Abstract
Insects respond to microbial infection by the rapid and transient expression of several genes encoding antibacterial peptides. In this paper we describe a powerful technique, two-dimensional difference gel electrophoresis, that, when combined with mass spectrometry, can be used to study the immune response of Drosophila melanogaster at the protein level. By comparatively analyzing the hemolymph proteome of 2,000 third-instar Drosophila larvae, we identified 10 differential proteins that appear in the fruit fly hemolymph very early after an immune-challenge with lipopolysaccharides. These proteins can be assigned to the immune response, because they are not induced after sterile injury. Reduction of intergel variability or quantification problems related to conventional two-dimensional electrophoresis and improvement of image analysis were achieved by the use of two fluorescent dyes to label the two different protein samples. Some of the immune-induced proteins, such as thioester-containing protein 2, can be assigned to specific aspects of the immune response; others were already reported as being involved in stress response. An immune-induced protein (CG18594) is homologous to a mammalian serine protease inhibitor that mediates the mitogen-activated protein kinase and the NF-κB signaling pathways. In addition, a number of proteins that had not been associated with the immune response before were isolated and identified, and some of these were still present in the hemolymph 4 h after injury. Determining the function of all of these immune-induced proteins represents an exciting challenge for increasing our knowledge of insect immunity.
Keywords: immune response proteins, two-dimensional difference gel electrophoresis, stress response, secretome, innate immunity
Insects are particularly resistant to microbial infections, although they do not have an acquired immune system that is capable of specifically recognizing and selectively eliminating foreign microorganisms and molecules (i.e., foreign antigens). The defense system of insects consists of different innate reactions. Innate immunity is based on the recognition of microbial molecules, such as lipopolysaccharides (LPS) and peptidoglycans, by specific receptors and the subsequent activation of the cellular response, which includes phagocytosis and encapsulation, and the humoral response. Immediately after septic injury, the insect fat body (a homologue of the mammalian liver) and some blood cells start to produce a battery of potent antimicrobial and antifungal peptides (1). These molecules are released into the blood, where they synergistically act to destroy the invading microorganisms. Many induced antimicrobial molecules are apparent in the hemolymph only 4 h after infection.
Innate immunity, which refers to the first-line host defense against the early phase of microbial infection, is an evolutionarily ancient defense mechanism. Quite recently this first-line defense received renewed attention. The similarities between the human and Drosophila immune cascades introduced Drosophila melanogaster as a model system to study the biochemical pathways and all the components involved in innate immunity. In the last few years, studies were focused on the expression of these antimicrobial molecules, of which seven have been characterized. At least two pathways are involved in the Drosophila immune response (2). Recent genetic and molecular work has led to a detailed characterization of these pathways. The Toll signaling pathway, which controls the defense against fungal or Gram-positive bacterial molecules, was identified because of its parallels with the cytokine-induced activation of NF-κB in mammals (3, 4). The immune deficiency pathway, which is involved in the expression of most of the antibacterial peptide genes, mediates the defense against Gram-negative infections (5, 6).
The humoral reactions involve several proteolytic cascades. The D. melanogaster genome contains 34 antimicrobial peptide-encoding genes belonging to eight families and a large number of putative protease-encoding genes (7). Lectins or other molecules playing a role in recognition, phagocytosis, or antimicrobial activity may be present in the hemolymph. The Drosophila genome possesses at least three Gram-negative bacteria-binding proteins (8), but they do not appear to be induced after infection. On the other hand, two uncharacterized genes encoding short proteins with partial similarity to Drosophila Gram-negative bacteria-binding proteins are up-regulated after septic injury (9). Furthermore, the Drosophila genome encodes at least 12 peptidoglycan recognition proteins; transcripts of several peptidoglycan recognition protein genes have been found in hemocytes (10). Transferrin genes, which are involved in iron transport and protection against iron overload in the diet, appear to have an additional role in innate immunity, because they are also induced during septic injury (9). Finally, a microarray study of the Drosophila immune response not only revealed the involvement of the above mentioned immune response genes but also showed the involvement of a large number of genes with unknown function in the immune response (9). Although the multifarious power of genomics may reveal several aspects of innate immunity and genetic approaches have elucidated many components belonging to the Drosophila immune response, there are still many missing factors. Upon the completion of the Drosophila genome, the proteomics approach offers possibilities for (i) uncovering additional components that support immune processes and (ii) revealing the functions of genes predicted to be involved in the immune response.
Our present knowledge of the Drosophila immune response is mainly based on studies showing that the involved genes are transcriptionally modified after infection. However, it is important to understand that mRNA-based approaches measure message abundances and not the actual proteins, the real mediators of physiological functions. In addition, mRNA-based approaches cannot be used for fluids, such as the hemolymph. Because hemolymph is important for insect survival against invading microorganisms, we have analyzed the earliest changes of the hemolymph proteome of Drosophila larvae after challenge with LPS or pricking with sterile needles. In contrast to studies using microarrays, in which the total gene expression profile after infection is analyzed, we are interested in how infection affects the secretome of already synthesized proteins in the hemolymph. We used two-dimensional difference gel electrophoresis (2D-DIGE), a fluorescence-based method that increases the power of the proteomics technique by allowing two different protein samples tagged with two distinct fluorescent dyes to be run on the same gel. Such an approach enables a rapid screening for differences in hemolymph protein profiles between naive and immune-challenged Drosophila. The differential hemolymph proteins were subsequently identified by scanning the Drosophila genome database with mass spectrometric data.
Materials and Methods
Animals. D. melanogaster were kept in 250-ml bottles containing 70 ml of water, 17 g of sucrose, 0.45 g of yeast, 0.9 g of agar, 0.5 ml of 8% Nipagin, and 0.36 ml of propionic acid.
