Abstract
The transcription factor GATA-1 and its cofactor, friend of GATA-1 (FOG-1), are essential for normal erythroid development. FOG-1 physically interacts with GATA-1 to augment or inhibit its activity. The mechanisms by which FOG-1 regulates GATA-1 function are unknown. By using an assay that is based on the phenotypic rescue of a GATA-1-null erythroid cell line, we found that a conditional form of GATA-1 (GATA-1-ER) strongly induced histone acetylation at the β-major globin promoter in vivo, consistent with previous results. In contrast, GATA-1 bearing a point mutation that impairs FOG-1 binding [GATA-1(V205M)-ER] failed to induce high levels of histone acetylation at this site. However, at DNase I-hypersensitive site (HS)3 of the β-globin locus control region, GATA-1-induced histone acetylation was FOG-1-independent. Because the V205M mutation does not disrupt GATA-1 binding to DNA templates in vitro, we were surprised to find that in vivo GATA-1(V205M)-ER fails to bind the β-globin promoter. However, at HS3, DNA binding by GATA-1 was FOG-1-independent, thus correlating histone acetylation with GATA-1 occupancy. Examination of additional GATA-1-dependent regulatory elements showed that the interaction with FOG-1 is required for GATA-1 occupancy at select sites, such as HS2, but is dispensable at others, including the FOG-1-independent GATA-1 target gene EKLF. Remarkably, at the GATA-2 gene, which is repressed by GATA-1, interaction with FOG-1 was dispensable for GATA-1 occupancy and was required for transcriptional inhibition and histone deacetylation. These results indicate that FOG-1 employs distinct mechanisms when cooperating with GATA-1 during transcriptional activation and repression.
Keywords: globin, chromatin, histones, acetylation
GATA transcription factors control diverse developmental processes in vertebrates and invertebrates (see refs. 1–4 for review). The majority of vertebrate GATA proteins contain two adjacent zinc fingers where the C-terminal finger is essential for DNA binding and the N-terminal finger stabilizes DNA binding on complex GATA elements (5). Most, if not all, double-finger GATA factors contact friend of GATA (FOG) proteins by means of their N-terminal zinc finger (6, 7). Although FOG proteins contain multiple zinc fingers, there is no evidence to indicate that they bind DNA directly. In mature erythroid cells, GATA-1 and FOG-1 are the predominant representatives of their respective families, and both are essential for normal erythroid development (6, 8, 9). Loss of GATA-1 through gene targeting leads to differentiation arrest and apoptosis in primitive and definitive erythroid precursor cells (10, 11). Similarly, lack of FOG-1 causes embryonic death as a result of failed erythropoiesis with a differentiation arrest at a stage similar to that observed with loss of GATA-1 (12). Direct physical interaction between GATA-1 and FOG-1 is required for their function during erythroid maturation (13), and patients with point mutations within the N-terminal zinc finger domain of GATA-1 that disrupt or reduce FOG-1 binding suffer from anemia and thrombocytopenia (14).
Although most studies of GATA-1 have focused on its role as a transcriptional activator, recent microarray experiments revealed that the number of genes repressed by GATA-1 is similar to that of GATA-1-activated genes, suggesting that gene repression by GATA-1 might play a similarly important role during terminal erythroid differentiation (15). For example, GATA-1-induced cellular maturation is accompanied by rapid repression of the c-myc and GATA-2 genes (13, 15). Forced expression of c-myc overrides GATA-1-induced cell cycle arrest, demonstrating the importance of c-myc gene repression by GATA-1 (15). Of note, c-myc (15) and GATA-2 (ref. 16 and this article) appear to be direct targets of GATA-1, and their repression requires interaction with FOG-1 (13). Although FOG-1 functionally synergizes with GATA-1 during the differentiation of erythroid cells, FOG-1 can either activate or inhibit GATA-1 activity in transfection-based assays depending on cell and promoter context (6, 7, 17, 18). Although the mechanisms by which FOG-1 augments or inhibits GATA-1 activity are unknown, the assumption has been that FOG-1 acts at a step following DNA binding of GATA-1.
GATA-1 interacts with several additional proteins to regulate erythroid gene expression (see ref. 19 for review). Among these is the acetyltransferase CREB (cAMP response element-binding protein)-binding protein (CBP), which can bind to either GATA-1 zinc finger and enhance the transcriptional activity of GATA-1 (20). Interference with CBP activity results in loss of erythroid differentiation and globin gene induction (20). Moreover, CBP acetylates GATA-1 at conserved lysine residues near each of the zinc fingers (20, 21), and mutations at these sites impair the ability of GATA-1 to induce erythroid maturation in vivo (20). An additional role for CBP recruitment by GATA-1 comes from studies showing that restoration of GATA-1 activity in GATA-1-deficient pro-erythroblasts (G1E cells) leads to increased histone H3 and H4 acetylation at the β-major globin gene promoter and the locus control region (LCR) (22, 23). Specifically, GATA-1 occupancy at these sites in vivo correlates with CBP recruitment, histone acetylation, and onset of globin gene transcription (23). Thus, GATA-1, together with CBP, contributes to the erythroid-specific histone acetylation patterns found at the murine β-globin locus (24–27) and, presumably, other GATA-1-dependent genes.
