nakao, A., Ebihara, N., Setoguchi, Y., Fukuchi, Y., Okumura, K., Ra, C., Ogawa, H. & Kanai, A. (2003) Cornea 22, 153–159. [DOI] [PubMed] [Google Scholar]
- 11.Brown, A. J., Sun, L., Feramisco, J. D., Brown, M. S. & Goldstein, J. L. (2002) Mol. Cell 10, 237–245. [DOI] [PubMed] [Google Scholar]
- 12.Kapoor, G. S., Atkins, B. A. & Mehta, K. D. (2002) Mol. Cell. Biochem. 236, 13–22. [DOI] [PubMed] [Google Scholar]
- 13.Seoane, J., Le, H. V. & Massague, J. (2002) Nature 419, 729–734. [DOI] [PubMed] [Google Scholar]
- 14.Im, H. J., Pittelkow, M. R. & Kumar, R. (2002) J. Biol. Chem. 277, 14612–14621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubreuil, R. R. & Grushko, T. (1998) BioEssays 20, 875–878. [DOI] [PubMed] [Google Scholar]