Abstract
A protein’s flexibility is well recognized to underlie its capacity to engage in critical functions, such as signal transduction, biomolecular transport and biochemical reactivity. Molecular recognition is also tightly linked to the dynamics of the binding partners, yet protein flexibility has largely been ignored by the growing field of structure-based drug design (SBDD). In combination with experimentally determined structures, a number of computational methods have been proposed to model protein movements, which may be important for small molecule binding. Such techniques have the ability to expose new binding site conformations, which may in turn recognize and lead to the discovery of more potent and selective drugs through molecular docking. In this article, we discuss various methods and focus on the Relaxed Complex Scheme (RCS), which uses Molecular Dynamics (MD) simulations to model full protein flexibility and enhance virtual screening programmes. We review practical applications of the RCS and use a recent study of the HIV-1 reverse transcriptase to illustrate the various phases of the scheme. We also discuss some encouraging developments, aimed at addressing current weaknesses of the RCS.
INTRODUCTION
With an ever-expanding repertoire of high-resolution protein structures available, the computational field of structure-based drug design (SBDD) is enjoying exposure to progressively more drug discovery studies [1,2]. SBDD methodologies are generally used in the generation and optimization of lead compounds, typically using docking algorithms to predict the position and affinity of a library of small molecules within a protein binding site [3–6]. Despite encouraging results from such virtual screening programmes (as discussed in [7]), one of the key limitations of current small molecule docking is the poor treatment of protein flexibility [8]. Such approaches usually represent the protein as a rigid structure, while the small molecules, with far fewer degrees of freedom, are subjected to conformational variation (giving rise to the term “rigid-protein flexible-ligand”). The use of a single “snapshot” of the protein has traditionally been convenient, both because it reduces the computational demands of virtual screening and because there are typically only a few experimental structures available for each target. Fortunately, advances in computer hardware and the optimization of docking software, coupled with the theoretical modelling of protein flexibility, have eased this compromise.
Our current understanding of the molecular recognition between a protein and a drug-like molecule involves a dynamic process, whereby both partners require a degree of structural plasticity to negotiate the binding/unbinding event [9,10]. This role for flexibility is not surprising, given that proteins use conformational rearrangements to carry out a wide range of functions, including reaction catalysis and protein-protein interactions. Proteins are therefore highly dynamic entities and at any one time exist in an ensemble of conformational substates, each of which is not equally populated and each of which a small molecule may be exposed to. It follows that the most popular protein substate is not necessarily the conformation which binds a small molecule and, in fact, the rarest conformers may be responsible for forming productive protein-ligand complexes. A single crystallographic protein structure is therefore merely a single point on this complex conformational landscape and an inadequate representation of the multiple conformations that may be critical for drug binding.
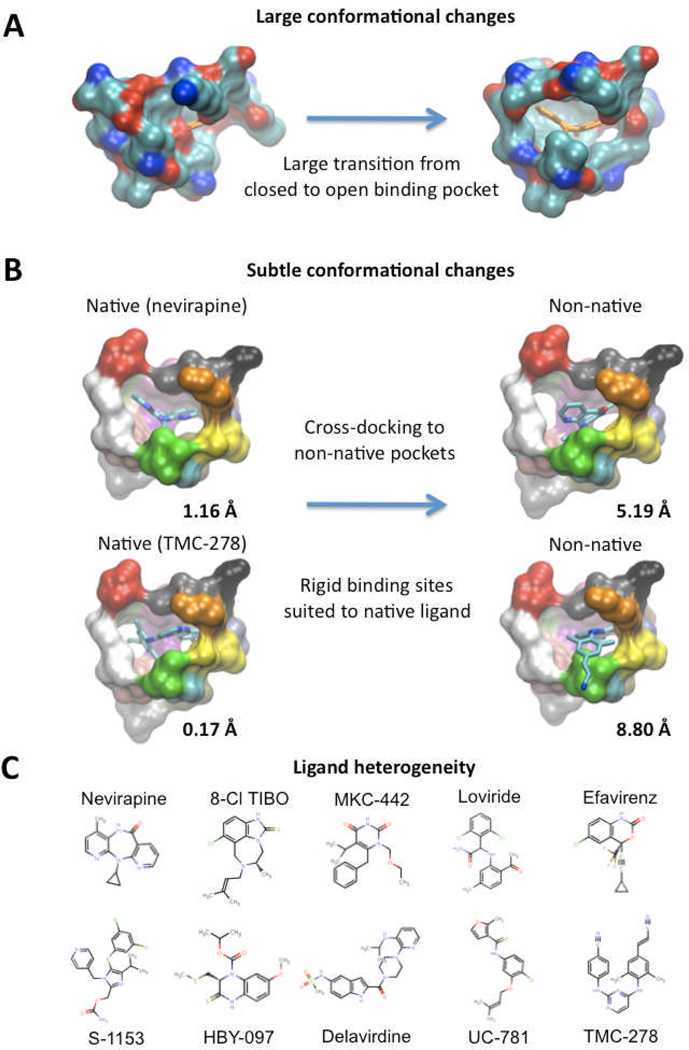
Figure 1 illustrates some aspects of the impact of binding site flexibility in the case of the non-nucleoside reverse transcriptase inhibitor (NNRTI) binding pocket of HIV-1 reverse transcriptase (HIV-1 RT) [11,12]. HIV-1 RT is a popular drug target for which substantial crystallographic structural data is available (over 100 structures) and which has revealed remarkable plasticity, both at the global and local level. In the absence of bound NNRTI, the binding pocket is completely occluded from the solvent and effectively collapsed. However, co-crystallized with NNRTIs, the pocket is captured in an open state, largely due to considerable torsional shifts of two key tyrosine residues, which flip out to accommodate the inhibitor (Figure 1A). Such a dramatic conformational reorganization is clearly vital to the formation of a productive binding site and therefore critical for successful virtual screening. Using single snapshots, such cryptic binding sites may either be buried or incompletely formed, requiring movements to recognize a ligand and be of use to docking software. Much more subtle motions, which are often neglected when using rigid structures, can also have an effect on the successful docking of small molecules. Figure 1B illustrates the sensitivity of molecular docking to modest sidechain shifts, which can modulate the shape and volume of the binding pocket. In this example, we show the docking of two diverse NNRTIs to both their own “native” pocket conformation and then to each other’s “non-native” pocket. While the ligand pose is accurate for the native pockets (as demonstrated by a low RMSD to the co-crystallized pose), the ligands are mis-docked in the non-native pocket, underscoring the need to introduce flexibility. Another important consideration is that a single protein binding site may interact with ligands of diverse structures and may therefore adopt a rather different shape in each case [13]. This concept of “single binding site: multiple ligands” is well suited to the case of the NNRTI binding pocket, which, as shown in Figure 1C, has been co-crystallized in the presence of ligands bearing quite distinct molecular scaffolds. Such behaviour further underscores the conformational heterogeneity of protein populations, which take on different guises and present multiple opportunities for a high affinity interaction with a variety of small molecules. The diversity of the NNRTI pocket highlights the problem of cross-docking, whereby binding specific pocket conformations are often tailored to specific ligands and less hospitable to other structures [14]. Clearly, a single snapshot of the binding pocket is highly restrictive and may choke computational screening efforts by reducing the chance of identifying a novel high-affinity drug scaffold.