Here we examined the role of FOG-1 during GATA-1-induced histone modifications by using the GATA-1-deficient erythroblast cell line G1E (28) stably expressing either GATA-1 fused to the ligand-binding domain of the estrogen receptor (ER) (GATA-1-ER) or GATA-1(V205M)-ER, which is defective for FOG-1 binding (14). We found that activation of GATA-1(V205M)-ER displayed diminished ability to induce histone acetylation at the β-globin promoter when compared with GATA-1-ER. In contrast, at HS3 of the LCR, GATA-1(V205M)-ER showed an activity comparable with GATA-1-ER. Surprisingly, GATA-1(V205M)-ER, which binds DNA normally in vitro, showed diminished association with the β-globin gene promoter and select other sites in vivo. Thus, one mechanism by which FOG-1 acts is to facilitate or stabilize GATA-1 binding to chromatinized DNA in vivo. However, when analyzed at the GATA-2 gene, which is repressed by GATA-1, FOG-1 did not regulate GATA-1 occupancy but instead provided corepressor activity. These results show that FOG-1 modulates GATA-1 activity by distinct mechanisms depending on promoter context.
Materials and Methods
Cell Culture. G1E cells were cultured as described (28). G1E cells expressing GATA-1-ER or GATA-1(V205M)-ER have been described (14). Cells were exposed to 1 μM estradiol (Sigma) where indicated.
Chromatin immunoprecipitation (ChIP) assays were performed as described (23). Briefly, 1 × 108 cells were crosslinked in 0.4% formaldehyde/PBS for 10 min at room temperature. Crosslinking was stopped with 0.125 M glycine. Chromatin was sonicated, precleared with irrelevant antibodies, and immunoprecipitated with indicated antibodies prebound to protein A or protein G beads. Beads were washed seven times, and the bound proteins were eluted into 100 mM NaHCO3/1% SDS. Crosslinks were reversed at 65°C for 4 h, and protein was digested with proteinase K (0.3 mg/ml) for 2 h at 45°C. DNA was purified by phenol/chloroform extraction and ethanol precipitation. [α-32P]dCTP-labeled PCR products were quantified by PhosphorImager analysis (23). PCR products were plotted as percentage of input chromatin. Although the absolute values expressed in percent of input might vary between experiments, the relative differences between samples of the same experiments are highly reproducible. Primer pairs used for PCR of the β-major promoter, HS2, HS3, and HS4 have been described (23, 25). Primer pairs for the EKLF upstream region are GATTTGAGGGGACTCCTTTTGC and AGGAGTGGACCAGGAAGGATAGA. GATA-2 primers, located ≈3 kb upstream of the 1S promoter, are GAGACCCGGCAAGGCATGAGC and GATAATCTGGAAGGCAGAGATAAG. In select experiments, real-time PCR (SYBR Green; Applied Biosystems) was used to confirm the data obtained with conventional PCR.
Antibodies. The antibodies used in this study and their suppliers were as follows: anti-diacetyl-H3 (acH3) (catalog no. 06-599) and anti-tetraacetyl-H4 (acH4) (catalog no. 06-866), Upstate Biotechnology, Lake Placid, NY; anti-GATA-1 (N6), Santa Cruz; and anti-ER (Ab10), Lab Vision Neomarkers (Fremont, CA). Anti-EKLF was a gift from James Bieker (Mount Sinai School of Medicine, New York).
Western Blotting. Thirty micrograms of nuclear extract was fractionated on a 10% polyacrylamide gel, transferred to a nitrocellulose membrane, and blotted with anti-GATA-1 (N6) and anti-EKLF.
Northern Blotting. Total RNA was isolated from G1E-GATA-1-ER and G1E-GATA-1(V205M)-ER cells at various time points with TRIzol (Invitrogen). Equal amounts of RNA were separated on an agarose gel, transferred to a nitrocellulose membrane, and probed with full-length 32P-labeled β-globin cDNA.
Results and Discussion
FOG-1 Requirement for GATA-1-Induced Histone Acetylation at Select Sites. In prior work we demonstrated that activation of GATA-1-ER in the GATA-1-null pro-erythroblast cell line G1E stimulated histone acetylation at the LCR and the β-major gene promoter (23). To examine whether FOG-1 contributes to this function of GATA-1, we used G1E cells stably expressing GATA-1(V205M)-ER, which is defective for FOG-1 binding. Valine-205 resides in the N-terminal finger on the surface opposite to that involved in DNA binding and contacts FOG-1 but not DNA (29). Replacement of V205 with methionine or glycine diminishes FOG-1 binding without affecting the interaction with DNA in vitro (13, 14, 30). Importantly, neither protein stability nor the interaction with other known GATA-1-binding proteins, such as CBP, is affected by mutations at this residue (ref. 13 and unpublished observations). In agreement with this interpretation is the finding that a compensatory mutation in FOG-1 that restores its binding to mutant GATA-1 rescues erythroid differentiation (13). Thus, the V205M substitution disrupts the FOG-1 interaction with a high degree of specificity.