Preparation of Protein Samples. In the first experiment, ≈160 third-instar larvae of D. melanogaster were pricked with needles dipped in a solution containing 0.3% LPS (Sigma; LPS from Escherichia coli serotype O55:B5) and 1% ethanol. Twenty-five minutes after induction of each larva, the hemolymph was collected with microcapillaries under a binocular microscope. The hemolymph (12 μl) was suspended in 55 μl of a lysis solution (4°C) consisting of 7 M urea, 2 M thiourea, 4% (wt/vol) 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), 40 mM Tris, 1% (wt/vol) DTT, and a mixture of protease inhibitors (Complete protease inhibitor, Roche Diagnostics). The suspension was sonicated and centrifuged (Biofuge 13, Heraeus) for 12 min at 13,000 rpm and 4°C. Next, the supernatant was desalted by dialysis. The small volume of our sample and the fact that collecting hemolymph is very labor-intensive made the use of microdialyzers (PlusOne Mini Dialysis Kit, Amersham Biosciences) necessary. The protein content in the supernatant was determined by Bradford's method (11). Generally, we collected ≈180 μg of protein. The control sample, containing hemolymph of ≈160 naive Drosophila larvae, was treated the same way as described above.
For each condition, 90 μg of the sample was labeled with propyl-Cy3 or methyl-Cy5. To determine and exclude nonspecific labeling, both a “forward” (test proteins labeled with Cy3, control proteins labeled with Cy5) and a “reverse” (test proteins labeled with Cy5, control proteins labeled with Cy3) labeling were done. The synthesis of the fluorescent dyes was performed as described by Van den Bergh et al. (12). Because only 1–2% of the lysine residues in the proteins are fluorescently marked, the solubility of the fluorescent proteins is not influenced during electrophoresis. After addition of the dyes, the hemolymph samples were incubated in the dark at room temperature for 45 min to improve labeling of the protein.
A second control experiment was performed to distinguish immune-induced proteins from injury- or stress-response proteins. For this purpose, ≈160 third-instar larvae of D. melanogaster were pricked with sterile needles. Twenty-five minutes after induction of each larva, the hemolymph was collected and treated the same way as described in the first experiment. The control sample contained hemolymph of ≈160 naive Drosophila larvae.
A third proteomic experiment was performed4hafter challenge with LPS to investigate whether the instantly released proteins were still present in the hemolymph. The experiments were conducted as described above.
Two-Dimensional Electrophoresis. For isoelectric focusing (IEF), the test sample (containing hemolymph of infected or sterile-pricked larvae) labeled with propyl-Cy3 and the control sample labeled with methyl-Cy5 were mixed and solubilized in rehydration solution {7 M urea/2 M thiourea/4% (wt/vol) 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate/orange G/0.3% immobilized pH gradient (IPG) buffer}. The final volume of the mixture was 350 μl. For the reverse labeling, the same instructions were followed during the protocol. These mixtures of sample-containing rehydration solution were applied in strip holder channels. The solubilized proteins were loaded onto 18-cm linear IPG strips, pH 4–7 (Amersham Biosciences). IEF was performed with the IPGphor system (Amersham Biosciences) at 20°C and 50 μA per IPG strip as follows: 5 h at 30 V, 5 h at 60 V, 1 h at 300 V, 1 h at 600 V, 1 h at 1,000 V, 1 h at 3,000 V, and 6–8 h at 8,000 V. The strips were focused for a total of 70,000 V·h. After IEF, the strips were equilibrated for two intervals of 15 min in equilibration buffer containing 6 M urea, 50 mM Tris·HCl, 0.07% SDS, and 30% glycerol, pH 7.6. For the first equilibration step, we added 1% DTT (wt/vol) to reduce cystine bridges. Thereafter the proteins were carbamidomethylated with 4% (wt/vol) iodoacetamide.
The equilibrated IPG strips were placed on top of a 1.5-mm-thick SDS/polyacrylamide gel [11.5% T (total monomer); 2.6% C (crosslinker)] and run in the Ettan Daltsix system (Amersham Biosciences). Electrophoresis was carried out at 20°C as follows: 1 h at 600 V, 8 mA per gel, and 10 W and overnight at 600 V, 16 mA per gel, and 15 W. After electrophoresis, the gels were rinsed in a solution of water containing 5% acetic acid and 50% methanol.
Gel Imaging and Image Analysis. For fluorescent gel imaging, we used a home-built imager as developed and described by Van den Bergh et al. (12). Briefly, the gels were illuminated with the excitation wavelength of Cy3 (540 ± 20 nm) immediately followed by the excitation wavelength of Cy5 (635 ± 20 nm). Both illuminations had an acquisition time of ≈10 min. Fluorescent imaging was done with a double wavelength band-pass emission filter (587.5 ± 18.5 nm; 695 ± 30 nm) and a liquid nitrogen-cooled charge-coupled device camera.
False-colored gel images were constructed by combining the protein patterns of the Cy3- and Cy5-labeled samples of the same gel. After adjusting the intensities of both images, we constructed an overlay image in which the yellow spots refer to proteins present in equal amounts in both gels and the reddish or greenish spots refer to proteins with a higher expression level in the Cy3- or Cy5-labeled condition, respectively.
imagemaster 2d elite 3.1 image analysis software (Amersham Biosciences) was used for quantitative analysis of the possibly differentially expressed proteins. The spots were detected manually on the images. Identical spots of two images of the same gel were placed in identical pixel positions. After choosing one reference gel, the spots were matched automatically across gels, and the background was subtracted manually. The spots were normalized by using the mean spot volume of three unchanging spots in the two conditions. A Wilcoxon matched-pairs signed-ranks test was performed to analyze whether the observed differences were statistically significant.