Figure 1.
Aspects of binding site conformational diversity, as exemplified by the HIV-1 RT NNRTI binding site. (A) The large transition from a closed to an open binding site, as shown by PDB structures 1DLO and 1VRT, respectively. Protein shown in surface representation and colored by atom type. The NNRTI Nevirapine is shown in orange stick representation and has been superimposed onto the closed structure to emphasize the occluded pocket. (B) The more subtle differences between the binding site conformations of two diverse NNRTI-bound structures can have a large impact on binding success. The left-hand side shows the NNRTIs Nevirapine and TMC-278 docked into their native pockets, while the right-hand side shows the same ligands docked into each other’s pocket. The RMSD between the docked pose and the co-crystallized pose is indicated. Protein shown in surface representation and colored by residue. NNRTIs shown in stick representation and colored by atom type. (C) A set of 10 NNRTI structures, illustrating the diverse range of scaffolds bound by the same binding pocket.
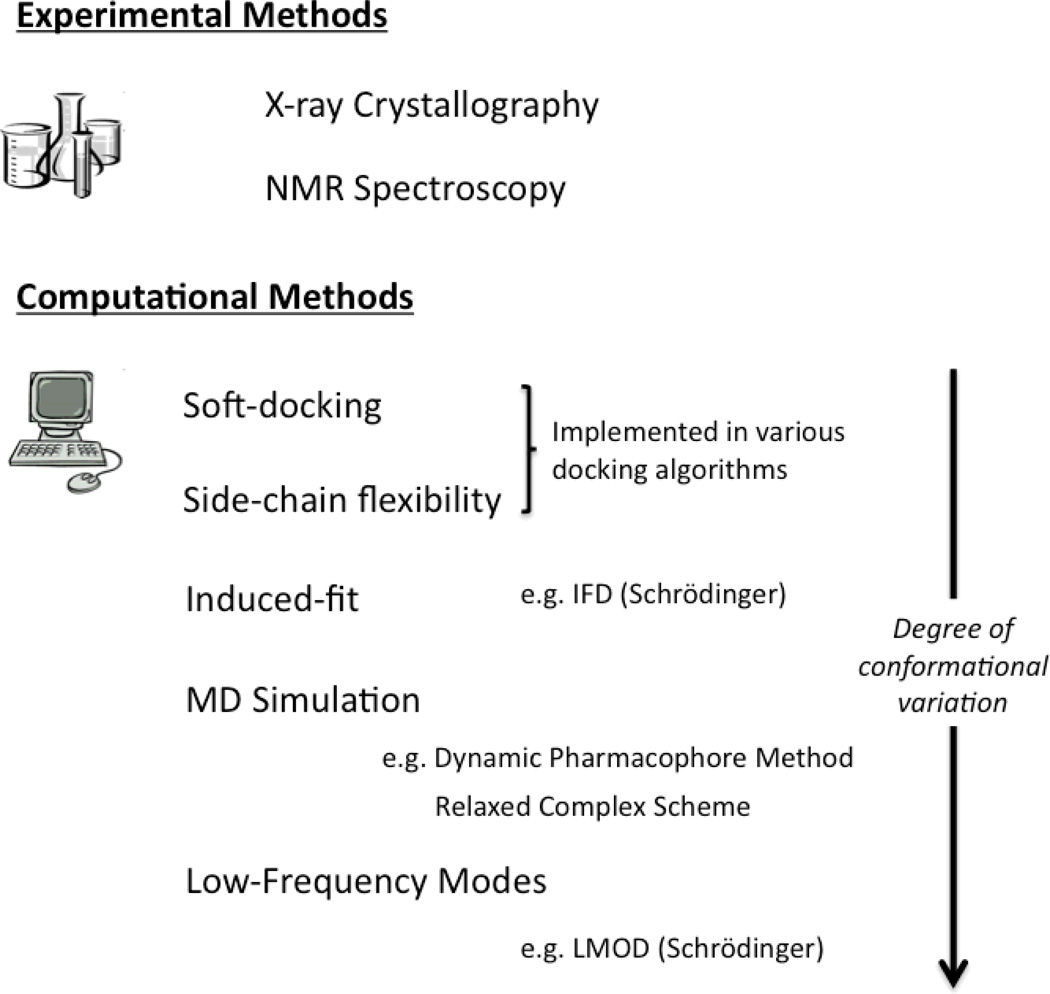
Several approaches have been adopted in order to better represent a dynamic and flexible binding site for molecular docking, each with varying degrees of practicality and conformational sampling (as reviewed in [15–19]), as summarized in Figure 2. While the field is still very much in its infancy, many studies have supported the conceptual appeal of a dynamic binding site with encouraging results for enrichment in virtual screening and correlations with biological activity [20–22]. At the experimental level, major advances in x-ray crystallography [23] and nuclear magnetic resonance (NMR) [24] have captured target proteins in a variety of poses (for example, HIV-1 protease and cyclin dependent kinase 2). Such ensembles have been used collectively in virtual screening studies, often generating results superior to those using a rigid representation of the protein [25,26]. Unfortunately, such datasets are generally a luxury enjoyed by relatively few drug targets and may still capture just a small proportion of the conformational variation of a protein. In particular, such experimental techniques may neglect some of the rarer protein poses and are typically extremely laborious. Recent advances such as time-resolved crystallography [27] and a focus on NMR, where structures are solved in solution and have demonstrated impressive results in a comparative study [28], may alleviate some of these issues. At the computational level, various methodologies have been proposed, which take a single snapshot of a protein and model flexibility around that substate (as reviewed in [17,19,29]). Perhaps the simplest implementation involves “soft docking” [30], whereby the docking algorithm permits some degree of overlap between the binding pocket and the ligand by reducing the penalty associated with steric clashes. This strategy is computationally cheap, although the binding pocket does not technically change shape and it may introduce a high rate of false positives if the tolerance is too high. Another approach involves the sampling of sidechain flexibility purely within the binding pocket, for example by simply switching the rotameric forms of each residue, or more rigorously by modulating torsional and rotational degrees of freedom (e.g. [31,32]). A weakness of this technique is the requirement to define precisely which degrees of freedom are important to ligand binding, which may or may not be well understood. Furthermore, while the focus on a subset of degrees of freedom keeps the conformational search more tractable, it obviously neglects backbone flexibility, which may be critical for ligand binding. A more recent development in the family of protocols which deal with local flexibility is the Induced Fit Docking (IFD) method [33]. IFD involves an iterative procedure whereby initial docked poses of the ligand (using a softened potential to permit some atomic overlap) are followed by relaxations of the protein to adapt to the ligand. While small backbone movements are catered for and the algorithm is computationally efficient, this method is likely unsuitable when more aggressive conformational changes take place and would therefore benefit from a diverse set of input protein structures. For a more intensive conformational search, molecular dynamics (MD) simulation has long been a popular choice and it has been argued that it provides the most accurate representation of protein motion [34]. MD simulation essentially charts the evolution of a protein structure from an input configuration, typically over a period of tens to hundreds of nanoseconds (ns) with current hardware capabilities [35,36]. The resulting “movie” of protein motion can then be processed to extract representative snapshots, which are then subjected to various ensemble-based docking schemes [37]. The key advantage of MD-generated conformers is the full flexibility which is modelled – all areas of the protein are free to move. Full, global backbone flexibility permits more dramatic conformational changes to take place and therefore a broader sampling of the conformational landscape is feasible. Another distinct advantage of MD simulation is that the receptor may move in a realistic, explicit solvent environment, whereas other techniques make rearrangements within the gas phase. While MD simulation is the most computationally demanding of the methods described, continued hardware advances (e.g. GPU processing [38]) and software enhancements are bound to improve the situation.