For our studies we chose two well characterized GATA-1(V205M)-ER clones and a control GATA-1-ER clone, all of which express GATA-1 proteins at levels similar to those found endogenously in non-gene-targeted erythroid cell lines (ref 14; Fig. 7A, which is published as supporting information on the PNAS web site; data not shown). Although both GATA-1(V205M)-ER clones produced virtually identical results, one clone was studied in greatest detail and is described in the following experiments. GATA-1(V205M)-ER failed to induce significant amounts of β-globin mRNA or repress GATA-2 (Fig. 7 B and C), consistent with previous results showing that FOG-1 binding is required for transcriptional activation and repression of a subset of GATA-1 target genes (13, 14).
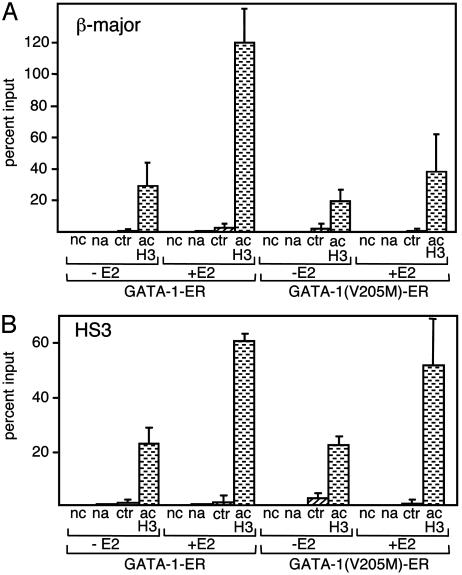
To determine whether FOG-1 is required for GATA-1-mediated increases in histone acetylation, we performed ChIP experiments with anti-acH3 antibodies and compared histone acetylation levels at the β-major globin promoter and the LCR in GATA-1(V205M)-ER and GATA-1-ER cells. As previously observed (23), estradiol treatment of GATA-1-ER cells triggered a pronounced increase in histone acetylation at both the β-major promoter and HS3 (Fig. 1A). In contrast, GATA-1(V205M)-ER was markedly impaired in its ability to induce histone acetylation at the β-major promoter (Fig. 1 A). Remarkably, at HS3, GATA-1(V205M)-ER elicited normal histone acetylation (Fig. 1B).
Fig. 1.
FOG-1 is required for GATA-1-induced histone acetylation at the β-major promoter but not at HS3. ChIP assays were performed with cells expressing GATA-1-ER or GATA-1(V205M)-ER before (–E2) and after (+E2) treatment with estradiol for 21 h. (A) Chromatin was immunoprecipitated with the anti-diacetyl H3 antibody (acH3) and analyzed with PCR primers spanning the β-major globin gene promoter. Results are plotted as percent of input chromatin. Controls include no chromatin (nc), no antibody (na), and nonimmune IgG (ctr). Results are averages of six independent experiments. (B) Experiments are as in A, but PCR primers are specific for HS3. Results are averages of four independent experiments. In all figures, error bars indicate SD.
A requirement for FOG-1 during GATA-1-induced histone acetylation at the β-major promoter was surprising, because the V205M substitution does not affect interaction between GATA-1 and CBP. This raised the possibility that FOG-1 might itself associate with acetyltransferase activity. Preliminary experiments failed to detect measurable histone acetylase activity in immunoprecipitation/acetylase assays with anti-FOG-1 antibodies (data not shown). Because acetylation of histone H3 at lysine-14 can be facilitated by prior phosphorylation of serine-10 (31, 32) we examined whether FOG-1 might associate with H3S10 kinase activity. In immunoprecipitation/kinase assays with H3 as a substrate, we failed to detect measurable histone kinase activity (data not shown). Thus, FOG-1 appears to facilitate GATA-1-induced histone acetylation by a different mechanism.
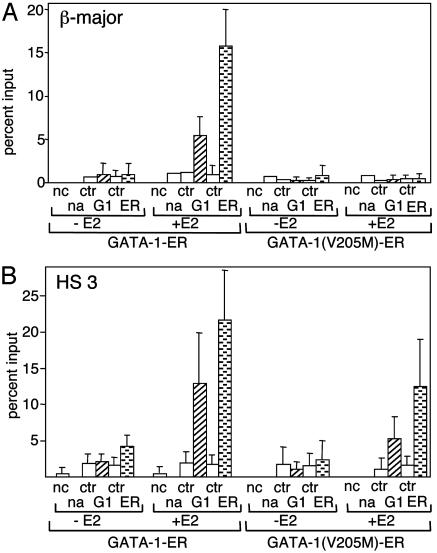
FOG-1 Is Required for GATA-1 Occupancy at the β-Globin Promoter but Not HS3 in Vivo. Previous reports showed that substitutions at valine-205 do not affect GATA-1 binding to naked DNA templates in vitro (13, 14). To test whether FOG-1 binding is required for GATA-1 to access chromatinized templates in vivo, we measured the presence of wild-type and mutant GATA-1-ER proteins at the β-major promoter and the LCR by using ChIP experiments with anti-GATA-1 antibodies. Remarkably, little or no GATA-1(V205M)-ER protein was detected at the β-major promoter, whereas GATA-1-ER was present at high levels (Fig. 2A). In contrast, GATA-1-ER and GATA-1(V205M)-ER bound HS3 with comparable efficiency, with the latter being only moderately reduced (Fig. 2B; for representative gels, see Fig. 8, which is published as supporting information on the PNAS web site). In control experiments, little or no occupancy of either GATA-1 fusion protein was detected at the inactive embryonic βH1-globin gene (Fig. 9, which is published as supporting information on the PNAS web site). Together, these results suggest that interaction with FOG-1 is essential for GATA-1 access or stable binding to the β-globin promoter but not to HS3 of the LCR.