Detection of Separated Proteins by Silver Staining and Trypsin Digestion. After 2D-DIGE imaging and analysis, the gels were post-stained to facilitate manual spot cutting by means of the silver nitrate method according to Schevchenko et al. (13). Differential spots were localized on the gel by comparing the silver-stained spot pattern with the 2D-DIGE protein pattern of the same gel. They were excised with a sterile scalpel in a laminar flow to prevent contamination with keratin. Identical spots in different gels were pooled to have sufficient amounts of protein for identification. The protocol followed to remove the silver ions and to digest the proteins is described in Vierstraete et al. (14). In brief, silver ions were removed with 30 mM potassium ferricyanide and 100 mM sodium thiosulfate (15). For dehydration and reswelling of the gel pieces, acetonitrile and 50 mM ammonium carbonate were used. Enzymatic digestion (overnight at 37°C) was performed with a solution containing 5 ng/μl trypsin (Promega) in a digestion buffer (50 mM ammonium carbonate/5 mM calcium chloride). The resulting peptides were extracted once with 80 μl of 50 mM ammonium carbonate (30 min) and two times with 80 μl of 50% acetonitrile and 5% formic acid (30 min).
MS. Mass spectrometry was performed on a Bruker (Billerica, MA) Reflex matrix-assisted laser desorption ionization time-of-flight mass spectrometer operated in the positive-ion mode. Instrument settings, calibration methods, and sample preparation were as described in Vierstraete et al. (14).
Proteins were identified in the Drosophila genome database through peptide mass fingerprinting by using the programs profound (The Rockefeller University, New York) and mascot (Matrix Science, London) (16, 17). One missed cleavage per peptide was allowed, and an initial mass tolerance of 0.1 Da was used in all searches. Carbamidomethylation of cysteines was set as a fixed modification.
In cases where peptide mass fingerprinting was not sufficient to identify the protein in a database, nano-liquid chromatography/mass spectrometry was performed as described (18). The fragmentation spectra of the tryptic peptides of a protein were subjected to a mascot search to identify the protein.
Results and Discussion
In the first experiment, three pools of D. melanogaster larval hemolymph proteins were collected 25 min after challenging with LPS and three pools of hemolymph proteins of unchallenged larvae were pairwise compared. In the control experiment, we compared three pools of Drosophila larval hemolymph proteins collected 25 min after challenge with sterile needles with three pools of hemolymph proteins of naive flies. The comparisons were consistently conducted on a single 2D-DIGE gel, elegantly excluding otherwise inevitable intergel variability. Each pool represents hemolymph proteins of ≈160 larvae; thus, ≈2,000 larvae were needed in the first two experiments. For the forward labeling, hemolymph proteins of pricked (LPS or sterile) larvae were marked with propyl-Cy3 and those from naive larvae were marked with methyl-Cy5. A reverse labeling was also performed to avoid false-positive results. In total, we ran 12 gels and obtained 24 images.
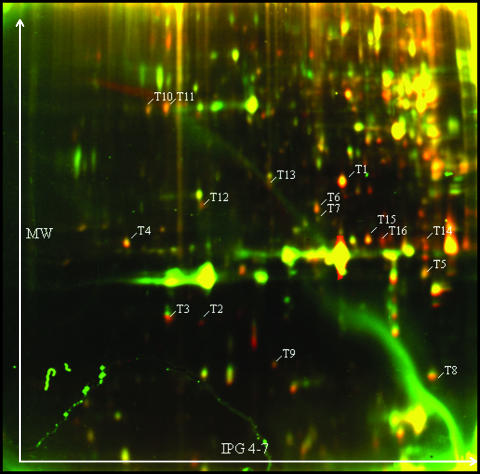
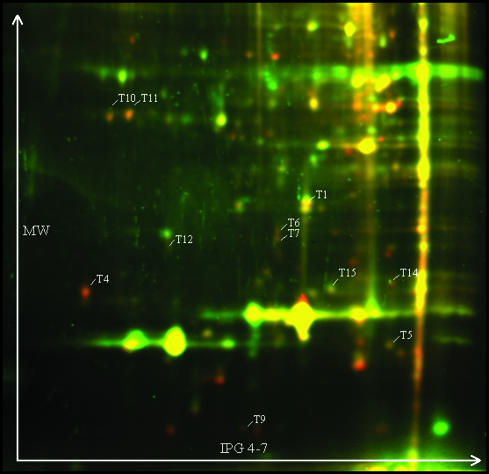
After imaging, the Cy3 and Cy5 images were false-colored in red and green, respectively, and an overlay image was obtained. The differential protein profiles of larval hemolymph proteins are shown in Figs. 1 and 2. Each figure represents the forward labeling (the reverse labeling is not shown). Only those spots that showed differential coloring (reddish or greenish) in both the forward and reverse labeling reaction were considered to contain differentially occurring proteins. In this way, 23 spots were found to contain differential proteins in the first experiment (LPS-challenged versus controls). A Wilcoxon matched-pairs signed-ranks test confirmed statistically significant differences in fluorescence level (P < 0.05) for 16 protein spots. All these proteins appeared more abundant in hemolymph of challenged Drosophila larvae. In the control experiment (sterile-pricked versus controls) we examined the differences in fluorescence level for these 16 protein spots. Five protein spots (T2, T3, T8, T13, and T16) could not be detected at all in the control experiment, which means that the corresponding proteins are secreted only after challenge with LPS or that the protein levels in naive and sterile-pricked larvae are very low. The Wilcoxon matched-pairs signed-ranks test confirmed statistically significant differences in fluorescence level (P < 0.05) for only 6 (of 11) protein spots.