Figure 2.
Overview of the various experimental and computational methods available for the representation of a flexible protein target.
The earliest use of MD simulation to model protein flexibility was incorporated into the Dynamic Pharmacophore Model (DPM), using a HIV target [39]. Diverse conformations of a HIV-1 integrase binding site were used to build a consensus pharmacophore based on the positions of bound probe molecules in different structures. The pharmacophore was then used to search a compound library, with impressive results for the experimentally tested compounds (one third of which were found to be inhibitory). The DPM has since been used to find novel inhibitors for the HIV-1 protease [40] and MDM2-p53 protein, using longer simulations [41]. A more recent area of activity employing MD simulation in drug discovery has seen the introduction of the Relaxed Complex Scheme (RCS), whereby multiple computer-generated conformations are subjected to virtual screens [42–44]. In the light of the computer-intensive nature of MD simulation, other methods have been developed in order to predict even larger scale conformational changes for drug docking (for example, due to loop motions and hinge-bending mechanisms). These methods, including applications to a DNA-ligand complex [45] and HIV-1 integrase [46], typically identify low-frequency modes of motion that are thought to be responsible for key functional movements (for example, through normal modes analysis or principal component analysis). Perturbation of the protein structure along these modes is then used to generate new conformers, and has been shown to improve both docking accuracy and enrichment in a virtual screening study [47].
In this article, we focus on the RCS, which provides a framework with which to address key issues in ensemble-based docking, which include the generation and selection of representative conformers and the scoring of compounds across the ensemble. We will summarize several successful applications of the RCS, along with a more detailed look at its application to the HIV-1 reverse transcriptase as an example, and some of the ongoing enhancements being developed.
THE RELAXED COMPLEX SCHEME
One of the motivations behind the RCS is the understanding that small molecules may bind to receptor conformations that occur relatively rarely and which may therefore elude traditional structure determination methods [44]. Inspired by the success of the DPM, the RCS provides a powerful tool for the discovery of new lead compounds at well-established drug binding sites for which structural data is available. The RCS provides solutions to three key problems in ensemble-based docking: 1) How to produce a diverse ensemble of structures; 2) How to select a reduced set of representative structures; 3) How to score a compound across a set of binding pocket poses.
The first phase of the RCS, and the most time-consuming, employs a long MD simulation to allow the receptor to explore new regions of its conformational space, under the guidance of an empirical force field [48]. A number of popular MD codes have been used with the RCS to date (including AMBER [49], NAMD [50], GROMACS [51]), which typically carry out conventional all-atom, explicit solvent simulations. The timescale of such simulations is linked to the extent of conformational sampling and applications of the RCS have ranged from 2 ns in 2002 through to ~0.5 µs today, with longer simulations on the horizon [52]. Furthermore, the RCS has been applied to progressively larger systems, ranging from ~23,000 atoms on FKBP to ~160,000 on HIV-1 RT. In addition to standard MD setup protocols, an important consideration in the configuration of RCS simulations is whether the binding pocket should be simulated in its ligand-free or ligand-bound state. While early implementations opted for unliganded systems, later work introduced known binders and this “holo” form has been preferential in certain cases and recognized to yield better enrichment [53–55]. While the optimal complexation state is likely to depend on the dynamics of the specific binding site in question, it is possible that a ligand-bound pocket provides a better conformation for docking as it embodies ligand-induced effects. Furthermore, some binding sites do not form a viable binding pocket in their unliganded state and may not develop an open conformation on timescales accessible to conventional MD simulation. We have found this to be the case with HIV-1 RT [56] and to address this issue, various methods have been implemented which aim to produce a viable binding site when only unliganded structures are available [57,58]. Another consideration when simulating protein-ligand complexes is the extent to which the particular ligand simulated biases the pocket conformation with its particular scaffold and volume. This “over-specialization” was recognized in previous work on co-crystallized complexes [54] and in such cases, it may be beneficial to simulate diverse known ligands, if available.
The output of a MD simulation is typically a set of thousands of snapshots, which must somehow be condensed into a more manageable number for the purposes of docking libraries of compounds. The task of selecting precisely which structures are used in the docking phase is challenging and pivotal to the success of successive steps in the RCS. An emerging theme in the field of protein-ligand docking is the notion that some structures are better than others at discriminating active from inactive compounds and would therefore be preferable to use in virtual screening [22,55,59,60]. Identifying these structures a priori is therefore highly desirable and although some general properties of successful structures have been investigated (e.g. conformational energy [60] and volume [59]), such predictions are still a “holy grail” and likely to vary by target. When active compounds are known for the target, a ligand-directed approach can be taken to filter the best structures a posteriori, although this adds computational effort as additional screens need to be performed [22,55]. As well as finding which receptors to dock to, another important question is how many to use from an ensemble. While there is obviously no hard-and-fast rule applicable to all targets, work by Rueda and co-workers has suggested that, with randomly selected conformers, 3–5 structures can be used to obtain better performance in comparison to a single structure [55]. In the RCS applications to date, various methods have been used to reduce the size of MD trajectories. The earliest work simply extracted snapshots at regular intervals, whereas later studies introduced clustering methods in order to eliminate conformational redundancy and further compress the size of the ensemble for larger compound libraries. The two most popular choices of algorithm have been the root mean-square deviation (RMSD) clustering and QR factorization methods, which have both been effective at capturing the structural diversity of the ensemble in a small dataset and thus improving the efficiency of the RCS [42].