Fig. 2.
FOG-1 is required for GATA-1 occupancy at the β-major promoter but not HS3. (A) ChIP assay as in Fig. 1 with anti-GATA-1 (G1) and anti-ER (ER) antibodies. Results are the averages of five independent experiments. (B) ChIP analysis as in A but with primers specific for HS3.
We considered the possibility that the failure to detect GATA-1(V205M)-ER at the β-major promoter was caused by occlusion of the epitope recognized by the anti-GATA-1 antibody, perhaps because of an altered protein environment at the inactive β-globin gene. To address this issue, we used an antibody against the ER portion of the GATA-1 fusion proteins for ChIP experiments. The results confirmed the virtual absence of GATA-1(V205M)-ER at the β-major promoter, whereas levels were comparable with GATA-1-ER at HS3 (Fig. 2). To rule out the possibility that reduced association of GATA-1(V205M)-ER with the β-major promoter was the result of clonal variation, we examined by ChIP an independently derived G1E cell clone expressing GATA-1(V205M)-ER and obtained essentially the same results (Fig. 10, which is published as supporting information on the PNAS web site).
At several sites, including HS3 and HS4 of the LCR, and EKLF (see below), we consistently observed slightly weaker signals with the anti-GATA-1 antibody compared with the anti-ER antibody. Although the reasons for this discrepancy are unclear, both data sets confirm the virtual absence of GATA-1(V205M)-ER from the β-major promoter. Furthermore, these results serve to stress the importance of using more than one antibody when assaying the presence or absence of a given protein by ChIP. Finally, because the V205M mutation does not disrupt the interaction of GATA-1 with CBP, the failure of GATA-1(V205M)-ER to induce high levels of histone acetylation at the β-major promoter is further (albeit indirect) evidence for the absence of GATA-1(V205M)-ER protein at this site.
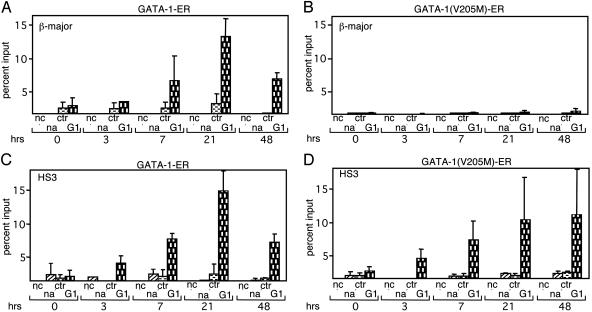
To examine whether the reduced association of GATA-1(V205M)-ER with the β-major promoter reflected altered kinetics of occupancy, we measured the presence of GATA-1 proteins in time course experiments. Cells expressing GATA-1-ER constructs were treated with estradiol for 3, 7, 21, and 48 h followed by ChIP analysis with anti-GATA-1 antibodies. Although GATA-1-ER was detected at the β-major promoter at 7 h after the addition of estradiol with maximal GATA-1-occupancy at 21 h (Fig. 3A), at no time point did we detect substantial amounts of GATA-1(V205M)-ER at the β-major promoter (Fig. 3B). In contrast, the kinetics and extent of occupancy at HS3 were similar between wild-type and mutant GATA-1-ER (Fig. 3 C and D). The same results were obtained with anti-ER antibodies (data not shown). At both the β-major promoter and HS3, a moderate decline in the presence of GATA-1-ER, but not GATA-1(V205M)-ER, was observed at 48 h after the addition of estradiol (Fig. 3). These data further substantiate the above results that FOG-1 is required for GATA-1 binding to the β-major promoter but not to HS3 of the LCR.
Fig. 3.
Kinetics of GATA-1 occupancy at the β-globin locus. ChIP experiments were performed at indicated time points after the addition of estradiol. Occupancy of GATA-1-ER (A) and GATA-1(V205M)-ER (B)atthe β-major promoter and at HS3 (C and D) is shown. The results of each time point are the averages of at least three independent experiments.