Fig. 1.
False-colored protein expression pattern of larval hemolymph proteins (Exp. 1). For the forward labeling, propyl-Cy3-labeled proteins of flies challenged with LPS are colored in red and methyl-Cy5-labeled proteins of naive flies are colored in green. IEF was performed with 18-cm IPG strips, pH 4–7. Numbers indicate spots with statistically significant differential fluorescence levels, which were further identified with mass spectrometry.
Fig. 2.
False-colored protein expression pattern of larval hemolymph proteins (Exp. 2). For forward labeling, propyl-Cy3-labeled proteins of flies challenged with sterile needles are colored in red and methyl-Cy5-labeled proteins of naive flies are colored in green. IEF was performed with 18-cm IPG strips, pH 4–7. Spots with statistically significant differential fluorescence levels in Exp. 1 are indicated; however, five spots (T2, T3, T8, T13, and T16) could not be detected at all in this control experiment.
Only those spots that have a significantly higher level of fluorescence in the LPS-challenged condition (Exp. 1) and that are not significantly differential in the sterile-pricked condition (Exp. 2) can be associated with the immune response, except for spot T15, for which the spot volume is significantly lower in sterile-challenged larvae; therefore, its corresponding protein is immune-induced as well.
Most of the proteins could be identified in the Drosophila genome database through peptide mass fingerprinting. Where required, nano-liquid chromatography/tandem mass spectrometry was used to confirm protein identity by partial amino acid sequencing, as we did for spots T1, T2, T3, and T8. A list of the identified proteins is given in Table 1. The ratio (T/C) of the normalized spot volume of two differently colored fluorescent signals in challenged (T) and naive (C) larvae is also presented in Table 1.
Table 1. List of statistically significant differentially expressed proteins identified in hemolymph of Drosophila third-instar larvae 25 min (Exp. 1) or 4 h (Exp. 3) after LPS challenge and 25 min after sterile challenge (Exp. 2).
| NCBI entry
|
Mass, kDa
|
T/C ± SE
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Spot | Protein name | Gene name | Accession no. | Family | p.m. | pl | Exp. 1 | Exp. 2 | Exp. 3 | ||
| T1 | Regucalcin homologue | CG1803 or regucalcin | Q9VYRT | SMP-30 | 18860103 | 22 | 6 | 33.65 | 3.38 ± 0.57 | — | 6.29 ± 2.41 |
| T2 | CG18594 protein | CG18594 | Q9VD01 | PBP | 24649019 | 3 | 5.7 | 19.61 | 4.64 ± 1.14 | — | — |
| T3 | RfaBp | CG11064 or RFABP | Q94907 | Vitellogenin_N | 7511958 | 3 | 8.1 | 372.65 | 2.29 ± 0.26 | — | — |
| T4 | LD03138p | CG6058 or ALD | Q8MQQ4 | Aldolase | 21711709 | 15 | 5.6 | 31.67 | 5.33 ± 1.34 | 2.23 ± 0.31 | — |
| T5 | GST 1-1 | CG10045 or GSTD1 | P20432 | GST | 478212 | 10 | 6.9 | 23.87 | 3.25 ± 0.66 | — | — |
| T6 | LD03138p | CG6058 or ALD | Q8MQQ4 | Aldolase | 21711709 | 13 | 5.6 | 31.67 | 3.51 ± 0.89 | 20.30 ± 15.28 | — |
| T7 | LD03138p | CG6058 or ALD | Q8MQQ4 | Aldolase | 21711709 | 7 | 5.6 | 31.67 | 3.08 ± 0.77 | 6.33 ± 3.34 | — |
| T8 | Twinstar protein | TSR or CADF or CG4254 | P42554 | Cofilin_ADF | 17136986 | 2 | 6.7 | 17.15 | 2.09 ± 0.18 | — | — |
| T9 | Ferritin 1 heavy-chain homolog | CG2216 or FER1 | O18410 | Ferritins | 6409189 | 6 | 5.6 | 23.15 | 3.46 ± 1.10 | — | 3.47 ± 1.51 |
| T10 | LD04994p | CG10067 or ACT57B | Q8SXN3 | Actins | 1952831 | 10 | 5.6 | 40.26 | 6.77 ± 4.28 | 1.97 ± 0.60 | 11.12 ± 3.51 |
| T11 | Actin-5C | ACT5C or CG4027 | P10987 | Actins | 156759 | 6 | 5.4 | 41.82 | 3.41 ± 0.76 | — | 10.13 ± 2.71 |
| T12 | LD03138p | CG6058 or ALD | Q8MQQ4 | Aldolase | 21711709 | 6 | 5.6 | 31.67 | 2.88 ± 0.57 | — | 1.62 ± 0.22 |
| T13 | GH08432p | CG7052 or TEP 2 | Q8MT94 | A2M | 21391954 | 10 | 5.6 | 88.63 | 2.80 ± 0.53 | — | — |
| T14 | CG6776 protein | CG6776 | Q9VSL2 | GST | 21355779 | 7 | 6.5 | 27.72 | 4.33 ± 1.17 | 4.05 ± 1.42 | — |
| T15 | GIP-like protein | CG2227 or GIP | P36951 | Hfi | 17530883 | 8 | 6.1 | 29.09 | 3.47 ± 0.74 | 0.65 ± 0.08 | — |
| T16 | Alcohol dehydrogenase | CG3481 or ADH | P00334 | adh_short | 17137714 | 7 | 6.9 | 27.63 | 11.36 ± 3.46 | — | — |
| T17 | CG1548 protein | CG1548 or CATHD | Q9V313 | Asp | 6685167 | 11 | 5.9 | 42.47 | — | — | 2.72 ± 0.73 |
| T18 | Enolase | ENO or CG17654 | P15007 | Enolase | 17137654 | 25 | 6.1 | 46.56 | — | — | 6.40 ± 3.74 |
| T19 | Fat body protein 2 | CG3763 or FBP2 | O61511 | adh_short | 3098532 | 14 | 6.2 | 29.02 | — | — | 15.56 ± 6.53 |
| T20 | Peroxiredoxin | CG1633 or jafrac1 | Q9V3P0 | AhpC/TSA | 17157991 | 6 | 5.5 | 21.74 | — | — | 11.39 ± 2.57 |
| T21 | Ferritin 1 heavy-chain homolog | CG2216 or FER1 | O18410 | Ferritins | 17933722 | 7 | 5.6 | 23.15 | — | — | 1.45 ± 0.21 |
| C23 | Larval serum protein 2 | CG6806 | Q9VTT8 | Hemocyanin | 17864446 | 12 | 5.9 | 83.35 | — | — | 0.35 ± 0.17 |