The third phase of the RCS involves protein-ligand docking of compound libraries against members of the sub-ensemble, with the ultimate selection of the top “hits” for further optimization and experimental testing. The evolution of computational power has seen a shift in the libraries of compounds used, with early work focusing on small sets of known inhibitors and later work screening vast datasets of drug-like compounds for new leads (e.g. those compiled by the NCI and ZINC). Also, various popular docking codes have been employed by the RCS (including Glide [61], Autodock [62] and Autodock Vina [63]), as well as a variety of scoring functions – for example, a combination of traditional empirical scoring with more rigorous methods such as MMGB/SA. For efficiency, a hierarchical approach is often taken, whereby an initial screen is performed on a small receptor ensemble (often incorporating experimental conformations) with a large compound library and the most promising binders are then re-ranked against a larger ensemble of MD conformers. A challenge in the ranking of compounds against an ensemble of receptor conformations is how to assess their overall performance. In the RCS, each compound is docked to each receptor structure and a histogram of binding free energies (BFEs) is obtained. A wide spectrum of BFEs is a typical hallmark of such histograms, which reflects the diversity of the structures and the fact that even a known active compound may not be able to bind to certain conformations. One solution is to simply take the mean BFE across the ensemble, to indicate compounds which perform well across multiple conformers. Another option is to weight the mean according to some property of each conformer – usually the population of the cluster it belongs to. Yet another possibility is to consider the minimum BFE across the ensemble, which may identify compounds which bind well to isolated, and potentially rare, conformers. Instead of performing an individual docking run on each member of the ensemble, some “grid-averaging” techniques have been used to consolidate the dynamic features of a binding pocket into a single docking run (as reviewed in [34]). While these have introduced significant efficiency benefits, it is unclear how well the averaging can represent larger conformational fluctuations and whether the average structure is physically realistic.
PREVIOUS APPLICATIONS OF THE RELAXED COMPLEX SCHEME
While a thorough review of previous applications of the RCS is beyond the scope of this article, we aim to briefly summarize the main features of some important studies, to illustrate the range of targets, simulation methods and key findings.
The first application of the RCS, in 2002, involved short 2 ns MD simulations of the FK506 binding protein (FKBP-12), which is involved in the immune system and for which high-affinity synthetic ligands are sought [43]. Initial work used Autodock to dock two known high-affinity compounds to unliganded MD snapshots and demonstrate the sensitivity of docking poses and energies to binding site plasticity. Also, a “double-ligand” docking approach was introduced, whereby multivalent compounds could be designed by linking multiple small fragments at different regions of the binding site. Later work on the same system used MM/PBSA to calculate more accurate binding free energies and thus enhance the ranking of the many protein-ligand complexes generated when using an ensemble of conformers [64].
Acetylcholinesterase (AChE) plays an important role in the central nervous system and is a key drug target for Alzheimer’s disease therapies. Kua and co-workers used snapshots from short 1 ns MD simulations of both the unliganded and ACh-bound protein to dock ACh and a selection of analogues [65]. A key finding of this work was the improvement in binding energies when ACh-bound conformers were used instead of the unliganded form. This was related to the “induced fit” effect of the inserted ligand, which yielded a binding site geometry capable of stronger protein-ligand interactions. Further work on this system used simulations of binding site mutants to elucidate the role of specific residues in ACh binding [66]. It was discovered that key wild-type residues confer particular shape and electrostatic properties, which are conducive to stronger binding energies than the mutants.
Babakhani and co-workers used the RCS with MD simulations of the acetylcholine binding protein (AChBP), which mimics the extracellular portion of the nicotinic acetylcholine receptor (nAChR) [67]. The nAChR constitutes an important pharmacological target for the cessation of smoking and drug addiction and through docking to a flexible representation of the AChBP, a number of NCI compounds were proposed. Many of these compounds exposed novel binding regions, which may afford them the desirable capacity to discriminate between the various nAChR receptor subtypes.
Protein Kinase A (PKA) is an important drug target for a range of diseases and for which a number of FDA-approved drugs have been developed. Wong and co-workers used 1 ns simulations of both balanol-bound and unliganded PKA to extract a series of diverse receptor structures, into which balanol was docked [68]. It was found that, while the liganded ensemble was able to consistently reproduce the co-crystallized pose of balanol, the unliganded set of structures yielded many mis-docked balanol conformations, likely due to the extra flexibility of the empty binding site. It was also noted that the lowest energy docking poses correlated well with the poses closest to the experimental conformation of balanol and thus might serve to help discriminate between good and bad poses.
Matrix metalloproteinases (MMPs) have recently become an interesting drug target due to their role in tissue remodelling, for example in tumour metastasis. A 30 ns MD simulation of MMP-2 was recently performed, with 3,000 snapshots used to dock a library of fragments, using the LUDI software [69]. Fragments which bound to different pocket subsites were identified and then linked to form composite compounds with predicted high affinity. An interesting aspect of this study was the use of 5 ns MD simulations of the top-scoring protein-ligand complexes, to assess induced-fit effects. Ligands were re-ranked based on their binding energy during the simulations and it was found that certain compounds fared better and some fared worse than their prediction based on docking alone. This suggests that short simulations of top complexes may be a useful way of improving binding energy predictions and is based on the problem of docking a compound into a pocket which was co-crystallized or simulated in the presence of a different ligand.
A range of neglected tropical diseases, caused by protozoan parasites, has been the subject of various drug discovery projects employing the RCS. Amaro and co-workers used a 20 ns MD simulation of the inhibitor-bound T. brucei RNA-editing ligase 1 (REL1) to re-rank a list of prospective compounds from an initial virtual screen [70]. It was found that many compounds, which scored relatively poorly against a single experimental structure, fared much better against the MD ensemble. Impressively, of the top 8 re-ranked NCI compounds experimentally tested for activity, 2 were confirmed low-micromolar inhibitors (with 2 further ~1 µM inhibitors discovered through a similarity search). Durrant and co-workers performed five 20 ns MD simulations of another T. brucei target, the UDP-galactose 4’-epimerase (TbGalE), for the purpose of enhancing a virtual screening study [71]. Of the top 26 compounds from the re-ranked screen which were experimentally tested, 16 were found to have activity at 100 µM, with a total of 14 low-micromolar compounds discovered after further testing of similar structures at lower concentrations. Interestingly, the tested compounds were selected based on both their ensemble-average and ensemble-best performance, suggesting that ligands binding tightly to a smaller number of conformers should also be considered. In a third line of work, cruzain, a T. cruzi protease targeted for the treatment of Chagas disease was subjected to five 20 ns simulations [72]. Diverse structures from this ensemble have then been used in a hierarchical RCS approach to re-score promising NCI compounds, the best of which have been considered for forthcoming experimental validation.