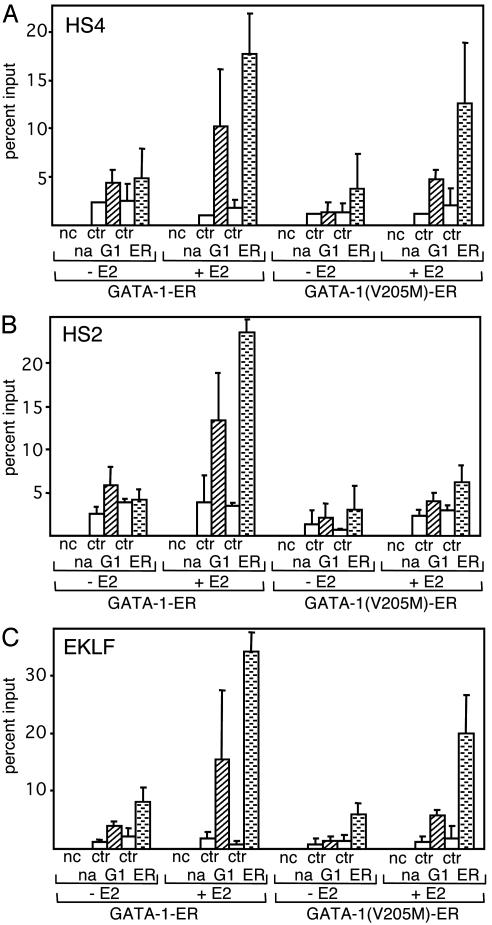
FOG-1 Requirement for GATA-1-ER Occupancy at Erythroid Regulatory Elements. To examine whether the V205M mutation reduces GATA-1 access to sites other than the β-major promoter, we examined HS2 and HS4 of the LCR, which, like HS3, are bound by GATA-1 in vivo (33–35). We found that although FOG-1 binding appears to be dispensable for GATA-1 access to HS4 (Fig. 4A), it is required for GATA-1 binding to HS2 (Fig. 4B). We next examined occupancy of GATA-1 at the EKLF gene, which is activated by GATA-1 (14, 36–38) in a FOG-1-independent manner (ref. 13; Fig. 7A). As expected, both wild-type and FOG-1-binding-defective GATA-1-ER fusion proteins were able to access the EKLF upstream enhancer, although the signal for GATA-1(V205M)-ER was somewhat reduced, especially when the anti-GATA-1 antibody was used. (Fig. 4C). Thus, at the EKLF gene, FOG-1 is not essential for GATA-1 occupancy and activity. Consistent with this finding is our observation that both wild-type and mutant GATA-1-ER proteins were able to trigger the same moderate increase in histone acetylation at this gene (data not shown).
Fig. 4.
FOG-1 is required for GATA-1 occupancy at select erythroid regulatory regions. For ChIP assay, anti-GATA-1 (G1) and anti-ER antibodies (ER) were used. Controls are as in Fig. 1. Cells were treated with estradiol (E2) for 21 h. Primer pairs were directed against HS4 (A), HS2 (B), and the upstream enhancer of the EKLF gene (C). Results are the averages of at least four independent experiments.
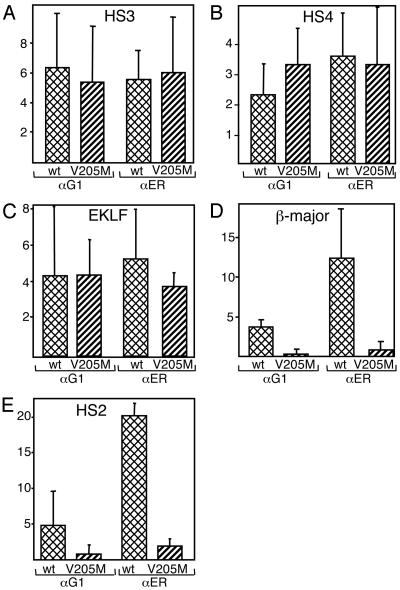
Despite similar expression of GATA-ER fusion proteins among the cell lines used in our studies, slight variations in protein levels or undefined clonal effects might account for some of the moderate differences in binding of GATA-ER fusion proteins to HS3, HS4, and the EKLF enhancer. To more precisely gauge GATA-1 access to its cognate elements in vivo, we plotted our data to indicate fold increase in occupancy after estradiol treatment. Fig. 5 shows a very similar increase in protein occupancy of wild-type and mutant proteins at HS3 (Fig. 5A), HS4 (Fig. 5B), and the EKLF gene (Fig. 5C), indicating that overall accessibility is comparable between wild-type and mutant proteins at these sites. In contrast, GATA-1(V205M)-ER was strongly impaired in accessing both the β-major promoter and HS2 (Fig. 5 D and E). Together, these data confirm that FOG-1 is required for GATA-1 access to certain regulatory elements in vivo while being dispensable at others. However, because the V205M mutation does not completely abrogate FOG-1 binding, we cannot rule out that FOG-1 might contribute to GATA-1(V205M)-ER occupancy at HS3 and HS4.
Fig. 5.
Increase in GATA-1 occupancy at select sites after estradiol treatment. Results from Figs. 2 and 4 are replotted to indicate fold increase in GATA-1 occupancy after estradiol treatment. Results are shown for HS3 (A), HS4 (B), the EKLF promoter (C), the β-major promoter (D), and HS2 (E). αG1, anti-GATA-1; αER, anti-ER.
Although enhancing GATA-1 access to erythroid regulatory elements is one mechanism by which FOG-1 cooperates with GATA-1, our data are not incompatible with additional functions for FOG-1. For example, upon recruitment to GATA-1-dependent genes, FOG-1 might provide coactivator function by forming contacts with components of the basal transcriptional machinery or by chromatin-mediated mechanisms.