Protein identity, gene name, SWISS-PROT or TrEMBL accession number, protein family, National Center for Biotechnology Information accession number, peptides matched (p.m.), theoretical isoelectric point (pl), and molecular mass are indicated. For each statistically significant differentially expressed protein, the average ratio (T/C) of the normalized spot volumes of the fluorescent signals in challenged (T) and naive (C) larvae is presented. The protein spot numbers (corresponding with Exps. 1 and 2) are indicated on the 2D-DIGE images in Figs. 1 and 2. SMP, senescence-marker protein; RfaBp, retinoid and fatty-acid-binding protein; cofilin_ADF, cofilin-actin depolymerizing factor; Hfi, hydroxypyruvate isomerase; GIP, transient receptor potential locus C protein (precursor); A2M, α2-macroglobulin; adh_short, short-chain alcohol dehydrogenase; Asp, aspartatic protease; AhpC/TSA, alkyl hydroxyperoxide reductase and thiol-specific antioxidant; —, no significant change.
Proteins Already Associated with the Immune Response. Spot T13, which contains thioester-containing protein (TEP) 2, which is known to be up-regulated after infection, illustrates the power and efficiency of the 2D-DIGE technique. The family of TEP genes is represented in many metazoa from Caenorhabditis elegans to humans. In the Drosophila genome, four TEP-encoding genes have been identified, and three of them (TEP 1, 2, and 4) are up-regulated during septic injury and fungal infection (9, 19). TEPs display substantial structural and functional similarities, including the highly conserved thioester motif, to both a central component of the mammalian complement system, factor C3, and a widespread protease inhibitor, α2-macroglobulin (20). In vertebrates, the complement system mediates inflammatory reactions, opsonization of microorganisms for phagocytosis, and direct killing of some pathogens. Anopheles gambiae TEP 1 serves as complement-like opsonin and promotes phagocytosis of some Gram-negative bacteria in a mosquito cell line (21). In Anopheles, particular TEPs were strongly induced during bacterial infection, and the parasite Plasmodium caused a sustained induction throughout its life cycle in the vector (22, 23). The presence in Drosophila and Anopheles of several proteins with structural characteristics similar to those of complement component C3 suggests a common evolutionary pathway.
We also observed an increase of actin-5C in the hemolymph of challenged larvae; this protein is one of the two cytoskeletal proteins. Courgeon et al. (24) already demonstrated the increase of actin synthesis after short hydrogen peroxide treatment or other stress situations such as anoxia or ethanol treatment. Later, functional genomic analysis of phagocytosis established the participation of actin cytoskeleton regulation proteins in innate immunity (25). In addition, we also observed a higher amount of the actin-binding protein Twinstar involved in actin polymerization and/or depolymerization.
Instantly Released Immune Proteins. The protein encoded by the CG18594 gene is much more abundant in larvae induced with LPS. This protein is homologous to the mammalian phosphatidylethanolamine-binding protein (PEBP). PEBP is expressed in a wide range of tissues but was originally isolated as a cytosolic 21- to 23-kDa protein from bovine brain and binds hydrophobic ligands, in particular phosphatidylethanolamine (26). Putative PEBP homologues have been identified in a variety of organisms, including Drosophila, C. elegans, and Saccharomyces cerevisiae; the parasites Plasmodium, Onchocerca volvulus, and Toxocara canis; and the flowering plants Arabidopsis and Antirrhinum (27). Here, we show that the homologous Drosophila CG18594 protein is actually expressed. Despite their widespread occurrence, in most cases the physiological role of these proteins is poorly understood. Recently, however, the mouse PEBP has been described as the prototype of a novel family of serine protease inhibitors with inhibitory activity against thrombin, neuropsin, and chymotrypsin (27). Interestingly, the human PEBP was described as a Raf-1 kinase inhibitor that modulates the mitogen-activated protein kinase-signaling cascade (28) as well as the signaling by the NF-κB pathway (29). NF-κB is required for the up-regulation of a large number of genes in response to inflammation, viral and bacterial infection, and other stress stimuli (30, 31). Our study suggests that the Drosophila homologue of mammalian PEBP (CG18594 protein) is an additional evolutionarily conserved mediator of the immune response, forming part of a similar pathway in Drosophila. The fact that the yeast homologue TFS1 (25 suppressor 1) acts as an inhibitor of proteolytic activity (32) is in favor of this hypothesis. Moreover, the increased presence of the Drosophila PEBP homologue in immune-challenged larvae is in complete agreement with our recent identification of 1-lysophosphatidylethanolamine as an antimicrobial compound in the housefly Musca domestica (33).