The avian influenza (H5N1) drug target, neuraminidase, has been the subject of two diverse RCS virtual screening studies, which drew an ensemble of receptor conformations from 40 ns MD simulations. In the first, an initial virtual screen of NCI compounds was performed against a combination of experimental and MD structures, followed by a re-ranking of the top compounds with a larger MD ensemble. In addition to the discovery of potential novel inhibitor structures, the docking work supported a previous study, which predicted the presence of multiple high-affinity sub-regions of the main binding site [53]. In the second study, the Autogrow algorithm was used to generate a set of novel compounds, by combining members of a fragment library with a core scaffold based on a known inhibitor [73]. By screening this library against experimental and MD conformations of neuraminidase, a final set of several drug-like inhibitors was proposed, with higher predicted affinity than the inhibitor they were based on.
HIV, and its associated drug targets, is an important field of activity for the RCS, with previous applications to the integrase and protease enzymes. A 2 ns simulation of HIV-1 integrase was used to extract an ensemble of conformers to which a known inhibitor, 5CITEP, was docked [74]. The key result from this study was the exposure of a second binding mode for 5CITEP, which was able to bind with higher predicted affinity to a “trench” next to the crystallized binding site. Significantly, this discovery eventually lead to the FDA-approval of the drug raltegravir and demonstrates that even relatively short MD simulations can sample critical protein rearrangements that are obscured in experimental structures. Another MD-based docking study of integrase was then used to elucidate the mechanism by which specific mutants affect raltegravir binding [75]. HIV-1 protease has also been simulated in both wild-type and mutant forms, with ensembles of 2,200 conformations, spanning 22 ns of simulation time, used in each case [76]. This work lead to a compilation of 23 potential new inhibitors, which were predicted to have greater affinity for the drug-resistance mutant than available protease inhibitors.
RELAXED COMPLEX SCHEME CASE STUDY: HIV-1 REVERSE TRANSCRIPTASE
To illustrate the various steps of a typical RCS workflow in more detail, we use a recent ensemble-based virtual screening study of HIV-1 RT as a case study [77], as summarized in Figure 3. While 17 FDA-approved drugs are currently available for RT (binding to both a catalytic and an allosteric site), the drug resistance which is characteristic of retroviruses means that new inhibitors are desperately sought. In this work, we employed a hierarchical virtual screening strategy, whereby a combination of experimental and MD simulation RT conformations was used to screen a library of compounds from the NCI. Firstly, a library of ~2,900 NCI compounds was compiled, composed of both the latest NCI diversity set and 1,500 NCI compounds from a similarity search of the latest-generation of NNRTIs. The library was then spiked with the structures of 20 diverse known active NNRTIs, which would serve as positive controls. In the first phase, an ensemble of 10 high-resolution crystallographic structures was selected, which capture RT bound to a diverse set of NNRTIs. Using the Glide Extra Precision (XP) docking algorithm, a virtual screen of the compound library was performed on all ten structures and all ligands were ranked based on their mean binding energy across the ensemble. As expected, the desirable ability of the docking algorithm to rank the known active compounds as the top hits varied by structure, despite the crystallographic structures reflecting relatively modest differences. It was found that, by using the mean docking score, the ability to discriminate known binders was slightly better than in the single best performing crystal structure. In the second phase, a reduced library of the highest-ranked 150 NCI compounds was selected for rescoring against a MD simulation ensemble. MD simulations were performed with RT complexed with 4 different NNRTIs (Nevirapine, APA, UC1, TBO). Diverse simulated ligands were chosen, so as to minimize the bias introduced by using a single inhibitor. By using a multi-copy approach, composed of individual 30 ns trajectories, a cumulative total simulation time of 480 ns was achieved. To extract representative structures from each system, an RMSD-based clustering approach was taken, whereby 30 diverse snapshots were taken from each system. The reduced compound library was then screened against all 120 MD conformers and re-ranked by their performance against this MD ensemble. In the final step, the top 50 re-scored compounds were reduced to a set of 20 with diverse scaffolds, by using a clustering algorithm. These novel compounds are currently undergoing experimental testing for anti-HIV activity with enzymatic and cell-based assays.
Figure 3.
Overview of an application of the RCS to the virtual screening of potential HIV-1 RT inhibitors. A hierarchical scheme was used, whereby an initial screen of experimental structures was used to filter out the most promising compounds, which were then re-screened against MD structures.
CONCLUSIONS AND FUTURE DEVELOPMENTS
The integration of receptor dynamics into SBDD programmes is both conceptually and practically appealing, as it has emerged that protein mobility plays a key role in molecular recognition. Receptor flexibility has been largely neglected in previous SBDD studies, largely due to the paucity of high-resolution experimental data, constraining docking algorithms to the “rigid-protein flexible-ligand” model. Various computational methods have now been proposed that introduce different degrees of flexibility to a protein structure and have enabled a new branch of SBDD known as ensemble-based docking. MD simulation based methods have thus far offered the most intensive treatment of full receptor dynamics and form an integral part of the RCS framework, which has been the focus of this article.
The RCS has been applied to a wide range of targets since its inception 8 years ago and has lead to the discovery of an FDA-approved drug, together with numerous potential and confirmed active lead compounds. It has emerged that many such compounds would have been either missed or overlooked in a rigid receptor scheme, which only represents a narrow portion of the complex protein conformational landscape. Furthermore, applications of the RCS are extending beyond the discovery of lead compounds at known sites to the exposure of entirely novel druggable binding pockets and binding modes, as has been shown for influenza neuraminidase [78], GPCRs [56], HIV-1 protease [79] and T. cruzi cruzain [72].
Despite these encouraging results, the RCS is still in its infancy and suffers from some limitations, which are currently being addressed through some encouraging developments. Firstly, the MD simulation phase at the core of the RCS is computationally demanding and can take of the order of weeks to months to generate typical trajectories in the tens to hundreds of ns range. Furthermore, even such timescales are unable to yield a complete sampling of conformational space for most pharmacologically relevant systems and therefore rarer events may be missed. To address these issues, a number of novel modified MD codes could be introduced to the RCS, with the aim of accelerating conformational sampling – for example, accelerated MD [80], replica exchange [81,82], conformational flooding [83] and implicit solvent MD [84]. At the hardware level, cloud computing and the combination of low latency supercomputers with highly parallelizable MD codes are two developments which will speed up such calculations. Another resource-demanding step inherent to all ensemble-based screening schemes is the docking phase, which needs to be repeated for each receptor conformation. This can be mitigated through the use of a hierarchical approach, whereby a smaller ensemble is used to condense a large compound library into a smaller one, which can then be re-ranked based on a larger ensemble. Also, some docking codes, such as Glide [61], are available in different degrees of precision: thus the cheaper lower-level docking algorithm can be used to filter out the most promising candidates and the more demanding, higher-precision docking can be saved for later stages of ranking. The selection of representative conformers from MD trajectories and scoring of compounds across multiple conformers are two other areas of ensemble-based docking which require attention and are currently being investigated.