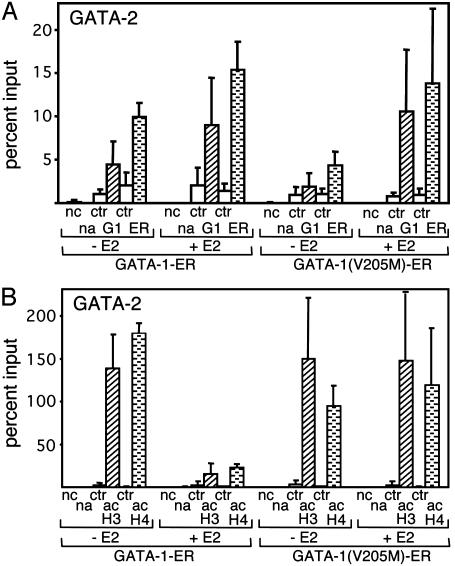
FOG-1 Can Function as Corepressor for GATA-1. Although it appears that gene repression plays an important role during GATA-1-regulated erythroid differentiation (15), the mechanism by which GATA-1 inhibits gene expression is unknown. However, the observation that GATA-1(V205M)-ER fails to repress the GATA-2 gene points to an important role for FOG-1 in this process (ref. 13 and Fig. 7C). Two mutually nonexclusive mechanisms might account for FOG-1 function. First, FOG-1 might cooperate with GATA-1 by enhancing GATA-1 access to the GATA-2 locus in a fashion similar to that observed at the β-globin promoter. Second, FOG-1 might endow GATA-1 with corepressor activity. In support of the latter possibility, it has been shown that FOG proteins can associate with members of the carboxyl-terminal binding protein (CtBP) family of corepressors (7, 39–42), although the relevance of these interactions to erythroid gene expression remains uncertain (43). To distinguish between these two possibilities, we examined occupancy by wild-type and mutant GATA-1-ER protein at the GATA-2 gene. ChIP assays with both anti-GATA-1 and anti-ER antibodies revealed that GATA-1(V205M)-ER bound the GATA-2 gene as efficiently as GATA-1-ER (Fig. 6A). Thus, FOG-1 is dispensable for GATA-1 binding to the GATA-2 gene but is required for gene repression. To examine whether repression is associated with histone deacetylation at the GATA-2 locus, ChIP experiments were performed with anti-acH3 and anti-acH4 antibodies. Although activation of GATA-1-ER triggered substantial deacetylation of histones H3 and H4, GATA-1(V205M)-ER had no effect (Fig. 6B). Together, these data suggest that FOG-1 is required for GATA-1-mediated gene repression and histone deacetylation but not for GATA-1 occupancy at the GATA-2 locus. FOG-1 might actively link GATA-1 to a histone deacetylase-containing repressor complex, or it might simply interfere with the action of acetylases. However, our findings do not rule out the possibility that FOG-1 might stimulate GATA-1 occupancy at other genes repressed by GATA-1.
Fig. 6.
FOG-1 can function as a corepressor for GATA-1. (A) GATA-1-ER and GATA-1(V205M)-ER occupancy at the GATA-2 gene. ChIP experiments were performed as described for Fig. 2. Results are the average of four independent experiments. (B) ChIP analysis of histone acetylation levels at the GATA-2 enhancer by using the acH3 and acH4 antibodies as in Fig. 1. Results are the averages of five acH3 and two acH4 independent experiments.
A challenging question remains as to how FOG-1 increases GATA-1 access to certain sites in chromatin. It is possible that FOG-1 associates with chromatin remodeling activity to facilitate access to chromatinized target genes. This model would imply that once GATA-1 is bound to its cognate elements FOG-1 might be dispensable for subsequent GATA-1 action. Such a transient requirement for FOG-1 activity might be one explanation for the relatively poor detection of FOG-1 at regulatory sites in the β-globin locus (data not shown). Of note, in parallel experiments in megakaryocytes, FOG-1 is readily detectable by ChIP at the megakaryocyte-restricted αIIb gene promoter (17), raising the possibility that FOG-1 functions by a mechanism that is distinct from that used at the β-globin gene locus. At sites where GATA-1 binding does not require FOG-1, such as HS3, neighboring proteins might provide an activity to render the local chromatin configuration permissive to GATA-1 binding. For example, previous studies have shown that GATA-1 binding to chromatinized templates in vitro is facilitated by prior binding and recruitment of remodeling activity by the transcription factor NF-E2 (44).
Another possibility is that FOG-1 enables or stabilizes GATA-1 association with the β-globin promoter by contributing to the physical interaction between the LCR and the β-globin promoter (45, 46). In this scenario, the communication between the LCR and the promoter in vivo depends on a large protein complex containing GATA-1 and FOG-1. In the absence of FOG-1, GATA-1 might fail to stabilize the LCR–promoter interaction, thus lowering its affinity for the promoter. An intriguing example of the interdependence between transcription factor binding to the LCR and the β-globin promoter is provided by the observation that association of EKLF with the LCR requires an intact β-globin promoter (47).
The absence of GATA-1(V205M)-ER at the β-major promoter is supported by ChIP with two different antibodies. Additionally, this result is consistent with the diminished induction of histone acetylation at this site. If GATA-1(V205M)-ER fails to occupy the β-major promoter, what triggers the small but consistently observed increase in histone acetylation (Fig. 1 A)? One possibility is that residual presence of GATA-1(V205M)-ER accounts for this activity. Alternatively, the transient up-regulation of EKLF by GATA-1(V205M)-ER (Fig. 7A) might lead to recruitment of histone acetylase activity to the β-major promoter (48).