The highest difference observed was for spot T16, which contains an alcohol dehydrogenase. Because of this remarkable increase after infection, we suggest that alcohol dehydrogenases might be involved in the Drosophila defense mechanism.
Another significant differentially occurring protein in infected larvae was retinoid and fatty-acid-binding protein (RfaBp). RfaBp plays a role in Drosophila development. It is interesting in this context that RfaBp is reported to be the only gene up-regulated during both hypoxia and anoxia.‡
We also found that regucalcin/senescence-marker protein was induced in the hemolymph during infection. Its sequence shows a high similarity with the anterior fat body protein of Sarcophaga peregrina. This anterior fat body protein is expressed in the anterior pair of fat body lobes of last-instar larvae and in larval hemocytes, and it interacts with the hexamerin receptor (34).
Furthermore, glutathione S-transferase, with its important physiological role in the detoxification of lipid peroxidase products in Drosophila and other organisms, was more abundant in hemolymph of infected larvae than in controls. Singh et al. (35) suggested that the enzyme plays a protective role against deleterious effects of oxidative stress. Glutathione transferases form one of the three major families that are primarily responsible for metabolic resistance to insecticides (36).
Also, the secretion of ferritin in hemolymph is up-regulated after infection. Ferritins are iron-storage proteins, and our previous study showed that they are abundantly present in hemolymph of Drosophila third-instar larvae (14). The presence of putative NF-κB-like binding sites (37) is in agreement with our results and indicates that insect ferritins, like vertebrate serum ferritin, play a role in the immune response. So far, only the induction of transferrin genes had been demonstrated after infection (9, 38).
Finally, the amount of glyoxylate-induced protein-like protein, which belongs to the family of hydroxypyruvate isomerase proteins, also increased after infection.
Instantly Released Wound Proteins. Pricking the larva with a needle causes a severe injury to the animal. Therefore, it is likely that some of the increased proteins have nothing to do with the immune response, but are only part of the stress/injury response. Those proteins, which are significantly more abundant in both the LPS-challenged flies as well as in the sterile-pricked flies, are probably players in the early stress/injury response. This is the case for a glutathione transferase encoded by the CG6776 gene and for actin-57B, which is encoded by one of the four muscle actin genes (39). Three other spots, which are also more abundant in hemolymph of sterile-pricked larvae, contain fructose-bisphosphate aldolases. These are enzymes involved in glycolysis. Studies performed in Drosophila larvae concerning the effect of dietary carbohydrates and ethanol on the expression of genes encoding aldolase demonstrated that nutritional conditions had little or no effect on transcript levels of fructose-bisphosphate aldolases (40). Therefore, it is not likely that the higher levels of fructose-bisphosphate aldolase are caused by an increased uptake of food after infection. Surprisingly, a fourth spot containing aldolase that is induced in LPS-challenged larvae is not significantly more abundant in sterile-pricked larvae. It is probable that the fluorescence signal is too close to the background to obtain a significant result.
Differential Proteins 4 h After Immune Challenge. In our third experiment (Table 1), we examined the hemolymph proteome 4 h after challenge with LPS. Significant differences in fluorescence level (P < 0.05) were detected for 11 protein spots. With respect to the 25-min time point, the amounts of regucalcin and actin-5C, which were already up-regulated in the hemolymph shortly after infection, further increased considerably. Ferritin is also still present in the hemolymph in equal amounts compared with the 25-min time point. In addition, a shifted spot, indicative of a posttranslationally modified form, was detected. Although some protein spots were no longer differential after 4 h, a few new differential spots were observed. A 15-fold increase was observed for fat body protein 2, an alcohol dehydrogenase similar to the one identified in the 25-min experiment (41). Another differential protein spot contains peroxiredoxin, which may play a role in eliminating peroxides generated during metabolism. Other enzymes more abundantly present 4 h after infection are enolase, which is involved in glycolysis, and an aspartic protease. Only one larval serum protein 2 was less abundant in the hemolymph 4 h after infection with LPS.
The data of the 4-h experiment suggest changes at the gene expression level for the identified differential proteins. However, microarray studies (9, 19) did not indicate changes in mRNA quantities for these proteins. This lack of change could be explained by the fact that these studies were performed on adults instead of larvae (present study).
Remarks. The actual pI of a protein may often differ from that calculated from the database-stored sequence, mainly because of posttranslational modifications. The molecular mass values observed on the 2D gel are approximate values, because the resolving power of the gel is limited. For instance, the apparent masses for both T3 and T13, are remarkably lower than the computed masses. It is plausible that T3 and T13 contain only a fragment of the identified protein, in particular because the identification of these two spots was based on tryptic peptides, all of which corresponded to the same part of the full amino acid sequence. Spots T13, T3, and C23 were identified based on tryptic peptides originating from regions between residues 488–779, 598–699, and 415–612, respectively. We ignore whether fragmentation was caused by in vivo biological processing or protease activity during sample preparation.
Conclusion
We have identified proteins in D. melanogaster that appear early in the hemolymph after immune-challenging with LPS. So far, mRNA-based approaches have revealed differential transcripts only after 1.5 h or longer. Our comparison of hemolymph protein profiles very early after immune challenge indicates that immune challenging also affects the release of specific proteins from their storage sites. This presently identified secretome is most likely the very first line humoral defense before the induction of the biosynthesis of immune response proteins/peptides.