ACKNOWLEDGEMENTS
This work has been supported in part by the National Science Foundation (NSF), the National Institutes of Health (NIH), the Howard Hughes Medical Institute (HHMI), the National Biomedical Computation Resource (NBCR), the Center for Theoretical Biological Physics (CTBP), San Diego Supercomputer Center (SDSC), and the NSF Supercomputer Centers.
REFERENCES
- 1.Gane PJ, Dean PM. Recent advances in structure-based rational drug design. Curr Opin Struct Biol. 2000;10:401–404. doi: 10.1016/s0959-440x(00)00105-6. [DOI] [PubMed] [Google Scholar]
- 2.Jorgensen WL. The many roles of computation in drug discovery. Science. 2004;303:1813–1818. doi: 10.1126/science.1096361. [DOI] [PubMed] [Google Scholar]
- 3.Jain AN. Virtual screening in lead discovery and optimization. Curr Opin Drug Discov Devel. 2004;7:396–403. [PubMed] [Google Scholar]
- 4.Jorgensen WL. Efficient drug lead discovery and optimization. Acc Chem Res. 2009;42:724–733. doi: 10.1021/ar800236t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rester U. From virtuality to reality - Virtual screening in lead discovery and lead optimization: a medicinal chemistry perspective. Curr Opin Drug Discov Devel. 2008;11:559–568. [PubMed] [Google Scholar]
- 6.Zoete V, Grosdidier A, Michielin O. Docking, virtual high throughput screening and in silico fragment-based drug design. J Cell Mol Med. 2009;13:238–248. doi: 10.1111/j.1582-4934.2008.00665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hou T, Xu X. Recent development and application of virtual screening in drug discovery: an overview. Curr Pharm Des. 2004;10:1011–1033. doi: 10.2174/1381612043452721. [DOI] [PubMed] [Google Scholar]
- 8.Teague SJ. Implications of protein flexibility for drug discovery. Nat Rev Drug Discov. 2003;2:527–541. doi: 10.1038/nrd1129. [DOI] [PubMed] [Google Scholar]
- 9.Forman-Kay JD. The 'dynamics' in the thermodynamics of binding. Nat Struct Biol. 1999;6:1086–1087. doi: 10.1038/70008. [DOI] [PubMed] [Google Scholar]
- 10.Verkhivker GM, Bouzida D, Gehlhaar DK, Rejto PA, Freer ST, Rose PW. Complexity and simplicity of ligand-macromolecule interactions: the energy landscape perspective. Curr Opin Struct Biol. 2002;12:197–203. doi: 10.1016/s0959-440x(02)00310-x. [DOI] [PubMed] [Google Scholar]
- 11.De Clercq E. Non-nucleoside reverse transcriptase inhibitors (NNRTIs): past, present, and future. Chem Biodivers. 2004;1:44–64. doi: 10.1002/cbdv.200490012. [DOI] [PubMed] [Google Scholar]
- 12.Sarafianos SG, Das K, Hughes SH, Arnold E. Taking aim at a moving target: designing drugs to inhibit drug-resistant HIV-1 reverse transcriptases. Curr Opin Struct Biol. 2004;14:716–730. doi: 10.1016/j.sbi.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 13.Ma B, Shatsky M, Wolfson HJ, Nussinov R. Multiple diverse ligands binding at a single protein site: a matter of pre-existing populations. Protein Sci. 2002;11:184–197. doi: 10.1110/ps.21302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ragno R, Frasca S, Manetti F, Brizzi A, Massa S. HIV-reverse transcriptase inhibition: inclusion of ligand-induced fit by cross-docking studies. J Med Chem. 2005;48:200–212. doi: 10.1021/jm0493921. [DOI] [PubMed] [Google Scholar]
- 15.Carlson HA. Protein flexibility and drug design: how to hit a moving target. Curr Opin Chem Biol. 2002;6:447–452. doi: 10.1016/s1367-5931(02)00341-1. [DOI] [PubMed] [Google Scholar]
- 16.Carlson HA, McCammon JA. Accommodating protein flexibility in computational drug design. Mol Pharmacol. 2000;57:213–218. [PubMed] [Google Scholar]
- 17.Cozzini P, Kellogg GE, Spyrakis F, Abraham DJ, Costantino G, Emerson A, Fanelli F, Gohlke H, Kuhn LA, Morris GM, Orozco M, Pertinhez TA, Rizzi M, Sotriffer CA. Target flexibility: an emerging consideration in drug discovery and design. J Med Chem. 2008;51:6237–6255. doi: 10.1021/jm800562d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henzler AM, Rarey M. In Pursuit of Fully Flexible Protein-Ligand Docking: Modeling the Bilateral Mechanism of Binding. Molecular Informatics. 2010;29:164–173. doi: 10.1002/minf.200900078. [DOI] [PubMed] [Google Scholar]
- 19.Totrov M, Abagyan R. Flexible ligand docking to multiple receptor conformations: a practical alternative. Curr Opin Struct Biol. 2008;18:178–184. doi: 10.1016/j.sbi.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cavasotto CN, Abagyan RA. Protein flexibility in ligand docking and virtual screening to protein kinases. J Mol Biol. 2004;337:209–225. doi: 10.1016/j.jmb.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 21.Popov VM, Yee WA, Anderson AC. Towards in silico lead optimization: scores from ensembles of protein/ligand conformations reliably correlate with biological activity. Proteins. 2007;66:375–387. doi: 10.1002/prot.21201. [DOI] [PubMed] [Google Scholar]
- 22.Yoon S, Welsh WJ. Identification of a minimal subset of receptor conformations for improved multiple conformation docking and two-step scoring. J Chem Inf Comput Sci. 2004;44:88–96. doi: 10.1021/ci0341619. [DOI] [PubMed] [Google Scholar]
- 23.Stewart L, Clark R, Behnke C. High-throughput crystallization and structure determination in drug discovery. Drug Discov Today. 2002;7:187–196. doi: 10.1016/s1359-6446(01)02121-3. [DOI] [PubMed] [Google Scholar]
- 24.Montelione GT, Zheng D, Huang YJ, Gunsalus KC, Szyperski T. Protein NMR spectroscopy in structural genomics. Nat Struct Biol. 2000;7 Suppl:982–985. doi: 10.1038/80768. [DOI] [PubMed] [Google Scholar]
- 25.Barril X, Morley SD. Unveiling the full potential of flexible receptor docking using multiple crystallographic structures. J Med Chem. 2005;48:4432–4443. doi: 10.1021/jm048972v. [DOI] [PubMed] [Google Scholar]
- 26.Huang SY, Zou X. Efficient molecular docking of NMR structures: application to HIV-1 protease. Protein Sci. 2007;16:43–51. doi: 10.1110/ps.062501507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schotte F, Lim M, Jackson TA, Smirnov AV, Soman J, Olson JS, Phillips GN, Jr., Wulff M, Anfinrud PA. Watching a protein as it functions with 150-ps time-resolved x-ray crystallography. Science. 2003;300:1944–1947. doi: 10.1126/science.1078797. [DOI] [PubMed] [Google Scholar]