In summary, our data reveal an unexpected function for FOG-1 during GATA-1-mediated gene activation by facilitating access to select regulatory sites in vivo. At a gene repressed by GATA-1, FOG-1 appears to employ a distinct mechanism by providing essential corepressor activity. Examination of the contextual requirements that specify the role of FOG-1 at a given gene will be a fertile area of research with implications for numerous tissues whose formation is controlled by GATA and FOG proteins.
Supplementary Material
Acknowledgments
We thank Merlin Crossley, Chris Vakoc, and Mitch Weiss for discussions and comments on the manuscript. This work was supported by National Institutes of Health Grants DK58044 and DK54937 (to G.A.B.), National Institutes of Health Training Grant T32GM008216 (to D.L.L.), and Cancer Research Institute Training Grant Predoctoral Emphasis Pathway in Tumor Immunology (to Y.-Y.C.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: FOG-1, friend of GATA-1; HS, DNase I-hypersensitive site; LCR, locus control region; CBP, cAMP response element-binding protein binding protein; ER, estrogen receptor; ChIP, chromatin immunoprecipitation; acH3, diacetyl-H3; acH4, tetraacetyl-H4.
References
- 1.Charron, F. & Nemer, M. (1999) Semin. Cell Dev. Biol. 10, 85–91. [DOI] [PubMed] [Google Scholar]
- 2.Molkentin, J. D. (2000) J. Biol. Chem. 275, 38949–38952. [DOI] [PubMed] [Google Scholar]
- 3.Patient, R. K. & McGhee, J. D. (2002) Curr. Opin. Genet. Dev. 12, 416–422. [DOI] [PubMed] [Google Scholar]
- 4.Weiss, M. J. & Orkin, S. H. (1995) Exp. Hematol. 23, 99–107. [PubMed] [Google Scholar]
- 5.Martin, D. I. & Orkin, S. H. (1990) Genes Dev. 4, 1886–1898. [DOI] [PubMed] [Google Scholar]
- 6.Tsang, A. P., Visvader, J. E., Turner, C. A., Fujiwara, Y., Yu, C., Weiss, M. J., Crossley, M. & Orkin, S. H. (1997) Cell 90, 109–119. [DOI] [PubMed] [Google Scholar]
- 7.Fox, A. H., Liew, C., Holmes, M., Kowalski, K., Mackay, J. & Crossley, M. (1999) EMBO J. 18, 2812–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pevny, L., Simon, M. C., Robertson, E., Klein, W. H., Tsai, S.-F., D'Agati, V., Orkin, S. H. & Costantini, F. (1991) Nature 349, 257–260. [DOI] [PubMed] [Google Scholar]
- 9.Fujiwara, Y., Browne, C. P., Cunniff, K., Goff, S. C. & Orkin, S. H. (1996) Proc. Natl. Acad. Sci. USA 93, 12355–12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiss, M. J. & Orkin, S. H. (1995) Proc. Natl. Acad. Sci. USA 92, 9623–9627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiss, M. J., Keller, G. & Orkin, S. H. (1994) Genes Dev. 8, 1184–1197. [DOI] [PubMed] [Google Scholar]
- 12.Tsang, A. P., Fujiwara, Y., Hom, D. B. & Orkin, S. H. (1998) Genes Dev. 12, 1176–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crispino, J. D., Lodish, M. B., MacKay, J. P. & Orkin, S. H. (1999) Mol. Cell 3, 219–228. [DOI] [PubMed] [Google Scholar]
- 14.Nichols, K. E., Crispino, J. D., Poncz, M., White, J. G., Orkin, S. H., Maris, J. M. & Weiss, M. J. (2000) Nat. Genet. 24, 266–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rylski, M., Welch, J. J., Chen, Y. Y., Letting, D. L., Diehl, J. A., Chodosh, L. A., Blobel, G. A. & Weiss, M. J. (2003) Mol. Cell. Biol. 23, 5031–5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grass, J. A., Boyer, M. E., Pal, S., Wu, J., Weiss, M. J. & Bresnick, E. H. (2003) Proc. Natl. Acad. Sci. USA 100, 8811–8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang, X., Crispino, J. D., Letting, D. L., Nakazawa, M., Poncz, M. & Blobel, G. A. (2002) EMBO J. 21, 5225–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaines, P., Geiger, J. N., Knudsen, G., Seshasayee, D. & Wojchowski, D. M. (2000) J. Biol. Chem. 275, 34114–34121. [DOI] [PubMed] [Google Scholar]
- 19.Cantor, A. B. & Orkin, S. H. (2002) Oncogene 21, 3368–3376. [DOI] [PubMed] [Google Scholar]
- 20.Blobel, G. A., Nakajima, T., Eckner, R., Montminy, M. & Orkin, S. H. (1998) Proc. Natl. Acad. Sci. USA 95, 2061–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boyes, J., Byfield, P., Nakatani, Y. & Ogryzko, V. (1998) Nature 396, 594–598. [DOI] [PubMed] [Google Scholar]