In addition to known or predicted immune response proteins (TEP 2, actin-5C, Twinstar, and ferritin), previously uncharacterized proteins involved in the immune system were identified in this study. These proteins include an alcohol dehydrogenase, retinoid and fatty-acid-binding protein, a regulcalcin homologue, glutathione S-transferase, a glyoxylate-induced protein-like protein, and a CG18594 protein, which has not been isolated or studied before in Drosophila. Its increased presence after infection, together with the identification of its putative affinity ligand (phosphatidylethanolamine) as an antimicrobial compound and the fact that its mammalian counterpart influences the signaling of the immune response mediator NF-κB, is strongly indicative for a role of CG18594 in the Drosophila immune response. Hence, we named this protein Drosophila instantly released immune protein or dIRIP.
We introduced an approach for studying the Drosophila immune response, and the presently identified proteins (such as TEP 2 and PEBP) validate the efficiency of this technique. Furthermore, the reproducibility and power of 2D-DIGE is confirmed by the second experiment, which shows that every increase in protein level triggered only by sterile pricking is also found in the hemolymph of LPS-challenged larvae. Five instantly released proteins are still differential 4 h after infection (some in higher concentrations), suggesting that their synthesis is induced after infection. Therefore, this technique will be useful for analyzing the changes in the cellular expression of immune proteins. The comparative proteomic analysis of flies in which immune key regulators are inactivated will also help to further characterize the role in innate immunity of the identified proteins. In addition, genetic mutants for each of the various identified immune proteins can be generated, and their phenotypes can be analyzed. Finally, the comparison of the present dataset with similar studies on other organisms will also be valuable.
With the described proteomic tool, we now have the opportunity to study the expression levels, structures, modifications, and interactions of immune response proteins not only in Drosophila but also in other species for which genome information is available. Finally, this study will lead to a comprehensive understanding of innate immunity.
Acknowledgments
We thank Filip Sas for assistance with sample preparation. This project was sponsored by the Flemish Science Foundation (Grants G.0187.00 and G.0175.02). G.B. is a postdoctoral fellow of the Flemish Science Foundation–Flanders, P.V. is junior research fellow of the Flemish Science Foundation, and G.V.d.B. is funded by the Research Council of the Katholieke Universiteit Leuven (OT-01/22).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: LPS, lipopolysaccharides; 2D-DIGE, 2D difference gel electrophoresis; IEF, isoelectric focusing; IPG, immobilized pH gradient; TEP, thioester-containing protein; PEBP, phosphatidylethanolamine-binding protein.
Footnotes
Xiao, Y. Y., White, K. P. & Haddad, G. G. (2002) Annu. Dros. Res. Conf. 43, 848B (abstr.).
References
- 1.Hultmark, D. (1993) Trends Genet. 9, 178–183. [DOI] [PubMed] [Google Scholar]
- 2.Lemaitre, B., Kromer-Metzger, E., Michaut, L., Nicolas, E., Meister, M., Georgel, P., Reichhart, J. M. & Hoffmann, J. A. (1995) Proc. Natl. Acad. Sci. USA 92, 9465–9469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lemaitre, B., Nicolas, E., Michaut, L., Reichhart, J. M. & Hoffmann, J. A. (1996) Cell 86, 973–983. [DOI] [PubMed] [Google Scholar]
- 4.Levashina, E. A., Langley, E., Green, C., Gubb, D., Ashburner, M., Hoffmann, J. A. & Reichhart, J. M. (1999) Science 285, 1917–1919. [DOI] [PubMed] [Google Scholar]
- 5.De Gregorio, E., Spellman, P. T., Tzou, P., Rubin, G. M. & Lemaitre, B. (2002) EMBO J. 21, 2568–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naitza, S., Rosse, C., Kappler, C., Georgel, P., Belvin, M., Gubb, D., Camonis, J., Hoffmann, J. A. & Reichhart, J. M. (2002) Immunity 17, 575–581. [DOI] [PubMed] [Google Scholar]
- 7.Hultmark, D. (2003) Curr. Opin. Immunol. 15, 12–19. [DOI] [PubMed] [Google Scholar]
- 8.Kim, Y. S., Ryu, J. H., Han, S. J., Choi, K. H., Nam, K. B., Jang, I. H., Lemaitre, B., Brey, P. T. & Lee, W. J. (2000) J. Biol. Chem. 275, 32721–32727. [DOI] [PubMed] [Google Scholar]