- 28.Damm KL, Carlson HA. Exploring experimental sources of multiple protein conformations in structure-based drug design. J Am Chem Soc. 2007;129:8225–8235. doi: 10.1021/ja0709728. [DOI] [PubMed] [Google Scholar]
- 29.Teodoro ML, Kavraki LE. Conformational flexibility models for the receptor in structure based drug design. Curr Pharm Des. 2003;9:1635–1648. doi: 10.2174/1381612033454595. [DOI] [PubMed] [Google Scholar]
- 30.Ferrari AM, Wei BQ, Costantino L, Shoichet BK. Soft docking and multiple receptor conformations in virtual screening. J Med Chem. 2004;47:5076–5084. doi: 10.1021/jm049756p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leach AR. Ligand docking to proteins with discrete side-chain flexibility. J Mol Biol. 1994;235:345–356. doi: 10.1016/s0022-2836(05)80038-5. [DOI] [PubMed] [Google Scholar]
- 32.Schaffer L, Verkhivker GM. Predicting structural effects in HIV-1 protease mutant complexes with flexible ligand docking and protein side-chain optimization. Proteins. 1998;33:295–310. doi: 10.1002/(sici)1097-0134(19981101)33:2<295::aid-prot12>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 33.Sherman W, Day T, Jacobson MP, Friesner RA, Farid R. Novel procedure for modeling ligand/receptor induced fit effects. J Med Chem. 2006;49:534–553. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- 34.Morra G, Genoni A, Neves MA, Merz KM, Jr., Colombo G. Molecular recognition and drug-lead identification: what can molecular simulations tell us? Curr Med Chem. 2010;17:25–41. doi: 10.2174/092986710789957797. [DOI] [PubMed] [Google Scholar]
- 35.Karplus M, McCammon JA. Molecular dynamics simulations of biomolecules. Nat Struct Biol. 2002;9:646–652. doi: 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- 36.Klepeis JL, Lindorff-Larsen K, Dror RO, Shaw DE. Long-timescale molecular dynamics simulations of protein structure and function. Curr Opin Struct Biol. 2009;19:120–127. doi: 10.1016/j.sbi.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 37.Alonso H, Bliznyuk AA, Gready JE. Combining Docking and Molecular Dynamic Simulations in Drug Design. ChemInform. 2006;37:531–568. doi: 10.1002/med.20067. [DOI] [PubMed] [Google Scholar]
- 38.Stone JE, Phillips JC, Freddolino PL, Hardy DJ, Trabuco LG, Schulten K. Accelerating molecular modeling applications with graphics processors. J Comput Chem. 2007;28:2618–2640. doi: 10.1002/jcc.20829. [DOI] [PubMed] [Google Scholar]
- 39.Carlson HA, Masukawa KM, Rubins K, Bushman FD, Jorgensen WL, Lins RD, Briggs JM, McCammon JA. Developing a dynamic pharmacophore model for HIV-1 integrase. J Med Chem. 2000;43:2100–2114. doi: 10.1021/jm990322h. [DOI] [PubMed] [Google Scholar]
- 40.Meagher KL, Carlson HA. Incorporating protein flexibility in structure-based drug discovery: using HIV-1 protease as a test case. J Am Chem Soc. 2004;126:13276–13281. doi: 10.1021/ja0469378. [DOI] [PubMed] [Google Scholar]
- 41.Bowman AL, Nikolovska-Coleska Z, Zhong H, Wang S, Carlson HA. Small molecule inhibitors of the MDM2-p53 interaction discovered by ensemble-based receptor models. J Am Chem Soc. 2007;129:12809–12814. doi: 10.1021/ja073687x. [DOI] [PubMed] [Google Scholar]
- 42.Amaro RE, Baron R, McCammon JA. An improved relaxed complex scheme for receptor flexibility in computer-aided drug design. J Comput Aided Mol Des. 2008;22:693–705. doi: 10.1007/s10822-007-9159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin JH, Perryman AL, Schames JR, McCammon JA. Computational drug design accommodating receptor flexibility: the relaxed complex scheme. J Am Chem Soc. 2002;124:5632–5633. doi: 10.1021/ja0260162. [DOI] [PubMed] [Google Scholar]
- 44.McCammon JA. Target flexibility in molecular recognition. Biochim Biophys Acta. 2005;1754:221–224. doi: 10.1016/j.bbapap.2005.07.041. [DOI] [PubMed] [Google Scholar]
- 45.Zacharias M, Sklenar H. Harmonic modes as variables to approximately account for receptor flexibility in ligand–receptor docking simulations: Application to DNA minor groove ligand complex. Journal of Computational Chemistry. 1999;20:287–300. [Google Scholar]
- 46.Keseru GM, Kolossvary I. Fully flexible low-mode docking: application to induced fit in HIV integrase. J Am Chem Soc. 2001;123:12708–12709. doi: 10.1021/ja0160086. [DOI] [PubMed] [Google Scholar]
- 47.Cavasotto CN, Kovacs JA, Abagyan RA. Representing receptor flexibility in ligand docking through relevant normal modes. J Am Chem Soc. 2005;127:9632–9640. doi: 10.1021/ja042260c. [DOI] [PubMed] [Google Scholar]
- 48.Mackerell AD. Empirical force fields for biological macromolecules: Overview and issues. Journal of Computational Chemistry. 2004;25:1584–1604. doi: 10.1002/jcc.20082. [DOI] [PubMed] [Google Scholar]
- 49.Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr., Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 52.Sanbonmatsu KY, Tung CS. High performance computing in biology: multimillion atom simulations of nanoscale systems. J Struct Biol. 2007;157:470–480. doi: 10.1016/j.jsb.2006.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheng LS, Amaro RE, Xu D, Li WW, Arzberger PW, McCammon JA. Ensemble-based virtual screening reveals potential novel antiviral compounds for avian influenza neuraminidase. J Med Chem. 2008;51:3878–3894. doi: 10.1021/jm8001197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McGovern SL, Shoichet BK. Information decay in molecular docking screens against holo, apo, and modeled conformations of enzymes. J Med Chem. 2003;46:2895–2907. doi: 10.1021/jm0300330. [DOI] [PubMed] [Google Scholar]