- 22.Kiekhaefer, C. M., Grass, J. A., Johnson, K. D., Boyer, M. E. & Bresnick, E. H. (2002) Proc. Natl. Acad. Sci. USA 99, 14309–14314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Letting, D. L., Rakowski, C., Weiss, M. J. & Blobel, G. A. (2003) Mol. Cell. Biol. 23, 1334–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schubeler, D., Francastel, C., Cimbora, D. M., Reik, A., Martin, D. I. & Groudine, M. (2000) Genes Dev. 14, 940–950. [PMC free article] [PubMed] [Google Scholar]
- 25.Forsberg, E. C., Downs, K. M., Christensen, H. M., Im, H., Nuzzi, P. A. & Bresnick, E. H. (2000) Proc. Natl. Acad. Sci. USA 97, 14494–14499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bulger, M., Schubeler, D., Bender, M. A., Hamilton, J., Farrell, C. M., Hardison, R. C. & Groudine, M. (2003) Mol. Cell. Biol. 23, 5234–5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bulger, M., Sawado, T., Schubeler, D. & Groudine, M. (2002) Curr. Opin. Genet. Dev. 12, 170–177. [DOI] [PubMed] [Google Scholar]
- 28.Weiss, M. J., Yu, C. & Orkin, S. H. (1997) Mol. Cell. Biol. 17, 1642–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kowalski, K., Czolij, R., King, G. F., Crossley, M. & Mackay, J. P. (1999) J. Biomol. NMR 13, 249–262. [DOI] [PubMed] [Google Scholar]
- 30.Newton, A., Mackay, J. & Crossley, M. (2001) J. Biol. Chem. 276, 35794–35801. [DOI] [PubMed] [Google Scholar]
- 31.Lo, W. S., Trievel, R. C., Rojas, J. R., Duggan, L., Hsu, J. Y., Allis, C. D., Marmorstein, R. & Berger, S. L. (2000) Mol. Cell 5, 917–926. [DOI] [PubMed] [Google Scholar]
- 32.Cheung, P., Tanner, K. G., Cheung, W. L., Sassone-Corsi, P., Denu, J. M. & Allis, C. D. (2000) Mol. Cell 5, 905–915. [DOI] [PubMed] [Google Scholar]
- 33.Johnson, K. D., Grass, J. A., Boyer, M. E., Kiekhaefer, C. M., Blobel, G. A., Weiss, M. J. & Bresnick, E. H. (2002) Proc. Natl. Acad. Sci. USA 99, 11760–11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horak, C. E., Mahajan, M. C., Luscombe, N. M., Gerstein, M., Weissman, S. M. & Snyder, M. (2002) Proc. Natl. Acad. Sci. USA 99, 2924–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duan, Z., Stamatoyannopoulos, G. & Li, Q. (2001) Mol. Cell. Biol. 21, 3083–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crossley, M., Tsang, A., Bieker, J. J. & Orkin, S. H. (1994) J. Biol. Chem. 269, 15440–15444. [PubMed] [Google Scholar]
- 37.Chen, X., Reitman, M. & Bieker, J. J. (1998) J. Biol. Chem. 273, 25031–25040. [DOI] [PubMed] [Google Scholar]
- 38.Anderson, K. P., Crable, S. C. & Lingrel, J. B. (1998) J. Biol. Chem. 273, 14347–14354. [DOI] [PubMed] [Google Scholar]
- 39.Turner, J. & Crossley, M. (1998) EMBO J. 17, 5129–5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deconinck, A. E., Mead, P. E., Tevosian, S. G., Crispino, J. D., Katz, S. G., Zon, L. I. & Orkin, S. H. (2000) Development (Cambridge, U.K.) 127, 2031–2040. [DOI] [PubMed] [Google Scholar]
- 41.Svensson, E. C., Huggins, G. S., Dardik, F. B., Polk, C. E. & Leiden, J. M. (2000) J. Biol. Chem. 275, 20762–20769. [DOI] [PubMed] [Google Scholar]
- 42.Fossett, N., Tevosian, S. G., Gajewski, K., Zhang, Q., Orkin, S. H. & Schulz, R. A. (2001) Proc. Natl. Acad. Sci. USA 98, 7342–7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katz, S. G., Cantor, A. B. & Orkin, S. H. (2002) Mol. Cell. Biol. 22, 3121–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Armstrong, J. A. & Emerson, B. M. (1996) Mol. Cell. Biol. 16, 5634–5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tolhuis, B., Palstra, R. J., Splinter, E., Grosveld, F. & de Laat, W. (2002) Mol. Cell 10, 1453–1465. [DOI] [PubMed] [Google Scholar]
- 46.Carter, D., Chakalova, L., Osborne, C. S., Dai, Y. F. & Fraser, P. (2002) Nat. Genet. 32, 623–626. [DOI] [PubMed] [Google Scholar]
- 47.Lee, J. S., Lee, C. H. & Chung, J. H. (1999) Proc. Natl. Acad. Sci. USA 96, 10051–10055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, W. & Bieker, J. J. (1998) Proc. Natl. Acad. Sci. USA 95, 9855–9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.