- 9.De Gregorio, E., Spellman, P. T., Rubin, G. M. & Lemaitre, B. (2001) Proc. Natl. Acad. Sci. USA 98, 12590–12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Werner, T., Liu, G., Kang, D., Ekengren, S., Steiner, H. & Hultmark, D. (2000) Proc. Natl. Acad. Sci. USA 97, 13772–13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bradford, M. M. (1976) Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 12.Van den Bergh, G., Clerens, S., Cnops, L., Vandesande, F. & Arckens, L. (2003) J. Neurochem. 85, 193–205. [DOI] [PubMed] [Google Scholar]
- 13.Schevchenko, A., Wilm, M., Vorm, O. & Mann, M. (1996) Anal. Chem. 68, 850–858. [DOI] [PubMed] [Google Scholar]
- 14.Vierstraete, E., Cerstiaens, A., Baggerman, G., Van den Bergh, G., De Loof, A. & Schoofs, L. (2003) Biochem. Biophys. Res. Commun. 304, 831–838. [DOI] [PubMed] [Google Scholar]
- 15.Gharahdaghi, F., Weinberg, C. R., Meagher, D. A., Imai, B. S. & Mische, S. M. (1999) Electrophoresis 20, 601–605. [DOI] [PubMed] [Google Scholar]
- 16.Zhang, W. & Chait, B. T. (2000) Anal. Chem. 72, 2482–2489. [DOI] [PubMed] [Google Scholar]
- 17.Perkins, D. N., Pappin, D. J., Creasy, D. M. & Cottrell, J. S. (1999) Electrophoresis 20, 3551–3567. [DOI] [PubMed] [Google Scholar]
- 18.Baggerman, G., Huybrechts, J., Clynen, E., Hens, K., Harthoorn, L., Van der Horst, D., Poulos, C., De Loof, A. & Schoofs, L. (2002) Peptides 23, 635–644. [DOI] [PubMed] [Google Scholar]
- 19.Irving, P., Troxler, L., Heuer, T. S., Belvin, M., Kopczynski, C., Reichhart, J. M., Hoffmann, J. A. & Hetru, C. (2001) Proc. Natl. Acad. Sci. USA 98, 15119–15124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lagueux, M., Perrodou, E., Levashina, E. A., Capovilla, M. & Hoffmann, J. A. (2000) Proc. Natl. Acad. Sci. USA 97, 11427–11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levashina, E. A., Moita, L. F., Blandin, S., Vriend, G., Lagueux, M. & Kafatos, F. C. (2001) Cell 104, 709–718. [DOI] [PubMed] [Google Scholar]
- 22.Oduol, F., Xu, J., Niare, O., Natarajan, R. & Vernick, K. D. (2000) Proc. Natl. Acad. Sci. USA 97, 11397–11402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christophides, G. K., Zdobnov, E., Barillas-Mury, C., Birney, E., Blandin, S., Blass, C., Brey, P. T., Collins, F. H., Danielli, A., Dimopoulos, G., et al. (2002) Science 298, 159–165. [DOI] [PubMed] [Google Scholar]
- 24.Courgeon, A. M., Maingourd, M., Maisonhaute, C., Montmory, C., Rollet, E., Tanguay, R. M. & Best-Belpomme, M. (1993) Exp. Cell Res. 204, 30–37. [DOI] [PubMed] [Google Scholar]
- 25.Ramet, M., Manfruelli, P., Pearson, A., Mathey-Prevot, B. & Ezekowitz, R. A. (2002) Nature 416, 644–648. [DOI] [PubMed] [Google Scholar]
- 26.Schoentgen, F. & Jolles, P. (1995) FEBS Lett. 369, 22–26. [DOI] [PubMed] [Google Scholar]
- 27.Hengst, U., Albrecht, H., Hess, D. & Monard, D. (2001) J. Biol. Chem. 276, 535–540. [DOI] [PubMed] [Google Scholar]
- 28.Yeung, K. C., Seitz, T., Li, S., Janosch, P., McFerran, B., Kaiser, C., Fee, F., Katsanakis, K. D., Rose, D. W., Mischak, H., et al. (1999) Nature 401, 173–177. [DOI] [PubMed] [Google Scholar]
- 29.Yeung, K. C., Rose, D. W., Dhillon, A. S., Yaros, D., Gustafsson, M., Chatterjee, D., McFerran, B., Wyche, J., Kolch, W. & Sedivy, J. M. (2001) Mol. Cell. Biol. 21, 7207–7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghosh, S., May, M. J. & Kopp, E. B. (1998) Annu. Rev. Immunol. 16, 225–260. [DOI] [PubMed] [Google Scholar]
- 31.Karin, M. (1998) Cancer J. Sci. Am. 4, S92–S99. [PubMed] [Google Scholar]
- 32.Bruun, A. W., Svendsen, I., Sorensen, S. O., Kielland-Brandt, M. C. & Winther, J. R. (1998) Biochemistry 37, 3351–3357. [DOI] [PubMed] [Google Scholar]
- 33.Meylaers, K., Clynen, E., Daloze, D., De Loof, A. & Schoofs, L. (2003) Insect Biochem. Mol. Biol. 34, 43–49. [DOI] [PubMed] [Google Scholar]
- 34.Hansen, I. A., Meyer, S. R., Schafer, I. & Scheller, K. (2002) Eur. J. Biochem. 269, 954–960. [DOI] [PubMed] [Google Scholar]
- 35.Singh, S. P., Coronella, J. A., Benes, H., Cochrane, B. J. & Zimniak, P. (2001) Eur. J. Biochem. 268, 2912–2923. [DOI] [PubMed] [Google Scholar]
- 36.Ranson, H., Claudianos, C., Ortelli, F., Abgrall, C., Hemingway, J., Sharakhova, M. V., Unger, M. F., Collins, F. H. & Feyereisen, R. (2002) Science 298, 179–181. [DOI] [PubMed] [Google Scholar]
- 37.Dunkov, B. C. & Georgieva, T. (1999) Cell Biol. 18, 937–944. [DOI] [PubMed] [Google Scholar]
- 38.Yoshiga, T., Georgieva, T., Dunkov, B. C., Harizanova, N., Ralchev, K. & Law, J. H. (1999) Eur. J. Biochem. 260, 414–420. [DOI] [PubMed] [Google Scholar]
- 39.Fyrberg, E. A., Mahaffey, J. W., Bond, B. J. & Davidson, N. (1983) Cell 33, 115–123. [DOI] [PubMed] [Google Scholar]
- 40.Lissemore, J. L., Baumgardner, C. A., Geer, B. W. & Sullivan, D. T. (1990) Biochem. Genet. 28, 615–630. [DOI] [PubMed] [Google Scholar]
- 41.Meghlaoui, G. K. & Veuille, M. (1997) Mol. Evol. 44, 23–32. [DOI] [PubMed] [Google Scholar]