- 55.Rueda M, Bottegoni G, Abagyan R. Recipes for the selection of experimental protein conformations for virtual screening. J Chem Inf Model. 2010;50:186–193. doi: 10.1021/ci9003943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ivetac A, McCammon JA. Mapping the druggable allosteric space of g-protein coupled receptors: a fragment-based molecular dynamics approach. Chem Biol Drug Des. 2010;76:201–217. doi: 10.1111/j.1747-0285.2010.01012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abagyan R, Kufareva I. The flexible pocketome engine for structural chemogenomics. Methods Mol Biol. 2009;575:249–279. doi: 10.1007/978-1-60761-274-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seeliger D, de Groot BL. Conformational transitions upon ligand binding: holo-structure prediction from apo conformations. PLoS Comput Biol. 2010;6:e1000634. doi: 10.1371/journal.pcbi.1000634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thomas MP, McInnes C, Fischer PM. Protein structures in virtual screening: a case study with CDK2. J Med Chem. 2006;49:92–104. doi: 10.1021/jm050554i. [DOI] [PubMed] [Google Scholar]
- 60.Wei BQ, Weaver LH, Ferrari AM, Matthews BW, Shoichet BK. Testing a flexible-receptor docking algorithm in a model binding site. J Mol Biol. 2004;337:1161–1182. doi: 10.1016/j.jmb.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 61.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 62.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin JH, Perryman AL, Schames JR, McCammon JA. The relaxed complex method: Accommodating receptor flexibility for drug design with an improved scoring scheme. Biopolymers. 2003;68:47–62. doi: 10.1002/bip.10218. [DOI] [PubMed] [Google Scholar]
- 65.Kua J, Zhang Y, McCammon JA. Studying enzyme binding specificity in acetylcholinesterase using a combined molecular dynamics and multiple docking approach. J Am Chem Soc. 2002;124:8260–8267. doi: 10.1021/ja020429l. [DOI] [PubMed] [Google Scholar]
- 66.Kua J, Zhang Y, Eslami AC, Butler JR, McCammon JA. Studying the roles of W86, E202, and Y337 in binding of acetylcholine to acetylcholinesterase using a combined molecular dynamics and multiple docking approach. Protein Sci. 2003;12:2675–2684. doi: 10.1110/ps.03318603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Babakhani A, Talley TT, Taylor P, McCammon JA. A virtual screening study of the acetylcholine binding protein using a relaxed-complex approach. Comput Biol Chem. 2009;33:160–170. doi: 10.1016/j.compbiolchem.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wong CF, Kua J, Zhang Y, Straatsma TP, McCammon JA. Molecular docking of balanol to dynamics snapshots of protein kinase A. Proteins. 2005;61:850–858. doi: 10.1002/prot.20688. [DOI] [PubMed] [Google Scholar]
- 69.Durrant JD, de Oliveira CA, McCammon JA. Including receptor flexibility and induced fit effects into the design of MMP-2 inhibitors. J Mol Recognit. 2009;23:173–182. doi: 10.1002/jmr.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Amaro RE, Schnaufer A, Interthal H, Hol W, Stuart KD, McCammon JA. Discovery of drug-like inhibitors of an essential RNA-editing ligase in Trypanosoma brucei. Proc Natl Acad Sci U S A. 2008;105:17278–17283. doi: 10.1073/pnas.0805820105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Durrant JD, Urbaniak MD, Ferguson MA, McCammon JA. Computer-aided identification of Trypanosoma brucei uridine diphosphate galactose 4'-epimerase inhibitors: toward the development of novel therapies for African sleeping sickness. J Med Chem. 2010;53:5025–5032. doi: 10.1021/jm100456a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Durrant JD, Keranen H, Wilson BA, McCammon JA. Computational identification of uncharacterized cruzain binding sites. PLoS Negl Trop Dis. 2010;4:e676. doi: 10.1371/journal.pntd.0000676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Durrant JD, McCammon JA. Potential drug-like inhibitors of Group 1 influenza neuraminidase identified through computer-aided drug design. Comput Biol Chem. 2010;34:97–105. doi: 10.1016/j.compbiolchem.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schames JR, Henchman RH, Siegel JS, Sotriffer CA, Ni H, McCammon JA. Discovery of a novel binding trench in HIV integrase. J Med Chem. 2004;47:1879–1881. doi: 10.1021/jm0341913. [DOI] [PubMed] [Google Scholar]
- 75.Perryman AL, Forli S, Morris GM, Burt C, Cheng Y, Palmer MJ, Whitby K, McCammon JA, Phillips C, Olson AJ. A dynamic model of HIV integrase inhibition and drug resistance. J Mol Biol. 2010;397:600–615. doi: 10.1016/j.jmb.2010.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Perryman AL, Lin JH, McCammon JA. Optimization and Computational Evaluation of a Series of Potential Active Site Inhibitors of the V82F/I84V Drug-resistant Mutant of HIV-1 Protease: an Application of the Relaxed Complex Method of Structure-based Drug Design. Chemical Biology & Drug Design. 2006;67:336–345. doi: 10.1111/j.1747-0285.2006.00382.x. [DOI] [PubMed] [Google Scholar]
- 77.Ivetac A, McCammon JA. In preparation. [Google Scholar]
- 78.Landon MR, Amaro RE, Baron R, Ngan CH, Ozonoff D, McCammon JA, Vajda S. Novel druggable hot spots in avian influenza neuraminidase H5N1 revealed by computational solvent mapping of a reduced and representative receptor ensemble. Chem Biol Drug Des. 2008;71:106–116. doi: 10.1111/j.1747-0285.2007.00614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Perryman AL, Lin JH, McCammon JA. HIV-1 protease molecular dynamics of a wild-type and of the V82F/I84V mutant: possible contributions to drug resistance and a potential new target site for drugs. Protein Sci. 2004;13:1108–1123. doi: 10.1110/ps.03468904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hamelberg D, Mongan J, McCammon JA. Accelerated molecular dynamics: a promising and efficient simulation method for biomolecules. J Chem Phys. 2004;120:11919–11929. doi: 10.1063/1.1755656. [DOI] [PubMed] [Google Scholar]
- 81.Zhou R. Replica exchange molecular dynamics method for protein folding simulation. Methods Mol Biol. 2006;350:205–223. doi: 10.1385/1-59745-189-4:205. [DOI] [PubMed] [Google Scholar]
- 82.Sugita Y, Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chem Phys Lett. 1999;314:141–151. [Google Scholar]
- 83.Grubmuller H. Predicting slow structural transitions in macromolecular systems: Conformational flooding. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics. 1995;52:2893–2906. doi: 10.1103/physreve.52.2893. [DOI] [PubMed] [Google Scholar]
- 84.Onufriev A. Annual Reports in Computational Chemistry. Volume 4. Elsevier; 2008. Implicit solvent models in molecular dynamics simulations: A brief overview; pp. 125–137. [Google Scholar]